Abstract
Objective
Activating mutations in the KRAS gene, found in approximately 53% of metastatic colorectal cancer (mCRC) cases, can render epidermal growth factor receptor (EGFR) inhibitors ineffective. Regional differences in these mutations have been reported. This is the first study which aims to describe the pattern of KRAS mutations in a Sri Lankan cohort of mCRC patients.
Results
The KRAS genotypes detected in mCRC patients which have been maintained in an anonymized database were retrospectively analyzed. Of the 108 colorectal tissue samples tested, 25 (23.0%) had KRAS mutations. Overall, there were 68 (63.0%) males and 40 (37.0%) females. Among the KRAS positive cases, there were 14 (56.0%) males and 11 (44.0%) females. Their age distribution ranged from 29 to 85 years with a median age of 61 years. There were 15 patients (60.0%) with point mutations in codon 12 while 10 (40.0%) had a single mutation in codon 13. The most common KRAS mutation identified was p.Gly13Asp (40.0%), followed by p.Gly12Val (24.0%). Other mutations included p.Gly12Cys (12.0%), p.Gly12Ser (12.0%), p.Gly12Asp (8.0%), and p.Gly12Arg (4.0%). The codon 13 mutation was a G>A transition (40.0%), while G>T transversions (32.0%), G>A transitions (24.0%), and G>C transversions (4.0%) were found in the codon 12 mutations. The frequency of KRAS mutations was similar to that reported for Asian patients. However, in contrast to several published studies, the G>A transition in codon 12 (c.35G>A; p.Gly12Asp), was not the most common mutation within codon 12 in our cohort. This may be a reflection of the genetic heterogeneity in the pattern of KRAS mutations in mCRC patients but valid conclusions cannot be drawn from these preliminary findings due to the small size of the study sample.
Keywords: Colorectal carcinoma, Epidermal growth factor receptor, Genotype, KRAS gene, Mutation
Introduction
In Sri Lanka, colorectal cancer (CRC) is one of the top five cancers accounting for almost 7.0% of all male cancers and 6.0% of all female cancers [1]. Recent advances in molecular targeted therapeutic approaches to CRC have identified the potential role of anti-epidermal growth factor receptor (EGFR) targeted therapies, cetuximab and panitumumab as adjuvant treatment in advanced disease in combination with cytotoxic chemotherapy [2, 3]. However, EGFR, the target of these drugs, which is overexpressed in approximately 50.0–80.0% of CRC, failed to predict a therapeutic response when used clinically. Research showed that the Kirsten rat sarcoma viral oncogene homolog (KRAS) gene, an important member of the EGFR signalling cascade, can acquire activating mutations in exon 2 codons 12 and 13 in approximately 35.0–45.0% of the CRC cases, rendering EGFR inhibitors ineffective [4, 5]. In 2009, the US Food and Drug Administration (FDA) approved EGFR-targeted monoclonal antibody therapy with cetuximab and panitumumab in patients with metastatic colorectal cancer (mCRC) along with analysis of KRAS mutation status, which is a predictive biological marker of resistance [6–8].
Somatic KRAS mutations in codons 12, 13 (exon 2), 59, 61 (exon 3), 117 and 146 (exon 4) are common hotspots. Approximately 53.0% of patients with mCRC have RAS mutations, with 42.0% having KRAS exon 2 mutations and 11.0% having KRAS non-exon 2 and NRAS mutations [3]. Other non hotspot mutations in codons 2, 3, 4, 63 and 154 occur less frequently in CRC, accounting for less than 5.0% of all mutation types [3, 4]. Most of the reported mutations are single nucleotide point mutations, particularly G>A transitions and G>T transversions [9]. According to published western studies, among the codon 12 mutations, p.Gly12Asp, and p.Gly12Val are the most frequent, while in codon 13, the substitution of glycine for aspartate (p.Gly13Asp) is the most common [3]. Some new and uncommon KRAS mutations that are found in codons 45, 69 and 80 have also been identified in Chinese patients with CRC, but their clinical significance has yet to be defined [10, 11].
It is essential to investigate the underlying mutations in the KRAS gene in mCRC patients to identify the prevailing patterns as regional differences in these mutations have been reported in different population groups. Such knowledge would be helpful in selecting the appropriate patients with mCRC for EGFR-inhibitor therapy and for developing cost-effective targeted mutation testing for predominant mutations in the local context. This is the first study which aims to describe the frequency and types of KRAS mutations among Sri Lankan patients with mCRC referred for KRAS mutation testing to the only genetics diagnostic center undertaking such testing in the country.
Main text
Methods
The sex, age and KRAS genotypes detected in mCRC patients referred from all parts of Sri Lanka for KRAS mutation testing to the Asiri Centre for Genomic and Regenerative Medicine, Colombo from January 2010 to December 2014 have been maintained in an anonymized database completely de-identified from the original patients. This database was retrospectively analysed. All individuals listed in the database with mCRC who had undergone KRAS mutation testing during the period of study specified above were included in the retrospective analysis.
Genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tissue sections from histologically confirmed primary colorectal tumors using QIAamp DNA FFPE Tissue Kits [Qiagen, Germany (Cat No./ID: 56404)] according to the manufacturer’s protocol. The samples were specifically chosen by a pathologist to include predominantly tumor cells without significant necrosis or inflammation. The sequences of codons 12 and 13 of the KRAS gene were genotyped by polymerase chain reaction amplification and direct sequencing according to the validated method previously described by De Roock et al. [8]. This is a well-established test to detect mutations in codons 12 and 13 of the KRAS gene. In addition, in the diagnostic assay, internal positive and negative control samples were used and those samples gave the expected result always, indicating that the method used was 100% sensitive and specific. The primers and conditions for thermal cycling are available on request from the corresponding author. Cycle sequencing was performed with a BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA, USA). The sequences were read following capillary electrophoresis on an ABI 3130 Genetic Analyzer (Applied Biosystems, USA).
Data were analysed with respect to age groups, gender, and the KRAS genotype using standard descriptive statistics. Associations between variables were tested through Fisher’s Exact Test. All p values below 0.05 were considered statistically significant.
Results
The colorectal tissue samples of 108 patients were tested. Of these, 25 (23.0%) tested positive for KRAS mutations. The remaining 83 (77.0%) samples had the wild-type allele. Overall, there were 68 (63.0%) males and 40 (37.0%) females. Among the KRAS positive cases, there were 14 (56.0%) males and 11 (44.0%) females. Their age distribution ranged from 29 to 85 years with a median age of 61 years. There were 15 patients (60.0%) with point mutations in codon 12 while 10 (40.0%) had a single mutation in codon 13. The most common KRAS mutation was NM_004985.3:c.38G>A; NP_004976.2: p.Gly13Asp (40.0%), followed by c.35G>T; p.Gly12Val (24.0%). The distribution of the different KRAS mutations identified in Sri Lankan mCRC patients is shown in Table 1. The G>A transitions in both codons 12 and 13 accounted for 64.0% of all the mutant KRAS cases. Figure 1 shows the percentage distribution of the mutant KRAS (G>A) transitions and (G>T) and (G>C) transversions identified in the Sri Lankan cohort.
Table 1.
Distribution of KRAS mutations in Sri Lankan colorectal cancer patients
| KRAS mutation type | Number of patients (n) | Percentage (%) |
|---|---|---|
| Codon 12 | ||
| p.Gly12Cys (c.34G>T) | 3 | 12.0 |
| p.Gly12Val (c.35G>T) | 5 | 20.0 |
| p.Gly12Val (c.35G>A) | 1 | 4.0 |
| p.Gly12Arg (c.34G>C) | 1 | 4.0 |
| p.Gly12Ser (c.34G>A) | 2 | 8.0 |
| p.Gly12Ser (c.35G>A) | 1 | 4.0 |
| p.Gly12Asp (c.35G>A) | 2 | 8.0 |
| Codon 13 | ||
| p.Gly13Asp (c.38G>A) | 10 | 40.0 |
| Total | 25 | 100.0 |
Fig. 1.
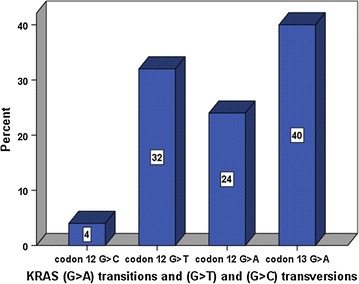
Percentage distribution of the mutant KRAS (G>A) transitions and (G>T) and (G>C) transversions in the study cohort
The KRAS mutation pattern according to gender correlated well with the overall KRAS mutant type. The p.Gly13Asp (c.38G>A) mutation had the highest frequency in both males (16.0%) and females (24.0%). The gender distribution of the KRAS genotypes is shown in Fig. 2. No statistically significant difference was found between the KRAS genotypes and the gender (p = 0.153, Fisher’s exact test). The highest percentage of mutant KRAS cases was found in the 50–59 years age group. The stratification of the mutant KRAS genotypes according to age groups is shown in Fig. 3. No statistically significant difference was found between the KRAS genotypes and the age groups (p = 0.981, Fisher’s exact test). Multiple mutations in the same individual were not detected in any of the patients in this cohort.
Fig. 2.
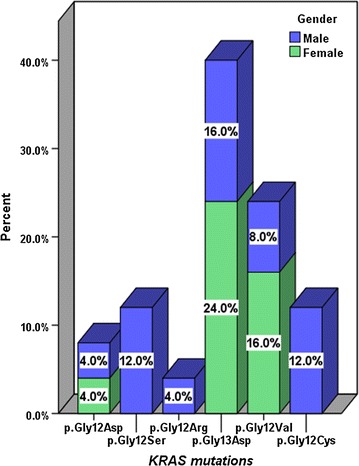
Percentage distribution of the KRAS mutations according to gender
Fig. 3.
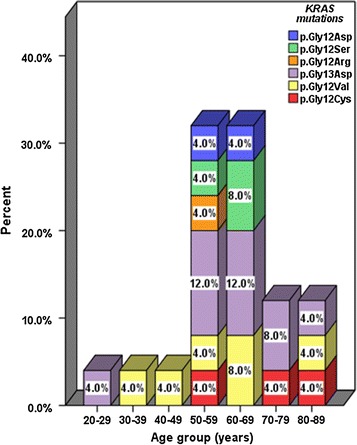
Percentage distribution of the mutant KRAS mutations according to age groups
Discussion
The KRAS mutation frequencies in Asian, European, and Latin American mCRC patients were reported to be 24.0, 36.0 and 40.0%, respectively [2]. The overall frequency (23.0%) observed in this study is in accordance with what is reported for Asian patients. It is unclear why a relatively lower frequency is observed in Asian patients. The difference of mutation status may result from various factors, such as the FFPE tissue samples, the percent of tumor cells, the extracted DNA quality, the testing methods and the testing target as well as different molecular pathogenetic mechanisms and environmental exposures [12].
Previous studies [2, 6, 7] have reported that the most common KRAS mutation types are p.Gly12Asp, p.Gly12Val and p.Gly13Asp, accounting for almost 70.0% of all mutations. Overall, a similar pattern was seen in this study where these 3 mutations accounted for 72.0% of all the KRAS mutations. The most common single mutation (40.0%) identified in our cohort was a G>A transition in codon 13 (c.38G>A; p.Gly13Asp), as opposed to mutations in codon 12 which have been reported in several Caucasian and Asian studies. Furthermore, our findings indicated a relatively lower frequency (8.0%) of the G>A transition in codon 12 (c.35G>A; p.Gly12Asp). However, it should be noted that our sample size was too small to draw meaningful conclusions regarding these variations. A larger study with sufficient numbers would be required to derive meaningful results to determine whether these variations are valid.
The frequency of KRAS mutations among 99 Japanese mCRC patients was reported to be 37.4% (37/99) [2]. Similar to our findings, the most prevalent mutation within codon 13 was the GGC→GAC (p.Gly13Asp) mutation found in 11 (29.7%) patients. However within codon 12, they obtained a higher frequency of the GGT→GAT (p.Gly12Asp) mutation found in 10 (27.0%) patients, followed by the GGT→GTT (p.Gly12Val) found in 8 (21.6%) patients. A similar pattern was reported in a Chinese study where KRAS mutations were detected in 33.3% (30/90) of the CRC tumor samples using the DNA sequencing method [3]. In a study conducted among Tunisian patients, KRAS somatic mutations were detected in the CRC tissue samples of 31.5% (16/51) patients [13]. According to their findings, 81.2% had a single mutation in codon 12 and 23.0% had a single mutation in codon 13. In contrast to our findings where transitions accounted for 64.0%, 81.3% of their mutations were transitions and 23.0% were transversions and the most common single mutation (50.0%) was a G>A transition in codon 12 (c.35G>A; p.Gly12Asp). A preponderance of the G>A transition in codon 12 (c.35G>A; p.Gly12Asp) has been reported in several other Caucasian and Asian studies [12, 14–19]. In contrast to these studies, the most common mutation identified in codon 12 in this study was p.Gly12Val (24.0%). A similar finding was observed in a retrospective study conducted among 299 patients with mCRC in India [7]. This may be a reflection of the genetic heterogeneity in the pattern of KRAS mutations in mCRC patients. However as stated earlier, valid conclusions cannot be drawn regarding these variations due to the limitations of this study.
Another factor that may influence the results is the method of detection and the sample type. Pyrosequencing, a real-time, non-electrophoretic, nucleotide-extension sequencing and next generation sequencing techniques, have been shown to be efficient in various applications in comparison to direct sequencing. A study demonstrated that pyrosequencing detected 17.9% of the KRAS mutations in patients with KRAS wild-type using direct sequencing alone (30 tumors among 168) [20]. It is believed that the presence of subclones harboring KRAS mutations within genetically heterogeneous tumors may explain the low-frequency KRAS mutations detected by the direct sequencing method [21]. It is important to note that approximately 20.0% of patients who test wild-type based on KRAS exon 2 analysis may actually harbour undetected extended RAS mutations in codons 59, 61 (exon 3), or codons 117 and 146 (exon 4) [22, 23].
Some studies have reported significant differences between the KRAS genotypes and clinicopathological variables such as age, gender, tumour location, tumour histopathology, and metastasis while other studies observed no significant effects [7, 11]. Similar to the findings in the Indian study reported by Veldore et al. [7], no significant correlation was found between the KRAS genotypes and the patients’ age and gender in this study. However, significantly higher mutation rates in well-differentiated tumours and tumours located at the distal end of the colon were observed in the Indian cohort.
Limitations
Our preliminary findings suggest a frequency of the KRAS mutation at 23.0% indicating that this testing is crucial for targeted therapy management in mCRC in Sri Lanka. Using KRAS testing to restrict the use of EGFR-inhibitor therapy would help to select the appropriate patients and avoid administration of unnecessary, ineffective and toxic treatments to patients who would not benefit from them. However, the present study had a number of limitations. Most importantly, it was a retrospective study, the sample size was small and clinico-pathological parameters of the tumour were not available for analysis. In addition, other biomarkers, such as KRAS exon 3 or 4, NRAS, or BRAF mutations were not evaluated. It is recommended that future studies be undertaken with larger samples to correlate the KRAS genotype with the clinicopathological parameters in Sri Lankan patients with mCRC. Analysis of other homologues such as NRAS and downstream signalling effectors such as BRAF, would also be valuable.
Authors’ contributions
NS drafted the manuscript, performed the statistical analysis and interpreted the data. KD critically revised and approved the final manuscript. MDEN carried out the molecular genetic studies and compiled the data. HHMP carried out the molecular genetic studies. VHWD conceived the study and critically revised the final manuscript. All authors read and approved the final manuscript.
Acknowledgements
Nil.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The primers and conditions for thermal cycling, and the datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable. This is a retrospective audit of an anonymized database completely de-identified from the original patients. For this type of study formal consent is not required.
Ethics approval and consent to participate
The protocol for the study confirms with the Code of Ethics of the World Medical Association (Declaration of Helsinki). This study was approved by the Ethics Review Committee of the Faculty of Medicine, University of Colombo, Sri Lanka [Protocol no. EC-15-027]. Written informed consent was obtained from the patients for genetic testing as part of standard care.
Funding
Nil.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- mCRC
metastatic colorectal cancer (mCRC)
- EGFR
epidermal growth factor receptor
- FFPE
formalin-fixed paraffin-embedded
Contributor Information
Nirmala Dushyanthi Sirisena, Phone: + 94 773499086, Email: nirmala@anat.cmb.ac.lk.
Kemal Deen, Email: radihan@mail.ewisl.net.
Dayupathi Eranda Nipunika Mandawala, Email: nipunika1@hotmail.com.
Pumindu Herath, Email: puminduherath@yahoo.com.
Vajira Harshadeva Weerabaddana Dissanayake, Email: vajira@anat.cmb.ac.lk.
References
- 1.Cancer Registry, National Cancer Control Programme. Sri Lanka cancer incidence data 2007; 2013.
- 2.Inoue Y, Saigusa S, Iwata T, Okugawa Y, Toiyama Y, Tanaka K, et al. The prognostic value of KRAS mutations in patients with colorectal cancer. Oncol Rep. 2012;28:1579–1584. doi: 10.3892/or.2012.1974. [DOI] [PubMed] [Google Scholar]
- 3.Tan C, Du X. KRAS mutation testing in metastatic colorectal cancer. World J Gastroenterol. 2012;18:5171–5180. doi: 10.3748/wjg.v18.i42.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harlé A, Filhine-Tresarrieu P, Husson M, Boidot R, Rouyer M, Dubois C, et al. Rare RAS mutations in metastatic colorectal cancer detected during routine RAS genotyping using next generation sequencing. Target Oncol. 2016;11:363–370. doi: 10.1007/s11523-015-0404-7. [DOI] [PubMed] [Google Scholar]
- 5.Yamane L, Scapulatempo-Neto C, Reis RM, Guimaraes D. Serrated pathway in colorectal carcinogenesis. World J Gastroenterol. 2014;20:2634–2640. doi: 10.3748/wjg.v20.i10.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behl AS, Goddard KA, Flottemesch TJ, Veenstra D, Meenan RT, Lin JS, et al. Cost-effectiveness analysis of screening for KRAS and BRAF mutations in metastatic colorectal cancer. J Natl Cancer Inst. 2012;104:1785–1795. doi: 10.1093/jnci/djs433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veldore VH, Rao MR, Prabhudesai SA, Tejaswi R, Kakara S, Pattanayak S, et al. Prevalence of KRAS mutations in metastatic colorectal cancer: a retrospective observational study from India. Indian J Cancer. 2014;51:531–537. doi: 10.4103/0019-509X.175371. [DOI] [PubMed] [Google Scholar]
- 8.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 9.Coppede F, Lopomo A, Spisni R, Migliore L. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World J Gastroenterol. 2014;20:943–956. doi: 10.3748/wjg.v20.i4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Ma BB. Colorectal cancer in Chinese patients: current and emerging treatment options. Oncotargets Ther. 2014;7:1817–1828. doi: 10.2147/OTT.S48409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, et al. Association of KRAS p. G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 12.Gil Ferreira C, Aran V, Zalcberg-Renault I, Victorino AP, Salem JH, Bonamino MH, et al. KRAS mutations: variable incidences in a Brazilian cohort of 8234 metastatic colorectal cancer patients. BMC Gastroenterol. 2014;14:73. doi: 10.1186/1471-230X-14-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aissi S, Buisine MP, Zerimech F, Kourda N, Moussa A, Manai M, et al. KRAS mutations in colorectal cancer from Tunisia: relationships with clinicopathologic variables and data on TP53 mutations and microsatellite instability. Mol Biol Rep. 2013;40:6107–6112. doi: 10.1007/s11033-013-2722-0. [DOI] [PubMed] [Google Scholar]
- 14.Elbjeirami WM, Sughayer MA. KRAS mutations and subtyping in colorectal cancer in Jordanian patients. Oncol Lett. 2012;4:705–710. doi: 10.3892/ol.2012.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurtado C, Encina G, Wielandt AM, Zarate AJ, Castro M, Carrillo K, et al. KRAS gene somatic mutations in Chilean patients with colorectal cancer. Rev Med Chile. 2014;142:1407–1414. doi: 10.4067/S0034-98872014001100007. [DOI] [PubMed] [Google Scholar]
- 16.Jakovljevic K, Malisic E, Cavic M, Krivokuca A, Dobricic J, Jankovic R. KRAS and BRAF mutations in Serbian patients with colorectal cancer. J BUON. 2012;17:575–580. [PubMed] [Google Scholar]
- 17.Marchoudi N, Amrani Hassani Joutei H, Jouali F, Fekkak J, Rhaissi H. Distribution of KRAS and BRAF mutations in Moroccan patients with advanced colorectal cancer. Pathol Biol. 2013;61:273–276. doi: 10.1016/j.patbio.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Mazurenko NN, Gagarin IM, Tsyganova IV, Mochal’nikova VV, Breder VV. The frequency and spectrum of KRAS mutations in metastatic colorectal cancer. Vopr Onkol. 2013;59:751–755. [PubMed] [Google Scholar]
- 19.Nakanishi R, Harada J, Tuul M, Zhao Y, Ando K, Saeki H, et al. Prognostic relevance of KRAS and BRAF mutations in Japanese patients with colorectal cancer. Int J Clin Oncol. 2013;18:1042–1048. doi: 10.1007/s10147-012-0501-x. [DOI] [PubMed] [Google Scholar]
- 20.Tougeron D, Lecomte T, Pagès JC, Villalva C, Collin C, Ferru A, et al. Effect of low-frequency KRAS mutations on the response to anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol. 2013;24:1267–1273. doi: 10.1093/annonc/mds620. [DOI] [PubMed] [Google Scholar]
- 21.Atreya CE, Corcoran RB, Kopetz S. Expanded RAS: refining the patient population. J Clin Oncol. 2015;33:682–685. doi: 10.1200/JCO.2014.58.9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hecht JR, Douillard JY, Schwartzberg L, Grothey A, Kopetz S, Rong A, et al. Extended RAS analysis for anti-epidermal growth factor therapy in patients with metastatic colorectal cancer. Cancer Treat Rev. 2015;41:653–659. doi: 10.1016/j.ctrv.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Sorich MJ, Wiese MD, Rowland A, Kichenadasse G, McKinnon RA, Karapetis CS. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Ann Oncol. 2015;26:13–21. doi: 10.1093/annonc/mdu378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The primers and conditions for thermal cycling, and the datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.


