Introduction
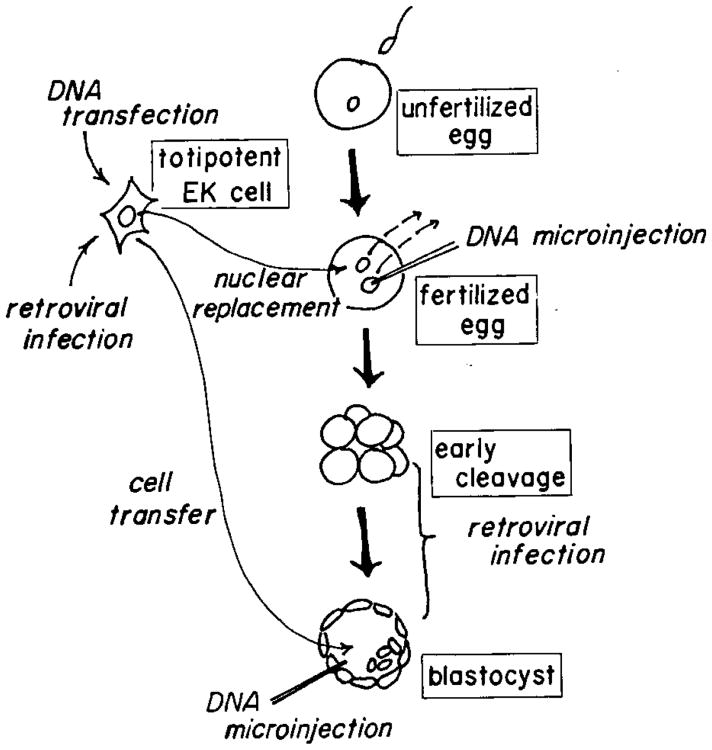
The ability to introduce functional genes into organisms provides a powerful tool for dissecting complex biological processes. Gene transfer is especially valuable in diploid organisms with long life cycles where classical genetic approaches are impractical. A variety of methods for introducing foreign DNA into both somatic and germ cells of mammals have been developed over the last 15 years (Figure 1). Of fundamental importance for germ-line transformation was the contribution of mammalian embryologists, who developed techniques for removing embryos, culturing them briefly in vitro, and returning them to foster mothers where normal embryogenesis could proceed (5). These techniques opened the way for combining embryonic cells from one animal with those of another to produce chimeric animals. They also provided a means of introducing cultured teratocarcinoma cells, which are derived from germ cells or early embryos, into developing embryos (67). At the same time, geneticists and virologists were developing methods for introducing selectable genes into tissue culture cells. These advances presented the possibility of transferring genes into teratocarcinoma cells and then introducing those cells into the blastocyst of developing embryos to produce mosaic animals (70). As an alternative to cell transfer, replacing the pronuclei of fertilized eggs with nuclei of genetically modified teratocarcinoma cells was suggested because of the success of nuclear transplantation in amphibians (28a). The ability to manipulate embryos also presented a way for infecting early embryos with intact viruses or viral DNAs. Experiments performed in the mid-1970s showed that infection of preimplantation embryos with murine leukemia virus (MuLV) resulted in mice with the retroviral DNA integrated into both somatic and germ cells (40). The dissection of the retroviral life cycle paved the way for genetically engineering these viruses to carry exogenous genes; these recombinant viruses could then be used to infect preimplantation embryos. Although each of these techniques has considerable potential for introducing foreign genes into the germ line, none of them has developed into a routine procedure with wide application.
Figure 1.
Methods of producing transgenic mice that have been devised or proposed. The most popular approach involves direct microinjection of a few hundred copies of linearized DNA into one of the pronuclei of a fertilized egg. DNA has also been injected into the cytoplasm, nuclei of two-cell eggs, or into the blastocoel cavity. Another strategy involves introducing DNA into totipotent teratocarcinoma cells and then mixing some of these cells with normal blastocyst cells to produce a chimeric mouse or using their nuclei to replace the pronuclei of fertilized eggs. Genetically engineered retroviruses are also being developed that carry foreign genes and can infect early embryos or tissue culture cells.
In contrast, a method that has been used extensively involves direct microinjection of DNA into the pronucleus. Techniques for injecting mRNA and then cloned genes, as they became available, were rapidly developed for the large amphibian eggs (29). Meanwhile, microinjection of viral or cellular genes into tissue culture cells and means of detecting their expression were mastered. During the late 1970s, these methods were adapted for microinjection of mRNA, and then DNA, into mouse eggs (6, 7). In late 1980, the first report describing transgenic mice that developed from microinjected eggs was published (27), and within the next few months four other groups reported similar success in stably integrating foreign DNA into the genome of the mouse (8, 21, 116, 117). Moreover, evidence suggested that at least some of the foreign genes could be expressed (8, 116, 117) and that the foreign genes were not only incorporated into somatic tissues but also into the germ line (21, 24, 79, 98). Furthermore, offspring of transgenic founder mice often continued to express the foreign genes (79). These facts, along with the observation that it was possible to produce significant amounts of biologically active gene products that would affect the physiology of the mouse (77), sparked considerable interest in this new approach to manipulating the genome of mammals.
Two minireviews (11, 78), as well as more extensive reviews dealing primarily with the more biological aspects of transgenic mouse experiments (12, 25, 115), have been published. In this review we focus on the molecular biology of DNA integration and subsequent expression of genes introduced into mice by the microinjection approach. We include data on genes that are expressed, as well as those that are not expressed, when introduced into the genome of developing mice. In addition, this review attempts, by means of the tables, to summarize the latest information regarding tissue-specific gene expression, regulated gene expression, oncogenesis, and insertional mutagenesis. Although many fascinating biological problems are currently being tackled with the aid of germ-line transformation, we describe only a few examples, mainly from our own work, for illustrative purposes. We also mention some of the more perplexing observations that have emerged from the study of gene expression in transgenic mice with the hope that they may stimulate discussion and, ultimately, experiments to help understand these unexpected findings.
Methods of Gene Transfer
Pronuclear Microinjection
The most prevalent technique for introducing genes into the germ line is direct microinjection of cloned DNA into pronuclei of fertilized eggs. During the six years since the first successful application of this approach, about fifty research groups have used the microinjection technique. Several thousand transgenic mice, as well as a few transgenic sheep, pigs, and rabbits (34), have been produced by this method. Pronuclear microinjection is conceptually straightforward, although it demands special equipment and technical skill, and has the additional feature that any cloned DNA can be used. Technical aspects of the microinjection procedure and some of the important parameters for optimizing integration of foreign DNA have been described (9, 26, 38). When linear DNA molecules are injected into pronuclei, about 25% of the mice that are born carry one or more copies of the injected DNA (9). These animals are generally called “transgenic,” and the foreign genes are often referred to as “trans genes.” Because integration usually occurs prior to DNA replication, about 70% of the transgenic mice carry the trans genes in all of their cells, including the germ cells. In the remaining 30% of the transgenic mice, integration apparently occurs after one or more rounds of replication; hence, the transgenes are in only a fraction of the cells. These mice usually show the same degree of mosaicism in somatic and germ cells, but in some mice the germ line is deficient (121).
The mechanism of integration of microinjected DNA is unknown. When more than one copy integrates, they usually all integrate at the same chromosomal site in a tandem, head-to-tail array, although other arrangements have been documented (25, 34, 83) and iontophoretic techniques have been reported to produce multiple integrants (55). The chromosomal site of integration is probably determined randomly: foreign DNA integrates on many different autosomes (50, 53) as well as on the X (56) and Y chromosomes (E. Wagner & M. LeMeur, unpublished observations). There is no evidence for site-directed integration (9). As a working hypothesis for the integration mechanism, we proposed (9) that randomly generated chromosomal breaks are the rate-limiting step for integration (to explain the predominance of single integration sites), that the ends of the injected DNA molecules initiate integration at these breaks (to account for the fivefold increase in integration efficiency with linear vs circular DNA), and that homologous recombination occurs among injected molecules (to explain the prevalence of tandem arrays) but not between the injected DNA and chromosomal sequences (possibly due to differences in chromatin structures between injected and chromosomal DNAs). Although this mechanism is plausible, evidence for it is scant. Some aspects of this model can be tested; for example, injecting two different MT-hGH (human growth hormone) genes with nonoverlapping deletions yielded some instances of homologous recombination between these genes: intact, recombinant MT-hGH genes (some of which produced functional hGH) were present in the transgenic mice that developed (81). Cloning the junction fragments between the transgenes and chromosomal DNA will also provide clues about the events that occur during integration. Although only a few junctions have been characterized so far (see section on insertional mutagenesis), deletions, duplications, and translocations of chromosomal sequences can clearly occur at the site of integration, and some junctions contain short novel DNA sequences found neither in the injected DNA nor in the vicinity of the integration site. These findings are reminiscent of observations made by studying breakpoints in naturally occurring chromosomal translocations (20), suggesting that similar mechanisms may operate during integration of foreign DNA. Indeed, a chromosomal translocation was recently reported in a transgenic mouse injected with an RSV-CAT construct (74).
In general, the foreign DNA is stably transmitted for many generations with no evidence of rearrangement. However, a few examples have been reported in which transgenes were either rearranged, partially deleted, or amplified (10, 79, 83, 94). In nonmammalian species, DNA injected into eggs can be replicated extrachromosomally (68, 90, 99). This replication is apparently rare in mammals (18, 122), although retention of extrachromosomal plasmid sequences has been reported in one transgenic mouse (27). Experiments with bovine papilloma virus, which normally replicates extrachromosomally, indicate that it integrates into the chromosomes of transgenic mice, but interestingly, extrachromosomal replication is evident in skin tumors that develop in these mice (52a). In another study, some unintegrated copies of bovine papilloma virus DNA were detected in total fetal DNA derived from microinjected eggs (our unpublished observations).
Other Approaches for Introducing DNA into the Germ Line
In addition to microinjection of pronuclei, foreign DNA has been incorporated into the genome of mice by microinjection of DNA into the cytoplasm (9, 36), into the nuclei of two-cell embryos (9), and into the blastocoel cavity (42). There is no clear advantage to any of these alternate routes of injection for making transgenic mice because the efficiency is lower and mosaicism becomes prevalent when DNA is injected at later stages of development.
Infection of preimplantation embryos with natural or genetically engineered retroviruses has been successful (40, 44, 111), and this technique is currently receiving considerable attention because it offers some useful advantages over microinjection for certain applications. Its principal advantages are: (a) the ease of inserting DNA into embryos (eight-cell embryos from which the zona pellucida has been removed are placed in a tissue culture.dish with fibroblasts producing the virus to allow infection, then they are transferred to foster mothers for continued development) and (b) the viral integration mechanism allows single copies of the retroviral DNA to insert into the chromosome such that genes cloned into these vectors are flanked by the retroviral long terminal repeats after integration. Its disadvantages are: (a) the added steps required to produce a retrovirus carrying the gene of interest, (b) the size limit of the foreign DNA insert and constraints due to splicing and termination signals that may lie within the DNA of interest, (c) the mosaicism of the founder animals that results because infection occurs after cell division begins, which necessitates outbreeding to establish pure lines suitable for analysis of gene expression, and (d) the possible interference of retroviral DNA sequences with expression of genes that it carries. Although some foreign genes have now been expressed in transgenic mice produced by this method (44, 111), whether tissue-specific enhancers can be expressed appropriately when flanked by retroviral LTRs is still unclear. Thus, although very promising for some applications, the general utility of this method is not yet established.
Another method of introducing genes into the germ line involves introducing DNA into totipotent teratocarcinoma cells or, more recently, into embryonic stem cells (3a), and then incorporating these cells into the blastocyst of developing embryos or aggregating them with eight-cell embryos (58, 96, 114; M. Evans, unpublished observations). The attractive feature of this approach is that one could introduce the foreign DNA into cultured cells by any desired means and then select or screen the cells for desirable characteristics prior to manipulating them into an embryo. A variation on this theme involves replacing the pronuclei of a fertilized egg with the nucleus from an embryonic stem cell. Although an attractive idea, current evidence suggests that only nuclei from very early mouse embryos are capable of supporting development (65); therefore, the feasibility of this approach is dubious. In addition, these methods are limited by the inability to select for appropriate expression of many interesting genes in cell culture, as well as by the difficulty in maintaining the normal diploid complement of chromosomes.
Gene Expression
One of the most important features of gene transfer into mammals is that most, but not all, of the genes examined are appropriately expressed. Table 1 summarizes the results obtained with a variety of natural genes, some of which have been marked to discriminate their expression from endogenous genes. Table 2 summarizes results obtained with hybrid genes in which the regulatory elements of one gene are fused to the structural elements of another gene.
Table 1. Expression of natural genes microinjected into mouse eggs.
| Genea | Spb | Vector | Expression | Tissues | Ref.c |
|---|---|---|---|---|---|
| α-Actin | r | —d | 6/6 | mu, he | (94) |
| α-Actin | r | pBR | 1/4 | te, th, lu | (94) |
| Amylase-2.2 (cosmid) | m | pJB | 2/3 | pa | (a) |
| Amylase-2.2. (minigene) | m | — | 2/2 | pa | (a) |
| αl(II) Collagen | h | cos 4 | ND/2 | (b) | |
| Eα, MHC class II | m | — | 2/2 | various | (124) |
| Eα, MHC class II | m | pBR | 4/4 | various | (85) |
| Eα, MHC class II | m | — | 1/1 | various | (54) |
| Eα, MHC class II | m | λ | 1/1 | various | (54) |
| E1A, ElBf | Adl2 | — | ND/4 | e | (c) |
| Elastase-1 | r | pBR | 5/5 | pa | (107) |
| α-Fetoprotein | m | pBR | 15/34 | ys, li | (51) |
| α-Fetoprotein | m | pBR | 12/12 | ys, li | (52) |
| β-Globin | h | — | 7/10 | ery | (d) |
| β-Globin | h | — | 15/20 | ery | (109) |
| β-Globin | h | pUC | 3/16 | ery | (109) |
| β-Globin | h | — | 3/3 | ery | (108) |
| β-Globin | m | — | 2/5 | ery | (d) |
| β-Globin | m | pBR | ?/6 | (e) | |
| β-Globin | rb | λ | 2/9 | te, mu | (53) |
| β-Globin | h | pBR | 0/2 | (98) | |
| β-Globin | rb | ? | 5/? | ery | (117) |
| β-Globin | h | cos 4 | 1/2 | ery | (b) |
| δ-β Globin | h | λ | 0/1 | (39) | |
| ε-Globin | h | — | 0/6 | (d) | |
| γG-Globin | h | — | 3/6 | ery | (16) |
| γA-Globin | h | — | 2/5 | ery | (d) |
| γA+γG-Globin | h | — | 4/7 | ery | (c) |
| α1, α2-Globin (14 kb) | h | — | 0/11 | (c) | |
| α1-Globin (10.4 kb) | m | pBR | 1/19 | br | (88) |
| α1-Globin (10.4 kb) | m | — | 0/17 | (88) | |
| α1-Globin (cosmid) | m | pBR | 3/5 | ery | (f) |
| Gonadotropin (α-subunit) | h | pBR | 4/4 | pit | (g) |
| Growth hormone | h | pBR | 0/6 | (113) | |
| Growth hormone | h | — | 0/8 | (33) | |
| Growth hormone | r | pBR | 0/8 | (33) | |
| Hepatitis surface antigen | HBV | — | 1/10 | various | (19) |
| Hepatitis surface antigen | HBV | pBR | 2/6 | li | (3) |
| Immunoglobulin κ (14 kb) | m | pBR | 6/6 | B | (102) |
| Immunoglobulin κ (4.5 kb) | m | — | 0/5 | (f) | |
| Immunoglobulin μ | m | pBR | 5/5 | B, T | (28) |
| Immunoglobulin μ(γl) | h | λ | 1/2 | B | (125) |
| Immunoglobulin μ+κ | m | pBR | 1/1 | B | (89) |
| Immunoglobulin μ+κ | m | SVneo | 3/3 | B | (g) |
| Insulin | h | — | 4/4 | β-cells | (14, 91a) |
| Insulin | h | pBR | 0/3 | (15, 110) | |
| H-2Ld | m | λ | 0/1 | (d) | |
| HLA, Cw3, MHC class I | h | — | 3/4 | various | (h) |
| SLA, MHC class I | P | — | 1/1 | various | (23) |
| c-mycf | h | pBR | 0/5 | (i) | |
| αl(I) Collagen (cosmid) | h | pBR | 2/2 | various | (j) |
| Myosin light chain-2 | r | — | 4/8 | mu | (93, k) |
| Ovalbumin | c | pBR | 0/12 | (c) | |
| P-element | d | pBR | 0/2 | (47) | |
| Protamine 1 | m | — | 4/4 | testis | (c) |
| T-cell receptor-β | m | λ | 0/5 | (c) | |
| Thymidine kinase | HSV | pBR | 1/5 | fetus | (116) |
| Thymidine kinase | HSV | pBR | 1/8 | li | (c) |
| Transferrin | c | pBR | 6/7 | various | (66) |
This table includes genes microinjected into mouse eggs in which the promoter and enhancer remain associated with the structural gene; however, the structural genes may be modified to facilitate their detection.
Abbreviations:-Sp, species; c, chicken; d, Drosophila; h, human; m, mouse; p, pig; r, rat; rb, rabbit; b, bacterial; v, viral; Ad 12, adenovirus 12; HBV, hepatitis B virus; HSV, herpes simplex virus, type 1. Tissues: br, brain; B, lymphocytes; β, pancreatic β-cells; cp, choroid plexus; he, cardiac muscle; ery, erythroid cells; ki, kidney; li, liver; lu, lung; mu, skeletal muscle; pa, exocrine pancreas; pit, pituitary; te, testis; th, thymus; T, thymocytes; ys, yolk sac.
Unpublished observations from: (a) L. Treisman, M. Rosenberg, S. Keller & M. Meisler; (b) R. Lovell-Badge; (c) ours; (d) F. Costantini; (e) C. Lo; (f) S. Rusconi; (g) N. Fox, L. Hwang & D. Solter; (h) O. Dill; (i) C. Babinet; (j) R. Jaenisch & A. Berns; (k) M. Shani.
—, all plasmid vector sequences removed prior to injection.
None of the four mice (2 females, 2 males) transmitted the transgene to offspring.
Note that genes that are oncogenic are listed in Table 4 and not included here.
Table 2. Expression of hybrid genes in transgenic mice.
| Gene (promoter/structural)a | Vector | Expression | Tissues | Coramentsb | Ref.c |
|---|---|---|---|---|---|
| mMT-I/rGH | —d | 6/7 | var | growth (high) | (77) |
| mMT-I/hGH | — | 23/33 | var | growth (high) | (82) |
| mMT-I/hGH | — | 7/10 | var | growth (high) | (30) |
| mMT-I/pGH (cDNA) | — | 2/11 | var | growth (very low) | (b) |
| mMT-MiGRF (minigene) | — | 11/14 | var | growth (high) | (31) |
| mMT-I/hα-globin | pBR | 0/9 | (li) | (h) | |
| mMT-I/bβ-Gal | pBR | 0/11 | (li) | (i) | |
| mMT-I/hPLA | pBR | 2/5 | var | no effect (high) | (81) |
| mMT-I/vSrcg | pBR | ND/4 | no tumors | (h) | |
| mMT-I/rSomatostatin | pBR | 8/11 | var | no effect | (59) |
| mMT-I/hFactor IX | pBR | 0/21 | (li) | (h) | |
| mMT-I/hFactor IX | — | 0/10 | (li) | (h) | |
| mMT-I/mPOMC | pBR | 0/6 | (li) | (h) | |
| mMT-I/mPOMC | — | 0/6 | (li) | (h) | |
| mMT-I/vAd2 E1A (13S cDNA) | pBR | 2/2 | var | spermatogenesisf | (d) |
| mMT-I/vTK | pBR | 6/9 | var | (very low mRNA) | (79) |
| mMT-I/hHPRT | pBR | 4/17 | var | (high in brain) | (103) |
| mMT-I/mMycg | pBR | ND/23 | no tumors | (1) | |
| mMT-I/hIGF-I (cDNA) | — | 5/21 | var | no effect (moderate) | (h) |
| mMT-I/hiGF-II (cDNA) | — | 2/17 | var | no effect (low) | (h) |
| mMT-I/vHBsAg | — | 2/6 | var | high serum protein | (19) |
| mMT-I/hTGF-α | — | 0/7 | (li) | (h) | |
| mMT-I/hMycg | pUC | 2/5 | li | (e) | |
| hMT-UA/hGH | — | 4/8 | var | growth (high) | (30) |
| hMT-IIA/cFosg | — | 2/7 | te, pa | (b) | |
| hMT-IIA/cFos(polyoma T)g | pBR | 1/2 | te | (b) | |
| hMT-IIA/cFos(SVneo)g | pBR | 1/3 | te | (b) | |
| hMT-IIA/cFosmod g | pBR | 1/1 | te/br | (b) | |
| hMT-ITA/cFosLTRg | pBR | 2/5 | var | (b) | |
| hMT-IIA/bCAT | pBR | 0/3 | (b) | ||
| hMT-HA/pGH (cDNA) | — | 12/19 | var | growth | (a) |
| mSAA-2/hGH | — | 4/9 | li | no growth (Table 3) | (h) |
| mMyc/mlgA (translocation) | λ | 0/3 | (f) | ||
| mα2(I)CoIlagen/bCAT | pBR | 7/8 | var | (48, 120) | |
| cTf/rbβ-globin | pBR | 0/7 | (d) | ||
| mH-2K(class I MHC)/hGH | pBR | 13/13 | var | growth | (c) |
| vTK/hGH | — | 1/12 | var | growth (very low) | (h) |
| vLTR(MCF13)/hGH | — | 3/7 | var | growth (moderate) | (h) |
| vLTR/hMycg | pBR | 0/5 | (g) | ||
| vRSV/bCAT | pBR | 5/9 | var | (74) | |
| vMTV/vTK | pBR | 2/3 | var | (low) | (87) |
| rElastase-1/bNeo | pBR | 4/4 | pa | G418r cells | (h) |
| rElastase- l/mMycg | pBR | ND/9 | no tumors | (h) | |
| rElastase- l/vSrcg | — | ND/14 | no tumors | (h) | |
| rElastase-1/hGH | pBR | 8/11 | pa | no growth (very high) | (72, 73) |
| rElastase-1/hGH | — | 8/10 | pa | no growth (very high) | (72, 73) |
| mAlbumin/hGH (12 kb) | — | 16/16 | li | growth (very high) | (h) |
| mAlbumin/vHBsAg | — | 8/8 | li | moderate serum protein | (h) |
| mAlbumin/hFactorlX | — | 0/12 | (li) | (h) | |
| mAlbumin/hiGF-I | — | 2/6 | li | (very low) | (h) |
| mAlbumin/hLDL receptor | — | 9/12 | li | (very low) | (h) |
| mTransferrin/hGH | — | 11/11 | li | growth (very high) | (h) |
| hGRF/mNGF | — | ?/10 | nd | (i) | |
| mαA-Crystallin/bCAT | — | 2/2 | lens | (75, 120) | |
| mβ-Globin/bGalK | pBR | 0/2 | (39) | ||
| mIgH-cTf/rbβ-Globin | pBR | 3/3 | sp | (d) | |
| mIgH-mIgκ/rbβ-globin | pBR | 4/5 | sp | (d) | |
| mAmylase-la P/vSV40 T-Agg | — | 1/2 | ut,br,sp,ki | (e) |
This table includes hybrid genes that have been tested by microinjection into mouse eggs. These genes have the promoter/enhancer of one gene fused to the structural region of another gene; The slash (/) separates the two elements. The species is indicated by a lower case letter (same as in Table 1) before the abbreviation of the gene. The gene abbreviations are: Ad2, adenovirus 2; CAT, chloramphenicol acetyl-transferase; Factor EX, blood clotting factor; GH, growth hormone; GRF, growth-hormone releasing factor; Gal, galactosidase; GalK, galactose kinase; HBsAg, hepatitis B surface antigen; HPRT, hypoxanthine phosphoribosyl transferase; IGF, insulinlike growth factor; Ig, immunoglobulin; LDL, low-density lipoprotein; LTR, long terminal repeat; MHC, major histocompatibility complex; MT, metallothionein-1; Neo, neomycin phosphotransferase; NGF, nerve growth factor; PL, placental lactogen; POMC, proopiomelanocortin; RSV, Rous sarcoma virus; SAA, serum amyloid; SV40, simian virus 40; TGF, tumor growth factor; Tf, transferrin.
The relative level of expression is denoted in parentheses in some cases.
Unpublished observations from: (a) R. Seamark; (b) U. Ruether; (c) C. Babinet; (d) M. LeMeur, (e) N. Fox, L. Hwang & D. Solter; (OS. Ross; (g) C. Cremisi & C. Babinet (EMBO J., In press); (h) R. D. Palmiter & R. L. Brinster (i) H. van der Putten, F. Botteri & R. Evans.
The frequency of expression (the number of transgenic mice that express the transgene/total number of transgenic mice produced). In some cases the expression has not yet been determined (ND).
Tissues where expression was detected. Abbreviations are: br, brain; ki, kidney; li, liver; mg, mammary gland; pa, pancreas; sp, spleen; te, testis; ut, uterus; var, various tissues. (Tissues in parentheses indicate where expression was examined when there was no expression).
Defects in spermatogenesis.
Note that the oncogenic gene constructs are included in Table 4, and they are not duplicated here.
Tissue-Specific Gene Expression
Animal development depends upon differential expression of many genes in various cells that make up that organism. Some genes are expressed exclusively in one cell type, others are expressed in only a few cell types, and some are expressed in most cells. Genes from each of these categories have been appropriately expressed when introduced into mice. The usual strategy for studying the structural requirements for tissue-specific gene expression in transgenic animals involves injecting DNA containing the gene of interest and assaying various tissues for specific mRNA or protein. In order to discriminate the products of the injected gene from those of the endogenous counterpart, the injected gene must be marked in some way. For example, the gene may be from a different species, it may be a “minigene” with some exons deleted, or it may be modified by inserting or deleting a few nucleotides. Another strategy is to construct hybrid genes in which the control elements of the gene of interest are used to direct the expression of a “reporter” gene. Reporter genes might code for an easily assayable enzyme not normally found in the mouse, a hormone that has a pronounced physiological effect, a polypeptide that is easily identified immunologically, or an oncogene that could lead to a tumor of the cell type in which it is expressed.
Genes Expressed Predominantly in One Cell Type
A number of genes normally expressed only in one cell type have been studied. The first examples of tissue-specific expression were obtained with an immunoglobulin κ gene (13, 102) and an elastase-I gene (107); subsequently, several globin genes, myosin light-chain-2, α-actin, α-crystallin, insulin, and several other genes have been expressed in a tissue-specific manner (see Tables 1 and 2). Transgenic mice have been produced in which each of these genes is expressed predominantly, if not exclusively, in cells where the endogenous gene is also expressed. As a general conclusion, it appears that if a tissue-specific gene is expressed at all, then it is expressed appropriately, despite the fact that it is integrated at a different chromosomal location. This finding implies that trans-acting proteins involved in establishing tissue-specific expression are capable of finding their cognate sequences and activating transcription at most chromosomal locations. However, the level of expression of a particular gene varies widely from one founder animal to another (17, 72, 107, 109), suggesting that chromosomal position can influence accessibility of the genes to transcription factors. Moreover, usually a few transgenic mice produced with any construct do not express the gene at all, which may be due to its integration into heterochromatin domains. The quantitative analysis of trans-gene expression is complicated because multiple copies of the genes are usually integrated in a tandem array, and no means exists of determining how many of these genes are functional templates for transcription. The poor correlation between gene copy number and expression (17, 72, 107, 109) suggests that only a few of the genes are expressed or that the entire array is very sensitive to chromosomal position. Nevertheless, for several different transgenes, the level of expression in some mice approaches or exceeds that of the endogenous genes, which indicates that optimal expression probably does not depend upon the normal chromosomal position. Another tentative conclusion is that the signals for tissue-specific expression, and their recognition appear to be evolutionarily conserved between mammals because no species specificity has been documented. For example, many human genes have been expressed in transgenic mice (see Tables 1 and 2).
Genes Expressed in Several Cell Types
Genes normally expressed in several cell types have also been tested in transgenic mice. For example, the immunoglobulin heavy-chain genes directs the synthesis of “sterile” transcripts in both B- and T-cells prior to rearrangement (28). The α-fetoprotein (AFP) gene is induced in yolk sac, fetal liver, and gastrointestinal cells; subsequently, its expression declines in all of these cell types (51, 52). The transferrin gene, which is expressed predominantly in liver, testis (Sertoli cells), and brain, is another example of a gene whose expression is restricted to just a few cell types (66). Each of these genes has been expressed in appropriate cells of transgenic mice. One might expect that localizing the enhancers for these genes might be more complex than for those expressed in only one cell type. Indeed, the enhancers of the AFP gene appear to be spread out over a region extending 7-kb 5′ of the gene (52). Similarly, a major enhancer element for the albumin gene lies 9–12 kb upstream of the promoter (our unpublished observations).
Genes Expressed in Many Cell Types
Genes transcribed from metallothionein (MT), collagen, and various viral promoters fall into this category. The endogenous mouse MT-I gene is expressed in virtually all cells, although at markedly different levels; furthermore, the responsiveness of this gene to the multitude of inducers (glucocorticoids, metals, inflammatory signals, interferon) varies among tissues (76). One might expect that expression of an MT fusion gene in transgenic mice would parallel that of the endogenous gene as long as all of the essential cis-acting regulatory elements were present. However, this expectation is not fully realized (82). Generally, expression of MT fusion genes is high in liver, intestine, kidney, heart, pancreas, and testis, but their level of expression varies widely among tissues and among founder animals. Furthermore, occasionally mice show unusually distorted expression relative to that of the endogenous MT-I gene; for example, very high expression in kidney or pancreas with virtually none in liver (82; our unpublished observations). Whether the normal tissue distribution of MT-I gene expression is a consequence of the various inducible elements (such as the metal regulatory elements) or whether an enhancer acts independently of these elements is unclear (76). The variable expression among mice and among tissues suggests that the MT-I promoter is very sensitive to chromosomal context. With an absolutely tissue-specific enhancer, the chromosomal context can only influence the level of expression in one cell type, whereas with a promoter/enhancer that functions in many different cell types, the chromosomal influence could vary in different cell types, and from one founder animal to another, depending upon the site of integration.
Enhancer Localization
The cis-acting DNA elements responsible for tissue-specific gene expression can be effectively localized by the use of transgenic mice. In fact, for genes expressed in cells without tissue culture counterparts, production of transgenic animals is the only current means of localizing the control elements. It is also the only means of demonstrating that an element specifies expression exclusively in the appropriate cell type and that an element allows correct developmental expression. Examples where the tissue-specific control elements (enhancers) lie just upstream of the promoter, many kilobases upstream from the promoter, or within the genes they regulate have been described. In some cases the promoter also behaves in a tissue-specific manner.
If the level of expression of a transgene in some of the transgenic animals approximates that of the endogenous gene, then all of the essential cis-acting elements are probably present; if not, then some important elements may have been deleted or destroyed in preparation of the construct or chromosomal position may be very important for that gene. Once normal expression is achieved, then mutational strategies can be devised to locate the cis-acting elements important for tissue-specific expression. This approach can be straightforward for delineating a single element, but can be very tricky (and tedious) with multiple elements that act in either a redundant or a cooperative manner. This strategy can also be problematical if the element (or elements) lie within the gene of interest, as the deletions may affect the transcripts being analyzed. Other useful clues to the location of important elements can be derived from DNA-sequence comparisons of related genes from different organisms or of different genes expressed coordinately in a tissue-specific manner. The location of DNase I–hypersensitive sites also provides a useful guide to enhancer localization. Formal proof that a tissue-specific enhancer element has been identified involves showing that it, or a synthetic derivative of it, can appropriately control a heterologous promoter, regardless of orientation (92).
As an example of how a relatively simple tissue-specific element was elucidated, we summarize our studies on the rat elastase-I gene. This gene codes for a serine protease expressed predominantly in the exocrine pancreas, where about 10,000 mRNA molecules accumulate per cell (61). When a DNA fragment containing the entire rat gene and 7.2 kb of the 5′ flanking sequence was tested in transgenic mice, many of the mice expressed 10,000–120,000 elastase mRNA molecules per pancreatic acinar cell (107). Deletions that removed all but 205 bp of the 5′ flanking sequence were also expressed in a pancreas-specific manner (72). This 205 bp of the 5′ flanking sequence was also able to direct the expression of hGH exclusively to the acinar cells of the pancreas, as demonstrated by mRNA quantitation and by detection of hGH with fluorescent antibodies (72, 73). Furthermore, none of the mice grew larger than normal, indicating that no cells capable of secreting hGH into the bloodstream synthesized significant amounts of hGH. Deletions that left only 72 bp of the 5′ flanking sequence were inactive. This finding implies that the DNA sequence located between −205 and −72 is necessary for the tissue-specific expression of the elastase-I gene. This sequence has all the properties of a tissue-specific enhancer element: it also functions when moved 3-kb upstream of its normal location or when inserted into an intron, and it can also activate heterologous promoters appropriately. The sequence within this region is homologous to sequences in other serine protease genes (such as chymotrypsin) also expressed in the exocrine pancreas and shown by cell transfection assays to promote expression specifically in a pancreatic acinar cell line (61). Furthermore, a DNase I–hypersensitive site maps to this region of the elastase-hGH gene (73). These experiments strongly suggest that this 133-bp region contains all of the information necessary to direct the normal level of expression of an associated structural gene exclusively to the acinar cells of the pancreas.
Developmental Expression
Where it has been analyzed, expression of transgenes appears to be activated at the appropriate developmental time, i.e. coordinately with the endogenous genes. This finding has been documented for human β-globin genes (62, 109), AFP (52), α-actin (94), elastase-hGH (our unpublished observations), αA-crystallin (75), α2(I)-collagen (48), and murine immunoglobulin genes (101, 118, 125). These results are most easily explained by the developmental timmg of the appearance of functional transacting proteins that recognize the associated tissue-specific enhancer elements of the transgenes. According to this view, developmental timing and tissue-specific expression are controlled by the same elements. However, enhancers may be required only to establish tissue-specific expression. Data obtained from certain B-cell variants suggest that the immunoglobulin enhancers may not be necessary for maintaining tissue-specific gene expression (49, 112).
Curiously, human fetal γG -globm genes are expressed early in embryogenesis, along with embryonic murine globin genes (16; our unpublished observations). These data suggest that the enhancer element (or elements) of the human fetal globin genes evolved from the same primordial gene that gave rise to murine embryonic globin genes, and that the trans-acting proteins continue to recognize similar DNA signals (16).
Modulation of Gene Expression
The rate of transcription of some genes can be modulated by environmental signals. This type of transcriptional regulation is also mediated by cis-acting elements that are frequently reiterated several times m the promoter region. Examples of well-characterized elements are those involved in glucocorticoid hormone action and in regulation by heavy metals (76, 84, 104). In the case of glucocorticoids, both the cis-acting elements and the trans-acting regulator protein (the glucocorticoid hormone receptor) have been identified, and their interaction has been extensively studied in vitro (84, 123a). Most of the other environmental signals probably act in a similar manner, by either directly or indirectly activating a DNA-binding protein that acts as a positive transcription factor when it is bound near the promoter. Table 3 lists those genes that respond to environmental signals after being introduced into the germ line of mice. In most cases it appears that if a transgene is expressed, then it is likely to respond to environmental signals in a manner analogous to that of the endogenous gene, as long as the cis-acting elements are included in the construct. The only notable exception is the mouse MT-I promoter, which does not respond to glucocorticoids even though the endogenous gene responds well (64).
Table 3. Modulation of gene expression in transgenic mice.
| Transgenea | Inducer | Tissue | Induction (fold) | Reference |
|---|---|---|---|---|
| mMT-I/TK | Cadmium | Liver | 10–50 | (79) |
| mMT-I/TK | LPSb | Liver | 5–200 | (22) |
| mMT-I/hGH | Zinc | Liver | 3–100 | (82) |
| hMT-IIA/hGH | Dexamethasone | Liver | 3–6 | (30) |
| SAA-2/hGH | LPSb | Liver | ∼100 | (c) |
| MMTV/TK | Dexamethasone | Testes | 2–10 | (87, d) |
| MMTV/TK | Dexamethasone | Mammary gland | 2–3 | (d) |
| MMTV/TK | Androgen | Testes | 2–3 | (d) |
| Transferrin | Estrogen | Liver | ∼2 | (32) |
| Insulin | Glucose | β cells | 2–20 | (14, 91a) |
| Insulin | Amino acids | β cells | –20 | (91a) |
| H2-Eα | γ-interferon | Macrophage | 5–100 | (54, 85) |
| Amylase-2.2 | Streptozotocin | Pancreas | ∼20 (inhibition) | (e) |
Abbreviations: mMT-I, mouse metallothionein-I promoter; hMT-HA, human metallothionein-HA promoter; MMTV, mouse mammary tumor virus long terminal repeat; SAA-2, serum amyloid-2; TK, thymidine kinase structural gene; hGH, human growth hormone structural gene; H2-Eα, murine class 13 histocompatibility gene.
LPS, lipopolysaccharide (bacterial endotoxin).
R. D. Palmiter, H. Rienhoff, R. Beringer & R. L. Brinster, unpublished data. No stimulation of growth in absence of induction.
K. Zaret, R. Hammer, H. Chen, K. Yamamoto & R. L. Brinster, unpublished data.
L. Treisman, M. Rosenberg, S. Keller & M. Meisler, unpublished data.
Unusual Aspects of Gene Expression in Transgenic Mice
Inappropriate Expression
Occasionally transgenes are expressed in inappropriate tissues; however, when this occurs the level of expression is usually very low. For example, in early experiments a low level of β-globin gene expression (less than 10 molecules of mRNA per cell) was observed in muscle of one mouse and testis of another (53), and none was observed in erythroid cells. Similarly, rat α-actin was expressed infrequently and in inappropriate tissues when plasmid vector sequences were included (94). A low level of rat elastase-I mRNA was detected in inappropriate tissues of some mice, but in this case a very high level expression (103–104-fold higher) usually occurred in the appropriate tissue (61, 107). Inappropriate expression might be due to integration near endogenous enhancers, whereas lack of expression in appropriate tissues may be due to prokaryotic vector sequences (see below).
Another example of aberrant expression involves the chicken transferrin gene, which is expressed in kidney, where the endogenous mouse transferrin mRNA is barely detectable (32). Perhaps in this case the tissue-specific signals have evolved differently in chickens and mice; this may contribute to the inappropriate expression of transferrin gene in the kidney of the mouse. In general, it is our impression that gene expression in transgenic mice resembles the endogenous counterpart much more closely than does that observed after gene transfer into tissue culture cells. Transient assays in tissue culture cells have been extremely useful for localizing tissue-specific enhancers; however, in most cases the level of expression in inappropriate cell types is quite high, in the range of 1 to 10% of that in the appropriate cell type.
Low Frequency of Expression
While it is clear that many genes can be expressed in transgenic mice in a reasonably normal manner, some genes are not expressed or are expressed with very low frequency. Some of our best-documented cases of nonexpression are those that used metallothionein fusion genes. Many transgenic mice were generated carrying the mouse MT-I promoter fused to β-galactosidase, human factor IX, or rat α-tumor growth factor (see Table 2), but in none of them could expression be detected in the liver. In each of these cases the gene construct was functional when introduced into tissue culture cells (our unpublished observations). In fact, the MT-β-galactosidase construct is very active when microinjected into fertilized eggs, and this activity persists for several days of development (105; R. Pedersen, unpublished observations). Thus, the lack of expression of these genes in adult liver suggests that something occurs during development rendering them incapable of being expressed. For other constructs, the frequency of expression is much lower than the 70% we usually observe. For example, the frequency of MT-hepatitis B surface-antigen gene expression is very low (19). In this case, substitution of the mouse albumin promoter/ enhancer for the mouse MT-I sequences allows a normal frequency of expression of the hepatitis B surface-antigen gene (see Table 2).
Prokaryotic vector sequences are inhibitory for some genes. For human β-globin, the presence of plasmid or λ phage vector sequences severely inhibits both the level and the frequency of expression (109); plasmid sequences also inhibit MT-hGH, α-actin, and AFP gene expression (30, 52, 94). The human α-globin gene has been difficult to express in transgenic mice. Even in the absence of plasmid sequences, a 14-kb fragment containing both α-globin genes was not expressed in any of 12 founder mice. This result was particularly surprising considering that α-globin genes are expressed without the aid of exogenous enhancers when transfected into a variety of nonerythroid cell lines. However, three transgenic mice carrying a murine cosmid spanning the α-globin locus did express the marked α-globin gene (88; S. Rusconi, unpublished observation). This observation suggests that either a larger piece of DNA is essential for expression or the human α-globin gene is expressed in a species-specific manner.
With a sensitive assay, one can observe the activity of genes expressed in only a few cells. For example, we have been unable to demonstrate expression of rat, bovine, or human growth hormone genes in the pituitary when they are transcribed from their own promoters. However, fusion of a bovine GH promoter to SV40 T-antigen gene results in pituitary tumors in transgenic mice (our unpublished observations). The use of a transforming gene is an exquisitely sensitive assay because expression of it in a single cell may be sufficient to promote tumorigenesis.
The lack of expression of some constructs may be due to cis-acting “silencer” sequences. In this context, silencers are broadly defined as DNA sequences that prevent expression; they are thought to be the negative counterpart of enhancers (4). Silencers may be either physiologically relevant or fortuitous. To explain the lack of expression of some genes in transgenic mice, but not in transfected cell cultures, we hypothesize that these silencer sequences are recognized by proteins present (or functional) only during development. Silencers might be sites for covalent modification (e.g. methylation), they might initiate condensation into an inactive chromatin conformation, or they might phase nucleosomes in an inappropriate manner. Perhaps in their normal context they interact with other elements, but in fusion genes they may be separated from their normal counterparts. Alternatively, expression of some genes may require the presence of certain sequences (e.g. topoisomerase-binding sites, nuclear matrix–attachment sites, or origins of replication) inadvertently excluded from those constructs not expressed.
We demonstrated cis-acting, inhibitory influences by combining a gene not expressed well with a gene expressed well on the same plasmid. For example, experiments with MT-src suggested that this gene was not expressed well, so various regions of the src construct were juxtaposed to an MT-hGH gene, and these constructs were microinjected into eggs and assayed in fetal livers of founder mice. When the entire v-src gene and 3′ flanking region was included, the level and frequency of expression of MT-hGH was severely inhibited. Analysis of deletion mutants indicated that a region 3′ of the v-src structural gene was responsible for the inhibition (our unpublished observations). This approach should allow the identification of silencer sequences.
Low-Level Expression
When testing many different structural genes fused to the same promoter/enhancer, one obtains markedly different levels of mRNA. For example, with the elastase promoter/enhancer fused to hGH, the average level of mRNA is about 10,000 molecules per cell. However, when the same promoter/enhancer is fused to genes coding for neomycin phosphotransferase, SV40 T-antigen, c-Ha-ras, or c-myc, the amount of mRNA produced is usually less than 100 molecules per cell (our unpublished observations). Of course, mRNA abundance is a function of the rate of synthesis and the rate of degradation, and both rates could vary from one construct to another. However, the differences are so great that we suspect that they are not caused by changes in mRNA stability alone. Thus, these results suggest that different structural genes influence the rate of transcription in an unsuspected way. The same phenomenon is observed when the mouse MT-I promoter is fused to different structural genes. With growth hormone, the levels of mRNA are high (thousands of molecules per cell) and comparable to MT mRNA levels, but when fused to a thymidine kinase (TK) gene, mRNA levels are very low. In this case we showed by nuclear “run-on” assay that the rate of transcription of the MT-TK fusion gene was less than 5 percent of the endogenous MT-I genes (83).
Gene expression in tissues from transgenic animals is sometimes lower than that expected based on transfection experiments in tissue culture. As an example, in transfection studies into baby hamster kidney cells, the SV40 and TK gene promoters are comparable to the MT-I promoter (uninduced). However, in transgenic mice, these promoters appear to be very weak. When the TK gene with its own promoter was introduced into mice, the viral TK enzyme activity was barely detectable (116). Likewise, a TK-hGH gene was expressed at low frequency, and very little mRNA could be detected (see Table 2). Although the SV40 T-antigen gene with its own promoter/enhancer clearly functions in mice by causing choroid plexus tumors (see below), the level of these transcripts is surprisingly low (10). These arguments suggest that the relative activity of various promoters observed in tissue culture is not necessarily manifested in transgenic mice.
Variable Expression in Offspring
Two types of variability are observed in offspring of transgenic mice: variability relative to the parent and variation among different offspring. In founder transgenic mice that are mosaic, one expects to observe higher levels of expression in the offspring than in the parent, and this increase usually corresponds to that expected based on the germ-line transmission frequency (61, 66, 73, our unpublished observations). In subsequent generations of these mice, expression is stable.
Variability among offspring is a more perplexing problem. We initially observed this type of variability in all lines of mice expressing MT-TK fusion genes. TK activity varied dramatically from one animal to another, and this variability persisted through several generations with no obvious trend (i.e. a low expressor could give rise to offspring with higher or lower expression levels, and vice versa). The extreme case was cessation of activity in offspring; when this was observed, expression never reappeared in subsequent generations (79). The same phenomenon has been observed with transgenic mice bearing MT-hepatitis B surface-antigen genes (19). Initially, we considered the possibilities that this variability might be due to different numbers of genes being expressed in the large tandem arrays carried by some of these mice or to effects of modifier genes that could be segregating as a result of using C57 × SJL hybrid parents. However, even in inbred mice with only two copies of the MT-TK gene, the level of expression varies widely among offspring. We also considered the possibility of variable methylation, but in one line with only two copies of the MT-TK gene, no detectable methylation occurred at any of the methylation-sensitive, restriction enzyme sites tested (including Hpall, Hhal, Smal, and Sstll), yet the activity varied 100-fold (83). Perhaps some genes are very sensitive to nucleosome phasing, and during meiosis (or very early in development) the chromatin configuration might be “reset” in a way that would be inherited. Alternatively, different degrees of mosaic expression may be involved. If, for example, these MT fusion genes were in chromosomal locations where their activation or commitment was variable, then the number of cells that actually express the gene might vary from mouse to mouse. Indeed, this type of mosaic expression (based on immunofluorescence of the transgene product) has occurred in offspring of founder mice carrying MT or albumin fusion genes (F. Chisari, personal communication).
Microinjected DNA is clearly subject to de novo methylation (79), and considerable data implicate DNA hypermethylation with gene inactivity. Thus, DNA methylation is a reasonable candidate to explain the variable expression among founder animals and among their offspring. However, the analysis has generally been complicated by the fact that most mice have multiple integrants, which makes it almost impossible to know whether the changes in methylation occur in the gene copies being expressed. The analysis of methylation and gene expression is much clearer in the case of single-copy retroviral integrants. In mice bearing MuLV retroviruses DNA methylation correlates well with lack of viral gene expression (45); furthermore, treatment of mice carrying these proviruses with the drug 5-azacytidine, which interferes with normal methylation, can activate the expression of these silent viral genomes (43).
Another type of variability is a consequence of genomic instability. Deletion of some copies from a large tandem array has been documented (79), and a few other transgene rearrangements have occasionally been observed (73). Although these rearrangements correlate with changes in activity, they are relatively rare.
Epistatic Effects of Joining Diverse DNA Segments
Only fragmentary information exists on how promoters, enhancers, silencers, matrix attachment sites, topoisomerase-binding sites, and origins of DNA replication (to name a few candidates) may be involved in regulation of transcription. It is therefore not surprising that when diverse DNA elements are combined and introduced into a developing organism, gene expression may occasionally be perturbed in unexpected ways. For example, when the MT-hGH gene was juxtaposed in an opposite transcriptional orientation to that of a human β-globin gene, no human β-globin expression was observed in erythroid cells; when the β-globin was excised and tested on its own, however, levels of expression approaching that of the endogenous β-globin gene were observed (108). MT-hGH expression was not observed in erythroid cells either, but it was unusually low in the liver and high in intestine (108). These observations suggest that the neighboring MT-hGH gene in some way suppressed human β-globin gene expression. These inhibitory effects could relate to the binding of specific regulatory proteins to the neighboring sequences, or they might reflect nonspecific inhibition comparable to that observed with plasmid DNA.
Another apparent epistatic effect involves the inhibitory effect of juxtaposing the elastase enhancer with the MT promoter of an MT-hGH gene. In most mice with this construct a large amount of hGH mRNA is produced in the pancreas, but none in tissues where the MT promoter usually functions well. Consequently, the mice failed to grow larger than normal. The elastase enhancer appears to have an inhibitory effect on MT promoter function in the nonpancreatic tissues. Curiously, in two lines of mice this inhibition was not fully established in founders, but was established in the next generation.
A striking example of genetic interaction is the novel expression of MT-rGH and MT-hGH genes in a specific set of neuronal cells that do not normally express either MT or GH genes (106). Perhaps in some cases tissue-specificity is determined by multiple enhancerlike elements acting in a combinatorial fashion, and combining elements from one gene with those from another one may produce novel combinations that specify expression in unpredicted cell types. This phenomenon is not likely to be due to positional effects because many different founder mice, presumably with different integration sites, show expression in the same set of neurons. Similarly, MT-somatostatin genes are frequently expressed in the gonadotroph cells of the pituitary, where neither gene is normally expressed (60).
Physiological Consequences of Transgene Expression
By using fusion genes the expression of genes coding for regulatory proteins may be redirected to novel cell types. By redirecting expression of secreted proteins to large organs such as the liver, the amount of circulating protein may be tremendously increased. In addition, the expression of a gene can be divorced from normal feedback control mechanisms. The ability to amplify the expression of a product or to express it prematurely as well as to redirect it to heterologous cell types has many potential applications for dissecting aspects of complex systems not amenable to analysis in vitro.
Growth Control
The growth of mammals is under complex genetic and hormonal control. The cascade of hormones starts with the neuropeptides, somatostatin and growth-hormone releasing factor (GRF), which inhibit or stimulate, respectively, synthesis of growth hormone (GH) by the somatotroph cells in the anterior pituitary. GH binds to receptors in the liver where it stimulates the production of insulinlike growth factor-I (IGF-I, also called somatomedin C), which is thought to stimulate cellular proliferation. Each of the structural genes in this cascade has been put under control of heterologous promoters (usually MT) to allow elevated and chronic production of each of these polypeptide hormones. The most obvious effect of excess production of rat, bovine, or human GH is a stimulation of growth that commences at about 3 weeks and plateaus at about 12 weeks when the mice are as much as twice their normal size (77, 82). Similarly, production of excess human GRF stimulates the somatotroph cells to produce excess GH which then elevates IGF-I mRNA and stimulates growth (31; our unpublished observations). Experiments are currently underway to assess the effects of excess production of IGF-I driven off MT-I or albumin promoters. Ectopic production of somatostatin has not revealed any effects on growth; however, in this case the processing of the precursor polypeptide was abnormal in most tissues (59).
In addition to its effects on growth, ectopic production of GH affects other physiological processes. Chronic production of GH feedback inhibits endogenous GH synthesis, and the somatotroph cells of the pituitary are undetectable by standard histological procedures (82). GH also impinges on the sexual differentiation of the liver. The mRNA for the major urinary protein (MUP), which is normally the most abundant mRNA in the liver of males, is suppressed to very low levels in transgenic mice expressing GH chronically (71). Other physiological consequences of ectopic GH production include inhibition of fertility in females (but not males), signs of premature aging, and increased efficiency of utilizing feed (our unpublished observations).
When GRF is expressed ectopically, it not only chronically enhances GH synthesis but also leads to hypertrophy and proliferation of somatotroph cells to such an extent that total DNA content of the pituitary increases about 5-fold and total mass increases almost 15-fold (30). Thus, GRF acts as a specific growth factor for somatotroph cells as well as a regulator of GH gene expression and GH secretion.
Growth of mice is stunted when they harbor foreign dihydrofolate reductase genes (23a). In addition to this curious growth defect, these mice manifest reduced fertility, pigmentation changes, and skeletal defects. These defects are transmitted as dominant traits in several transgenic lines and are presumably due to aberrations in folate metabolism.
Cancer
Several oncogene constructs have been introduced into mice that elicit tumorigenic or other pathological changes; the data are summarized in Table 4. Mice that harbor the SV40 enhancers and early region (coding for large and small T-antigens) reproducibly develop choroid plexus tumors (10, 80, 95). These tumors are derived from the epithelial (ependymal) cells that line the ventricles of the brain. Choroid plexus tumors also develop when only an intact large T-antigen can be expressed; in addition, expression of a mutant form of T-antigen that fails to localize in the nucleus (due to an amino acid change in the nuclear transport sequence) is tumorigenic (our unpublished observations). However, deletion of the SV40 enhancers reduces the incidence of tumorigenesis, and those tumors that do develop are rarely of choroid plexus origin. We discovered quite fortuitously that when an enhancerless SV40 T-antigen gene is juxtaposed to an MT-hGH gene, transgenic mice frequently develop peripheral neuropathy (apparently due to a defect in myelination by the Schwann cells) as well as hepatocellular carcinomas and insulinomas (transformation of pancreatic β cells). This redirection of SV40 T-antigen expression to Schwann cells, hepatic cells, and β cells is thought to reflect the action of enhancerlike elements in the MT-hGH gene when the SV40 enhancers are removed (69, 80). Neuropathy is also observed in transgenic mice bearing the early region of the human JC papovavirus. These mice manifest hypomyelination in the brain and spinal cord and exhibit a shaking disorder similar to mutant jimpy and quaking mice (95a).
Table 4. Expression of transforming genes in transgenic mice.
| Transgene | Site of pathology | Reference* |
|---|---|---|
| SV40 early region | Choroid plexus | (10, 80, 95) |
| SV40 large T-antigen | Choroid plexus | (80) |
| MThGH-SV40(Δ enhancer) | Hepatocytes, pancreatic β cells, & Schwann cells | (69, 80) |
| Insulin/SV40 early region | Pancreatic β cells | (35) |
| Elastase/SV40 early region | Pancreatic acinar cells | (73) |
| Albumin/SV40 early region | Hepatocytes | (a) |
| bGH/SV40 early region | Pituitary (somatotroph cells?) | (a) |
| Prolactin/SV40 early region | Pituitary (lactotroph cells) | (b) |
| Crystallin/SV40 early region | Lens | (c) |
| Adeno E1A/SV40 early region | Glial cells | (d) |
| α-Glycotropin/SV40 early | Pituitary | (e) |
| GRF/SV40 early region | Thymus | (f) |
| Elastase/c-Ha-ras (EJ) | Pancreatic acinar cells | (a) |
| MMTV/c-myc | Mammary epithelial cells | (97) |
| LTR/c-myc | Thymocytes | (1) |
| SV40/c-myc | Lymphocytes, renal cells & fibroblasts | (1) |
| Ig(μ)/c-myc | Pre-B and B-cells | (1) |
| lg(κ)/c-myc | B-cells | (1) |
| Papilloma virus (bovine) | Skin fibropapilloma | (g) |
Unpublished observations: (a) ours, (b) G. Rosenfeld, (c) H. Westphal, (d) F. Kelly, (e) P. Mellon & D. Hanahan, (f) D. Hanahan, (g) F. Botteri, H. van der Putten & R. Evans.
Unpublished observations: (a) ours, (b) G. Rosenfeld, (c) H. Westphal, (d) F. Kelly, (e) P. Mellon & D. Hanahan, (f) F. Botteri, H. van der Putten & R. Evans; (g) A.-C. Andres, C.-A. Schonenberger, B. Groner, L. Hennighausen, M. LeMeur & P. Gerlinger.
Fusion of the promoter/enhancer region of various tissue-specific genes to SV 40 T-antigen has lead to tumorigenesis in the cell type specified by the enhancer. For example, the promoter/enhancer region from the insulin gene directs tumors exclusively to the β cells of the pancreatic islets (35), whereas the promoter/enhancer region of the elastase-I gene directs tumors to the acinar cells of the pancreas (73). In similar experiments, tumorigenesis has been directed to lactotroph cells, somatotroph cells, and lens; see Table 4 for other examples.
In another set of experiments, the c-myc gene was fused in a variety of ways to the long terminal repeat (LTR) of the mouse mammary tumor virus (MMTV). Transgenic females in one line characteristically developed mammary carcinomas during their second or third pregnancy (97). In another line, the mice developed testicular and lymphocytic tumors, in addition to breast tumors (53a, 97). However, in both lines tumors never developed in some tissues, e.g. the pancreas and salivary gland where MMTV-myc was expressed. Injection of constructs in which the c-myc gene was associated with enhancers from immunoglobulin heavy-chain or light-chain genes resulted in transgenic mice that develop pre-B or B-cell lymphomas (1).
In all of these cases, the tumors developed after the mice were several months old; the evidence suggests that the tumors were clonal in origin. In some cases, immunological evidence shows that the transforming genes are expressed in all of the cells appropriate for that enhancer (35; our unpublished observations). In one elastase-SV40 line the T-antigen genes are not apparently expressed in all of the acinar cells, but become activated in a few cells that then expand clonally (73). Tumors usually develop earlier in those lines of mice where all of the cells express the T-antigen gene. Expression of these transforming genes increases the probability of transformation (in many cases to nearly 100%, but expression alone is apparently insufficient for tumorigenesis. In these experimental systems, tumorigenesis appears to depend on other genetic events in addition to T-antigen (or c-myc) expression. The relatively late onset of these tumors has the advantage that one can usually develop lines of mice in which tumorigenesis is inherited as a dominant trait. These lines will provide a rich source of material for investigating the events associated with oncogenesis.
Another useful aspect of these experiments is that one may be able to develop transformed cell lines that retain differentiated functions. Cell lines that continue to express low levels of elastase or prolactin have been established from tumors of appropriate transgenic mice (73, G. Rosenfeld, personal communication).
In contrast to the previous examples, when an oncogenic form of human c-Ha-ras (with a glycine-to-valine substitution at amino acid 12) was fused to the elastase-I enhancer/promoter, transgenic mice were born with pancreatic neoplasms and died several days later. In this case, all of the differentiating pancreatic acinar cells become transformed within a day or so of their appearance during fetal development. Hence, expression of this oncogene within differentiating pancreatic acinar cells appears sufficient to transform these cells and promote tumorigenesis. Control mice bearing the normal human c-Ha-ras proto-oncogene have not developed tumors, although the level of gene expression is comparable to that of the mutant form (our unpublished observations).
See reference 35a for a more extensive discussion of oncogenesis in transgenic mice.
Immunoglobulin Gene Rearrangement
When functionally rearranged immunoglobulin genes are introduced into mice, they are expressed in a cell-specific manner and at appropriate developmental times, i.e. Ig κ light-chain genes are expressed exclusively in B-cells whereas Ig μ heavy-chain genes are expressed in pre-B, B- and T-cells. B-cells normally produce only one functional immunoglobulin, although they have the potential to produce several. This finding suggests that a mechanism exists to prevent further rearrangement once a functional anti-body has been produced. Thus, one might anticipate that introduction of a functionally rearranged immunoglobulin gene might affect the rearrangement process. Three groups have investigated this question. Expression of a Ig μ heavy-chain transgene inhibits the rearrangement of endogenous Ig μ. genes, and expression of a functional Ig κ light-chain transgene, along with the endogenous heavy-chain gene (but not alone), inhibits the rearrangement of endogenous Ig κ genes (86, 118). In mice where the transgene was not expressed or where the level of expression was low, the endogenous genes did rearrange (86, 89, 118). These data support the idea that production of a functional immunoglobulin is an important signal that terminates further rearrangement of the immunoglobulin genes. Complex abnormalities in immune repertoire generation have also been observed in mice bearing functional heavy- and light-chain transgenes (119). These observations emphasize that the control of antibody gene rearrangement and expression is complex. More extensive reviews of these results have appeared (2, 100).
Correction of Genetic Defects
The ability to introduce functional genes into the germ line of animals suggests the possibility of using this technique to correct genetic defects. However, in the absence of techniques to target genes efficiently by homologous recombination, it is only possible to introduce a functional gene, not to replace a mutant gene with a normal one. Therefore, in subsequent generations the mutant and functional genes segregate independently. Nevertheless, two examples of this sort of experiment have been published.
In the first example, an MT-rGH gene was introduced into an inbred line of dwarf mice, carrying the mutation little. The primary genetic defect in these mice appears to be a defect in GRF receptors; consequently the somatotroph cells cannot be stimulated to produce adequate amounts of GH. However, little mice respond to exogenous GH, which stimulates their growth and corrects male infertility. Several transgenic lines of little were produced that expressed the MT-rGH gene in the liver, and they grew to about three times the size of their little litter mates (35). In addition, male fertility was restored. However, females manifested reduced fertility, as was observed in all previous transgenic mice expressing rat or human GH genes (30).
In the second example, a genetic defect in an immune response was corrected. The immune-response genes of the major histocompatibility gene complex encode class II molecules, heterodimeric membrane proteins of A (Aα and Aβ chains) and E (Eα and Eβ chains) types. Foreign antigens are presented to T-cells in association with these class II proteins on the surface of macrophages and B-cells. Thus, a deficiency in these class II genes can restrict the ability of macrophages to present certain antigens. Indeed, mice of the H-2s and H-2b haplotypes have a deletion that removes the promoter and first exon of the Eα gene. Consequently they do not transcribe Eα mRNA. The Eβ gene product is also deficient because it cannot assemble properly in the absence of Eα. Three groups introduced a functional copy of the Eα gene into transgenic mice harboring this deletion, and in each case they were able to correct the iJllillune-response deficiency (54, 85, 124). The Eα protein was synthesized and functionally complexed with the Eβ protein on the membrane. Unlike the original mutant mice, the transgenic mice were able to present the synthetic antigen, poly (glutamic acid, lysine, phenylalanine) to T-cells. As roenti.oned earlier, these Eα transgenes were regulated by γ-interferon in macrophages. One curious observation is that males failed to transmit the trans gene to offspring when it contained about 30 kb of 5′ flanking sequence (85) but transmitted it normally when a smaller fragment containing only the Eα gene was present (54). Females transmitted both genes normally, suggesting that some sequences (genes?) within the upstream region may have a deleterious effect on spermatogenesis.
Insertional Mutagenesis
The site at which the foreign DNA inserts into the host chromosome is L probably random, although there may be some bias, e.g. foreign DNA may integrate preferentially into “open” chromatin. One can anticipate that the foreign DNA would occasionally disrupt the function of an endogenous gene or possibly activate a gene by providing an enhancer/promoter, thereby producing recessive or dominant mutations. Most insertional mutations are probably recessive, and hence would be found only upon inbreeding of transgenic offspring. Table 5 lists 11 examples of recessive insertional mutations revealed by inbreedmg 153 transgenic mice produced by microinjection or retroviral infection. Thus, the overall frequency of insertional mutation appears to be about 7%; this number is undoubtedly an underestimate because some recessive mutations may have subtle phenotypes, and some dominant mutations are probably lost because they cause embryonic lethality. The frequency of insertional mutation appears to be higher with the microinjection technique than with retroviral insertion; this difference might result from the fact that large regions of chromosomal DNA may be deleted or duplicated during integration of rnicroinjected DNA (see below), whereas retroviruses insert cleanly, with only a few base pairs of host DNA duplicated at the site of integration. The method of microinjection may also influence the mutation frequency.
Table 5. Incidence of insertional mutation.
| Number of TM bred to homozygositya | Number of homozygote TM with genetic defects due to insertional mutation | Number of failures to develop homozygous lines due to insertional mutation | Reference |
|---|---|---|---|
| 31 (μ) | 0 | 0 | (f) |
| 1 (μ) | 0 | 0 | (110) |
| 2 (μ) | 0 | lc line 4 | (63) |
| 17 (μ) | 0 | lc HB58 | (g) |
| 9 (μ) | 0 | lc line 106 | (h) |
| 9 (μ) | 0 | lc CV4 | (94) |
| 4 (μ) | 0 | 2c HUGH/3, HUGH/4 | (113) |
| 5 (μ) | 0 | ld MyK-103 | (83) |
| 4 (μ) | 1b IdHd | 0 | (123) |
| 20 (μ) | lb syfp−N | 0 | (74) |
| 40 (r) | 0 | lc Mov-34 | (i) |
| 0 (r*) | 0 | lc Mov-13 | (41, 91) |
| Total | |||
| 142 | 2 | 9 |
Transgenic mice (TM) were produced either by microinjection (μ) of DNA into pronuclei of fertilized eggs or by retroviral infection of preimplantation (r) or postimplantation (r*) embryos.
Limb deformity.
Recessive embryonic lethal.
Defect in spermatogenesis.
Embryonic lethal; insert in αl(I) collagen gene.
Unpublished data obtained from: D. Hanahan (8 lines), J. Jami (1 line), C. Lo (2 lines), S. Ross (4 lines), K. Raphael (1 line), E. Wagner (7 lines), F. Chisari (7 lines), Meisler (1 line).
F. Costantini, unpublished data.
M. LeMeur, unpublished data.
P. Soriano & R. Janeisch, unpublished data.
Eight of the insertional mutations cause embryonic lethality, but only in the case of Mov-13 (which is due to a retroviral insert) has the endogenous gene been identified. The retroviral insert and flanking DNA from Mov-13 mice were isolated from a lambda library. Unique probes from the flanking DNA identified abundant 5.2- and 6.5-kb transcripts present in fibroblasts, but not in epithelial or lymphoid cells. These, and a few other clues, suggested that a collagen gene was affected (91). Sequence analysis showed that the retroviral DNA inserted into the first intron of the αl(I) collagen gene (41). Mice homozygous for this insert arrest around day 12 of development owing to progressive necrosis of erythropoietic and mesenchymal cells, followed by rupture of major blood vessels (57). Curiously, the collagen gene carrying the insert is not transcribed even though the promoter is apparently intact, perhaps because the promoter region is extensively methylated at times when the normal collagen gene promoter is not (37). Previous work showed that retroviral genomes tend to be heavily methylated and inactivated when they are introduced into preimplantation embryos (45, 46); these data suggest that the flanking DNA may be subject to this methylation phenomenon as well.
One insertional mutation (called line 4) disrupts development prior to implantation, at approximately day 5 (63); all of the other embryonic lethals analyzed in any detail disrupt development after implantation. In none of these cases has the insertion or preinsertion DNA been isolated; thus, the nature of the genes affected is unknown.
Two of the insertional mutations affect limb development. One, designated ldHd, arose in a line of mice bearing MMTV-myc genes; in homozygotes the radius and ulna of the forelimb, and tibia and fibula of the hindlimb, are fused. In addition, many of the bones in the feet are fused or missing (123). The transgene and adjoining DNA sequences have been cloned, revealing that approximately one kilobase of DNA was deleted during the integration event. Unique probes from the flanking region were used to show that this insertion lies on chromosome 2 and that it is allelic to two previously identified mutations affecting limb development. These probes should ultimately allow characterization of the normal gene and its protein product. The other limb deformity, syfp–N, appears to be less severe (74).
Another insertional mutation affecting adults is designated MyK-103; the foreign DNA insert (which includes an inverted repeat of an MT-TK gene) is transmitted normally by females but never by males. The males are fertile and sire litters of normal size. These observations suggest that sperm carrying the insert are infertile. Hence, we proposed that the insert disrupts a gene that must be expressed postmeiotically in developing sperm (83). The insert has been mapped to chromosome 6. The foreign DNA insert with >15 kb of flanking mouse DNA from each junction, as well as the normal allele, have been isolated; approximately 5 kb of mouse DNA was duplicated at the site of integration (our unpublished observations). Hybridization of unique DNA probes isolated from the flanking mouse DNA to Northern blots has not yet revealed any RNA transcripts from testis or several other tissues. Thus, we have not been able to identify the gene presumably disrupted in the MyK-103 mice.
Prospects
Transgenic animals will continue to provide valuable opportunities for addressing fundamental questions of developmental gene expression, immune surveillance, neurobiology, and oncogenesis. Considering the rate at which new genes are being isolated and their controlling elements defined, it seems inevitable that there will be a tremendous increase in the generation of transgenic animals to study these questions. Some important areas for future experiments are noted below:
Improvements in the efficiency and ease of introducing genes into the germ line of animals.
Development of a simple means of introducing single copies of genes but without the limitations of current retroviral vectors.
Discovery of methods for insulating transgenes from the effects of neighboring chromatin.
Development of techniques for homologous recombination, so that endogenous genes can be deleted or replaced.
Improvements in methods for phenotypic inactivation of gene expression (e.g. development of effective antisense constructs).
Development of inducible promoters that will permit experiments in which gene expression can be tightly controlled.
While these are laudable goals, their realization requires extensive experimentation. In the meantime, current techniques will undoubtedly be used to define in more detail many enhancer (and silencer) sequences, to redirect or amplify the expression of genes involved in many physiological processes, to isolate important genes (by virtue of insertional tags), and to uncover many unsuspected developmental processes. These endeavors, plus the application of current techniques to the genetic improvement of farm animals, the production of medically useful products and generation of animal models of human genetic disease, bode well for an exciting era of transgenic research ahead.
Acknowledgments
We are grateful for our colleagues, collaborators, and research assistants who made our contribution to this field possible. We also appreciate their constructive suggestions during the preparation of this review. We extend our thanks to the many scientists who provided unpublished data incorporated in the tables. This work was supported in part by grants from the National Institutes of Health and National Science Foundation.
Contributor Information
Richard D. Palmiter, Howard Hughes Medical Institute, SL-15, Department of Biochemistry, University of Washington, Seattle, Washington 98195
Ralph L. Brinster, Laboratory of Reproductive Physiology, School of Veterinary Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104
Literature Cited
- 1.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–38. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 2.Alt F, Blackwell TK, Yancopoulos GD. Immunoglobulin genes in transgenic mice. Trends Genet. 1985;1:231–36. [Google Scholar]
- 3.Babinet C, Farza H, Morello D, Hadchouel M, Pourcel C. Specific expression of hepatitis B surface antigen (HBsAg) in transgenic mice. Science. 1985;230:1160–63. doi: 10.1126/science.3865370. [DOI] [PubMed] [Google Scholar]
- 3a.Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cells. Nature. 1984;309:255–58. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 4.Brand AH, Breeden L, Abraham J, Stemglanz R, Nasmyth K. Characterization of a “silencer“ in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985;41:41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- 5.Brinster RL. Cultivation of the mammalian embryo. In: Rothblat G, Cristafalo V, editors. Growth, Nutrition and Metabolism of Cells in Culture. Vol. 2. New York: Academic; 1972. pp. 251–86. [Google Scholar]
- 6.Brinster RL, Chen HY, Trumbauer ME, Avarbock MR. Translation of globin messenger RNA by the mouse ovum. Nature. 1980;283:499–501. doi: 10.1038/283499a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinster RL, Chen HY, Trumbauer ME. Mouse oocytes transcribe injected Xenopus SS RNA genes. Science. 1981;211:396–98. doi: 10.1126/science.7194505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinster RL, Chen HY, Trumbauer ME, Senear AW, Warren R, Palmiter RD. Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell. 1981;27:223–31. doi: 10.1016/0092-8674(81)90376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinster RL, Chen HY, Trumbauer ME, Yagle MK, Palmiter RD. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc Natl Acad Sci USA. 1985;82:4438–42. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brinster RL, Chen HY, Messing A, van Dyke T, Levine A1, Palmiter RD. Transgenic mice harboring SV40 T-antigen genes develop characteristic brain tumors. Cell. 1984;37:367–79. doi: 10.1016/0092-8674(84)90367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinster RL, Palmiter RD. Induction of foreign genes in animals. Trends Biochem Sci. 1982;7:438–40. [Google Scholar]
- 12.Brinster RL, Palmiter RD. Harvey Lectures, Series. Vol. 80. New Yark: Liss; 1986. Introduction of genes into the germ line of animals; pp. 1–38. [PMC free article] [PubMed] [Google Scholar]
- 13.Brinster RL, Ritchie KA, Hammer RE, O'Brien RL, Arp B, Storb U. Expression of a microinjected immunoglobulin gene in the spleen of transgenic mice. Nature. 1983;306:332–36. doi: 10.1038/306332a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bucchini D, Ripoche MA, Stinnakre MG, Desbois P, Lores P, et al. Pancreatic expression of human insulin gene in transgenic mice. Proc Natl Acad Sci USA. 1986;83:2511–15. doi: 10.1073/pnas.83.8.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burki K, Ullrich A. Transplantation of the human insulin gene into fertilized mouse eggs. EMBO J. 1982;1:127–31. doi: 10.1002/j.1460-2075.1982.tb01135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chada K, Magrarn J, Costantini F. An embryonic pattern of expression of human fetal globin gene in transgenic mice. Nature. 1986;319:685–88. doi: 10.1038/319685a0. [DOI] [PubMed] [Google Scholar]
- 17.Chada K, Magrarn J, Raphael K, Radice G, Lacy E, Costantini F. Specific expression of a foreign β-globin gene in erythroid cells of transgenic mice. Nature. 1985;314:377–80. doi: 10.1038/314377a0. [DOI] [PubMed] [Google Scholar]
- 18.Chen HY, Trumbauer ME, Ebert KM, Palmiter RD, Brinster RL. Developmental changes in the response of mouse eggs to injected genes. In: Bogorad L, editor. Molecular Developmental Biology 43rd Symp Soc Dev Biol. New York: Liss; 1986. pp. 149–59.pp. 164 [Google Scholar]
- 19.Chisari FV, Pinkert CA, Milich DR, Filippi P, McLachlan A, et al. A transgenic mouse model of the chronic hepatitis B surface antigen carrier state. Science. 1985;230:1157–60. doi: 10.1126/science.3865369. [DOI] [PubMed] [Google Scholar]
- 20.Cory S. Activation of cellular oncogenes in hemopoietic cells by chromosome translocation. Adv Cancer Res. 1986 doi: 10.1016/s0065-230x(08)60200-6. In press. [DOI] [PubMed] [Google Scholar]
- 21.Costantini F, Lacy E. Introduction of a rabbit β-globin gene into the mouse germ line. Nature. 1981;294:92–94. doi: 10.1038/294092a0. [DOI] [PubMed] [Google Scholar]
- 22.Dumam DM, Hoffman JS, Quaife CJ, Benditt EP, Chen HY, et al. Induction of mouse metallothionein-I mRNA by bacterial endotoxin is independent of metals and glucocorticoid hormones. Proc Natl Acad Sci USA. 1984;81:1053–56. doi: 10.1073/pnas.81.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frels WJ, Bluestone JA, Hodes RJ, Capecchi MR, Singer DS. Expression of a microinjected porcine class I major histocompatibility complex gene in transgenic mice. Science. 1985;228:577–80. doi: 10.1126/science.3885396. [DOI] [PubMed] [Google Scholar]
- 23a.Gordon JW. A foreign dihydrofolate reductase gene in transgenic mice acts as a dominant mutation. Mol Cell Biol. 1986;6:2158–67. doi: 10.1128/mcb.6.6.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon JW, Ruddle FH. Integration and stable germ line transmission of genes injected into mouse pronuclei. Science. 1981;214:1244–46. doi: 10.1126/science.6272397. [DOI] [PubMed] [Google Scholar]
- 25.Gordon JW, Ruddle FH. DNA-mediated genetic transformation of mouse embryos and bone marrow—a review. Gene. 1985;33:121–36. doi: 10.1016/0378-1119(85)90087-3. [DOI] [PubMed] [Google Scholar]
- 26.Gordon JW, Ruddle FH. Gene transfer into mouse embryos: production of transgenic mice by pronuclear injection. Meth Enzymol. 1983;101:411–33. doi: 10.1016/0076-6879(83)01031-9. [DOI] [PubMed] [Google Scholar]
- 27.Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA, Ruddle FH. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc Natl Acad Sci USA. 1980;77:7380–84. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grosschedl R, Weaver D, Baltimore D, Costantini F. Introduction of a μ immunoglobulin gene into the mouse germ line: specific expression in lymphoid cells and synthesis of functional antibody. Cell. 1984;38:647–58. doi: 10.1016/0092-8674(84)90259-9. [DOI] [PubMed] [Google Scholar]
- 28a.Gurdon J. Adult frogs derived from nuclei of single somatic cells. Dev Biol. 1962;4:256–70. doi: 10.1016/0012-1606(62)90043-x. [DOI] [PubMed] [Google Scholar]
- 29.Gurdon JB, Melton DA. Gene transfer in amphibian eggs and oocytes. Ann Rev Genet. 1981;15:189–218. doi: 10.1146/annurev.ge.15.120181.001201. [DOI] [PubMed] [Google Scholar]
- 30.Hammer RE, Brinster RL, Palmiter RD. Use of gene transfer to mcrease animal growth. Cold Spring Harbor Symp Quant Biol. 1985;50:379–87. doi: 10.1101/sqb.1985.050.01.048. [DOI] [PubMed] [Google Scholar]
- 31.Hammer RE, Brinster RL, Rosenfeld MG, Evans RE, Mayo KE. Expression of human growth hormone-releasing factor in transgenic mice results in increased somatic growth. Nature. 1985;315:413–16. doi: 10.1038/315413a0. [DOI] [PubMed] [Google Scholar]
- 32.Hammer RE, Idzerda RL, Brinster RL, McKnight GS. Estrogen regulation of the avian transferrin gene in transgenic mice. Mol Cell Biol. 1986 doi: 10.1128/mcb.6.4.1010. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammer RE, Palmiter RD, Brinster RL. Partial correction of murine hereditary disorder by germ-line mcorpora!Jon of a new gene. Nature. 1984;311:65–67. doi: 10.1038/311065a0. [DOI] [PubMed] [Google Scholar]
- 34.Hammer RE, Pursel VG, Rexroad CE, Jr, Wall RJ, Bolt DJ, et al. Production of transgemc rabbits, sheep and pigs by microinjection. Nature. 1985;315:680–83. doi: 10.1038/315680a0. [DOI] [PubMed] [Google Scholar]
- 35.Hanahan D. Heritable formation of pancreatic β-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature. 1985;315:115–22. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- 35a.Hanahan D. Oncogenesis in transgenic mice. In: Graf T, Kahn P, editors. Oncogenes and Growth Control. Heidelberg: Springer Verlag; 1986. In press. [Google Scholar]
- 36.Harbers K, Jahner D, Jaenisch R. Microinjection of cloned retroviral genomes into mouse zygotes: integration and expression in the animal. Nature. 1981;293:540–42. doi: 10.1038/293540a0. [DOI] [PubMed] [Google Scholar]
- 37.Hartung S, Jaenisch R, Breindl ;M. Retrovirus insertion inactivates mouse αl(I) collagen gene by blocking initiation of transcription. Nature. 1986;320:365–67. doi: 10.1038/320365a0. [DOI] [PubMed] [Google Scholar]
- 38.Hogan BLM, Costantini F, Lacy E. Manipulation of the Mouse Embryo: a Laboratory Manual. Cold Spring Harbor, New York: Cold Spnng Harbor Lab; 1986. In press. [Google Scholar]
- 39.Humphries RK, Berg P, DiPietro J, Bernstein S, Baur A, et al. Transfer of human and murine globmgene sequences into transgenic mice. Am J Hum Genet. 1985;37:295–310. [PMC free article] [PubMed] [Google Scholar]
- 40.Jaenisch R. Germ line integration and Mendelian transmission of the exogenous Moloney leukemia virus. Proc Natl Acad Sci USA. 1976;73:1260–64. doi: 10.1073/pnas.73.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaenisch R, Breindl M, Harbers K, Jahner D, Lohler J. Retroviruses and insertional mutagenesis. Cold Spring Harbor Symp Quant Biol. 1985;50:439–45. doi: 10.1101/sqb.1985.050.01.055. [DOI] [PubMed] [Google Scholar]
- 42.Jaenisch R, Mintz B. Simian virus 40 sequences in DNA of healthy adult mice derived from preimplantation blastocysts injected with viral DNA. Proc Natl Acad Sci USA. 1974;71:1250–54. doi: 10.1073/pnas.71.4.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaenisch R, Schnieke A, Harbers K. Treatment of mice with 5-azacytidine efficiently activates silent retroviral genomes in different tissues. Proc Natl Acad Sci USA. 1985;82:1451–55. doi: 10.1073/pnas.82.5.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jahner D, Haase K, Mulligan R, Jaenisch R. Insertion of the bacterial gpt gene into the germ line of mice by retroviral infection. Proc Natl Acad Sci USA. 1985;82:6927–31. doi: 10.1073/pnas.82.20.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jahner D, Jaenisch R. Chromosomal position and specific demethylation in enhancer sequences of germ-line transmitted retroviral genomes during mouse development. Mol Cell Biol. 1985;5:2212–20. doi: 10.1128/mcb.5.9.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jahner D, Stuhlmann H, Stewart CL, Harbers K, Lohler J, et al. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–28. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- 47.Khillan JS, Overbeek PA, Westphal H. Drosophila P element integration in the mouse. Dev Biol. 1985;109:247–50. doi: 10.1016/0012-1606(85)90365-3. [DOI] [PubMed] [Google Scholar]
- 48.Khillan JS, Schmidt A, Overbeek PA, deCrombrugghe B, Westphal H. Developmental and tissue-specific expression by the α2 type 1 collagen promoter in transgenic mice. Proc Natl Acad Sci USA. 1986;83:725–29. doi: 10.1073/pnas.83.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein S, Sablitzky F, Radbruch A. Deletion of the IgH enhancer does not reduce immunoglobulin heavy chain production of a hybridoma IgD class switch variant. EMBO J. 1984;3:2473–76. doi: 10.1002/j.1460-2075.1984.tb02158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krumlauf R, Chapman VM, Hammer RE, Brinster R, Tilghman SM. Differential expression of α-fetoprotein genes on the inactive X chromosome in extraembryonic and somatic tissues of transgenic mice. Nature. 1985;319:224–26. doi: 10.1038/319224a0. [DOI] [PubMed] [Google Scholar]
- 51.Krumlauf R, Hammer RE, Tilghman SM, Brinster RL. Developmental regulation of α-fetoprotein genes in transgenic mice. Mol Cell Biol. 1985;5:1639–48. doi: 10.1128/mcb.5.7.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krumlauf R, Hammer RE, Brinster RL, Chapman VM, Tilghman SM. Regulated expression of α-fetoprotein genes in transgenic mice. Cold Spring Harbor Symp Quant Biol. 1985;50:371–78. doi: 10.1101/sqb.1985.050.01.047. [DOI] [PubMed] [Google Scholar]
- 52a.Lacey M, Alpert S, Hanahan D. The bovine papilloma virus genome elicits skin tumors in transgenic mice. Nature. 1986;332:609–12. doi: 10.1038/322609a0. [DOI] [PubMed] [Google Scholar]
- 53.Lacy E, Roberts S, Evans EP, Burtenshaw MD, Costantini F. A foreign β-globin gene in transgenic mice: integration at abnormal chromosomal positions and expression in inappropriate tissues. Cell. 1983;34:343–58. doi: 10.1016/0092-8674(83)90369-0. [DOI] [PubMed] [Google Scholar]
- 53a.Lederf A, Pattengale PK, Kuo A, Stewart TA, Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell. 1986;45:485–95. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- 54.LeMeur M, Gerhnger P, Benoist C, Mathis D. Correcting a immune-response deficiency by creating Eα gene transgenic mice. Nature. 1985;316:38–42. doi: 10.1038/316038a0. [DOI] [PubMed] [Google Scholar]
- 55.Lo C. Transformation by iontophoretic microinjection of DNA: multiple integrations without tandem insertions. Mol Cell Biol. 1983;3:1803–14. doi: 10.1128/mcb.3.10.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lo C. Localization of low abundance DNA sequences in tissue sections by in situ hybridization. J Cell Sci. 1986;8:143–62. doi: 10.1242/jcs.81.1.143. [DOI] [PubMed] [Google Scholar]
- 57.Lohler JR, Timpl R, Jaenisch R. Embryonic lethal mutation in mouse collagen I gene causes rupture of blood vessels and is associated with erythropoietic and mesenchymal cell death. Cell. 1984;38:597–605. doi: 10.1016/0092-8674(84)90514-2. [DOI] [PubMed] [Google Scholar]
- 58.Lovell-Badge RH, Bygraye AE, Bradley A, Robertson E, Evans MJ, Cheah KSE. Transformation of embryonic stem cells with the human type-II collagen gene and its expression in chimeric mice. Cold Spring Harb Symp Quant Biol. 1985;50:707–11. doi: 10.1101/sqb.1985.050.01.087. [DOI] [PubMed] [Google Scholar]
- 59.Low MJ, Hammer RE, Goodman RH, Habener JF, Palmiter RD, Brinster RL. Tissue-specific posttranslational processing of pre-prosomatostatin encoded by a metal-lothionein-somatostatin fusion gene in transgenic mice. Cell. 1985;41:211–19. doi: 10.1016/0092-8674(85)90075-3. [DOI] [PubMed] [Google Scholar]
- 60.Low MJ, Lechran RM, Hammer RE, Brinster RL, Habener JF, et al. Gonadotroph-specific expression of metallothionein fusion genes in pituitaries of transgenic mice. Science. 1986;231:1002–4. doi: 10.1126/science.2868526. [DOI] [PubMed] [Google Scholar]
- 61.MacDonald RJ, Hammer RE, Swift GH, Ornitz DM, Davis BP, et al. Tissue-specific expression of pancreatic genes in transgenic mice. N Y Acad Sci. 1985 doi: 10.1111/j.1749-6632.1986.tb15527.x. In press. [DOI] [PubMed] [Google Scholar]
- 62.Magram J, Chada K, Costantini F. Developmental regulation of a cloned β-globin gene in transgenic mice. Nature. 1985;315:338–40. doi: 10.1038/315338a0. [DOI] [PubMed] [Google Scholar]
- 63.Mark WH, Signorelli K, Lacy E. An insertional mutation in a transgenic mouse line results in developmental arrest at day 5 of gestation. Cold Spring Harbor Symp Quant Biol. 1985;50:453–63. doi: 10.1101/sqb.1985.050.01.057. [DOI] [PubMed] [Google Scholar]
- 64.Mayo KE, Palmiter RD. Glucocorticoid regulation of metallothionein gene expression. Biochem Actions of Hormones. 1985;12:69–88. [Google Scholar]
- 65.McGrath J, Solter D. Nuclear transfer in mammalian embryos: genomic requirements for successful development. In: Costantini F, Jaenisch R, editors. Banbury Report 20: Genetic Manipulation of the Early Embryo. Cold Spring Harbor, New York: Cold Spring Harbor Lab; 1985. pp. 31–42.pp. 289 [Google Scholar]
- 66.McKnight GS, Hammer RE, Kuenzel EA, Brinster RL. Expression of the chicken transferrin gene in transgenic mice. Cell. 1983;34:335–41. doi: 10.1016/0092-8674(83)90368-9. [DOI] [PubMed] [Google Scholar]
- 67.McLaren A. Mammalian Chimaeras. Cambridge: Cambridge Univ Press; 1976. [Google Scholar]
- 68.McMahon AP, Flytzanis CN, Hough-Evans BR, Katula KS, Britten RJ, Davidson EH. Introduction of cloned DNA into sea urchin egg cytoplasm: replication and persistence during embryogenesis. Dev Biol. 1985;108:420–30. doi: 10.1016/0012-1606(85)90045-4. [DOI] [PubMed] [Google Scholar]
- 69.Messing A, Chen HY, Palmiter RD, Brinster RL. Peripheral neuropathies, hepatocellular carcinomas and islet cell adenomas in transgenic mice. Nature. 1985;316:461–63. doi: 10.1038/316461a0. [DOI] [PubMed] [Google Scholar]
- 70.Mintz B, Cronmiller C. METT-1: A karyotypically normal in vitro line of developmentally totipotent mouse teratocarcinoma cells. Som Cell Genet. 1981;7:489–505. doi: 10.1007/BF01542992. [DOI] [PubMed] [Google Scholar]
- 71.Norstedt G, Palmiter RD. Secretory rhythm of growth hormone regulates sexual differentiation of mouse liver. Cell. 1984;36:805–12. doi: 10.1016/0092-8674(84)90030-8. [DOI] [PubMed] [Google Scholar]
- 72.Ornitz DM, Palmiter RD, Hammer RE, Brinster RL, Swift GH, MacDonald RJ. Specific expression of an elastase-human growth hormone fusion gene in pancreatic acinar cells of transgenic mice. Nature. 1985;313:600–3. doi: 10.1038/313600a0. [DOI] [PubMed] [Google Scholar]
- 73.Ornitz DM, Palmiter RD, Messing A, Hammer RE, Pinkert CA, Brinster RL. Elastase 1 promoter directs expression of human growth hormone and SV40 T-antigen genes to pancreatic acinar cells in transgenic mice. Cold Spring Harb Symp Quant Biol. 1985;50:399–409. doi: 10.1101/sqb.1985.050.01.050. [DOI] [PubMed] [Google Scholar]
- 74.Overbeek PA, Lai SP, van Quill KR, Westphal H. Tissue-specific expression in transgenic mice of a fused gene containing RSV terminal sequences. Science. 1986;231:1574–77. doi: 10.1126/science.3006249. [DOI] [PubMed] [Google Scholar]
- 75.Overbeek PA, Chepelinsky A, Khillan JS, Piatigorsky J, Westphal H. Lens-specific expression and developmental regulation of the bacterial chloramphenicol acetyltrans-ferase gene driven by the murine αA-crystallin promoter in transgenic mice. Proc Natl Acad Sci USA. 1985;82:7815–19. doi: 10.1073/pnas.82.23.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palmiter RD. Molecular biology of metallothionein gene expression. In: Kagi JHR, Kojima Y, editors. 2nd Int Meet Metallothionein and other Low Mol Weight Metal-Binding Proteins. Basel: Birkhauser-Verlag; 1986. In press. [Google Scholar]
- 77.Palmiter RD, Brinster RL, Hammer RE, Trumbauer ME, Rosenfeld MG, et al. Dramatic growth of mice that develop from eggs microinjected with metallothionein-growth hormone fusion genes. Nature. 1982;300:611–15. doi: 10.1038/300611a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palmiter RD, Brinster RL. Transgenic mice. Cell. 1985;41:343–45. doi: 10.1016/s0092-8674(85)80004-0. [DOI] [PubMed] [Google Scholar]
- 79.Palmiter RD, Chen HY, Brinster RL. Differential regulation of metallothionein-thymidine kinase fusion genes in transgenic mice and their offspring. Cell. 1982;29:701–10. doi: 10.1016/0092-8674(82)90186-6. [DOI] [PubMed] [Google Scholar]
- 80.Palmiter RD, Chen HY, Messing A, Brinster RL. SV40 enhancer and large T-antigen are instrumental in development of choroid plexus tumors in transgenic mice. Nature. 1985;316:457–60. doi: 10.1038/316457a0. [DOI] [PubMed] [Google Scholar]
- 81.Palmiter RD, Hammer RE, Brinster RL. Expression of growth hormone genes in transgenic mice. 1985:123–32. See Ref 65. [PMC free article] [PubMed] [Google Scholar]
- 82.Palmiter RD, Norstedt G, Gelinas RE, Hammer RE, Brinster RL. Metallothionein-human GH fusion genes stimulate growth of mice. Science. 1983;222:809–14. doi: 10.1126/science.6356363. [DOI] [PubMed] [Google Scholar]
- 83.Palmiter RD, Wilkie TM, Chen HY, Brinster RL. Transmission distortion and mosaicism in an unusual transgenic mouse pedigree. Cell. 1983;36:869–77. doi: 10.1016/0092-8674(84)90036-9. [DOI] [PubMed] [Google Scholar]
- 84.Payvar F, DeFranco D, Firestone GL, Edgar B, Wrange O, et al. Sequence-specific binding of glucocorticoid receptor of MTV DNA at sites within and upstream of the transcribed region. Cell. 1983;35:381–92. doi: 10.1016/0092-8674(83)90171-x. [DOI] [PubMed] [Google Scholar]
- 85.Pinkert CA, Widera G, Cowing C, Heber-Katz E, Palmiter RD, et al. Tissue-specific, inducible and functional expression of the Eαd MHC class II gene in transgenic mice. EMBO J. 1985;4:2225–30. doi: 10.1002/j.1460-2075.1985.tb03918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ritchie KA, Brinster RL, Storb U. Allelic exclusion and control of endogenous immunoglobulin gene rearrangement in κ transgenic mice. Nature. 1984;312:517–20. doi: 10.1038/312517a0. [DOI] [PubMed] [Google Scholar]
- 87.Ross SR, Solter D. Glucocorticoid regulation of mouse mammary tumor virus sequences in transgenic mice. Proc Natl Acad Sci USA. 1985;82:5880–84. doi: 10.1073/pnas.82.17.5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rusconi S. Gene transfer in living organisms. In: Schell JS, Starlinger R, editors. The Impact of Gene Transfer Techniques in Eukaryotic Cell Biology. Berlin: Springer Verlag; 1984. pp. 134–52. [Google Scholar]
- 89.Rusconi S, Kohler G. Transmission and expression of a specific pair of rearranged immunoglobulin μ and κ genes in a transgenic mouse line. Nature. 1985;314:330–34. doi: 10.1038/314330a0. [DOI] [PubMed] [Google Scholar]
- 90.Rusconi S, Schaffner W. Transformation of frog embryos with a rabbit β-globin gene. Proc Natl Acad Sci USA. 1981;78:5051–55. doi: 10.1073/pnas.78.8.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schnieke A, Harbers K, Jaenisch R. Embryonic lethal mutation in mice induced by retrovirus insertion into the αl(I) collagen gene. Nature. 1983;304:315–20. doi: 10.1038/304315a0. [DOI] [PubMed] [Google Scholar]
- 91a.Selden RF, Skoskiewicz MJ, Howie KB, Russell PS, Goodman HM. Regulation of human insulin gene in transgenic mice. Nature. 1986;321:525–28. doi: 10.1038/321525a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Serfling E, Jasin M, Schaffner W. Enhancers and eukaryotic gene transcription. Trends Genet. 1985;1:224–30. [Google Scholar]
- 93.Shani M. Tissue-specific expression of rat myosin light-chain 2 gene in transgenic mice. Nature. 1985;314:283–86. doi: 10.1038/314283a0. [DOI] [PubMed] [Google Scholar]
- 94.Shani M. Tissue-specific and developmentally regulated expression of a chimeric actin/globin gene in transgenic mice. Mol Cell Biol. 1986;6:2624–31. doi: 10.1128/mcb.6.7.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Small JA, Blair DG, Showalter SD, Scangos GA. Analysis of a transgenic mouse containing simian virus 40 and v-myc sequences. Mol Cell Biol. 1985;5:642–48. doi: 10.1128/mcb.5.4.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95a.Small J, Scangos GA, Cork L, Jay G, Khoury G. The early region of human papovavirus JC induces dysmyelination in transgenic mice. Cell. 1986;46:13–18. doi: 10.1016/0092-8674(86)90855-x. [DOI] [PubMed] [Google Scholar]
- 96.Stewart CL, Vanek M, Wagner EF. Expression of foreign genes from retroviral vectors in mouse teratocarcinoma chimaeras. EMBO J. 1985;4:3701–9. doi: 10.1002/j.1460-2075.1985.tb04138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38:627–37. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- 98.Stewart TA, Wagner EF, Mintz B. Human β-globin gene sequences injected into mouse eggs, retained in adults, and transmitted to progeny. Science. 1982;217:1046–48. doi: 10.1126/science.6287575. [DOI] [PubMed] [Google Scholar]
- 99.Stinchcomb DT, Shaw JE, Carr SH, Hirsch D. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol Cell Biol. 1985;5:3484–96. doi: 10.1128/mcb.5.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Storb U. Regulation of immunoglobulin gene expression: Use of transgenic mice. Ann Rev Immunol. 1987;5 doi: 10.1146/annurev.iy.05.040187.001055. In press. [DOI] [PubMed] [Google Scholar]
- 101.Storb U, Denis KA, Brinster RL, Witte ON. Pre-B cells in κ-transgenic mice. Nature. 1985;316:356–58. doi: 10.1038/316356a0. [DOI] [PubMed] [Google Scholar]
- 102.Storb U, O'Brien RL, McMullen MD, Gollahon KA, Brinster RL. High expression of cloned immunoglobulin κ gene in transgenic mice is restricted to B lymphocytes. Nature. 1984;310:238–41. doi: 10.1038/310238a0. [DOI] [PubMed] [Google Scholar]
- 103.Stout JT, Chen HY, Brennand J, Caskey CT, Brinster RL. Expression of human HPRT in the central nervous system of transgenic mice. Nature. 1985;317:250–52. doi: 10.1038/317250a0. [DOI] [PubMed] [Google Scholar]
- 104.Stuart GW, Searle PF, Palmiter RD. Identification of multiple metal regulatory elements in mouse metallothionein-I promoter by assaying synthetic sequences. Nature. 1985;317:828–31. doi: 10.1038/317828a0. [DOI] [PubMed] [Google Scholar]
- 105.Stuart GW, Searle PF, Chen HY, Brinster RL, Palmiter RD. A 12-base-pair DNA motif that is repeated several times in metallothionein gene promoters confers metal regulation to a heterologous gene. Proc Natl Acad Sci USA. 1984;81:7318–22. doi: 10.1073/pnas.81.23.7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Swanson LW, Simmons DM, Arriza J, Hammer R, Brinster RL, et al. Novel developmental specificity in the nervous system of transgenic animals expressing growth hormone fusion genes. Nature. 1985;317:363–66. doi: 10.1038/317363a0. [DOI] [PubMed] [Google Scholar]
- 107.Swift GH, Hammer RE, MacDonald RJ, Brinster RL. Tissue-specific expression of the rat pancreatic elastase 1 gene in transgenic mice. Cell. 1984;38:639–46. doi: 10.1016/0092-8674(84)90258-7. [DOI] [PubMed] [Google Scholar]
- 108.Townes TM, Chen HY, Lingrel JB, Palmiter RD, Brinster RL. Expression of human β-globin genes in transgenic mice: effects of a flanking metallothionein-human growth hormone fusion gene. Mol Cell Biol. 1985;5:1977–83. doi: 10.1128/mcb.5.8.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Townes TM, Lingrel JB, Chen HY, Brinster RL, Palmiter RD. Erythroid-specific expression of human β-globin genes in transgenic mice. EMBO J. 1985;4:1715–23. doi: 10.1002/j.1460-2075.1985.tb03841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.van der Putten H, Botteri FM, Illmensee K. Developmental fate of a human insulin gene in transgenic mice. Mol Gen Genet. 1984;198:128–38. doi: 10.1007/BF00328712. [DOI] [PubMed] [Google Scholar]
- 111.van der Putten H, Botteri FM, Miller AD, Rosenfeld MG, Fan H, et al. Efficient insertion of genes into the mouse germ line via retroviral vectors. Proc Natl Acad Sci USA. 1985;82:6148–52. doi: 10.1073/pnas.82.18.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wabl MR, Burrows PD. Expression of immunoglobulin heavy chain at high level in the absence of a proposed immunoglobulin enhancer element in cis. Proc Natl Acad Sci USA. 1984;81:2452–55. doi: 10.1073/pnas.81.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wagner EF, Covarrubias L, Stewart TA, Mintz B. Prenatal lethalities in mice homozygous for human growth hormone gene sequences integrated in the germ line. Cell. 1983;35:647–55. doi: 10.1016/0092-8674(83)90097-1. [DOI] [PubMed] [Google Scholar]
- 114.Wagner EF, Keller G, Gilboa E, Ruther U, Stewart CL. Gene transfer into murine stem cells and mice using retroviral vectors. Cold Spring Harbor Symp Quant Biol. 1985;50:691–700. doi: 10.1101/sqb.1985.050.01.085. [DOI] [PubMed] [Google Scholar]
- 115.Wagner EF, Stewart CL. Integration and expression of genes introduced into mouse embryos. In: Rossant J, Pedersen RA, editors. Experimental Approaches to Mammalian Embryonic Development. Cambridge Univ Press; 1985. In press. [Google Scholar]
- 116.Wagner EF, Stewart TA, Mintz B. The human β-globin gene and a functional thymidine kinase gene in developing mice. Proc Natl Acad Sci USA. 1981;78:5016–20. doi: 10.1073/pnas.78.8.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wagner TE, Hoppe PC, Jollick JD, Scholl DR, Hodinka RL, Gault JB. Microinjection of a rabbit β-globin gene in zygotes and its subsequent expression in adult mice and their offspring. Proc Natl Acad Sci USA. 1981;78:6376–80. doi: 10.1073/pnas.78.10.6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Weaver D, Costantini F, Isamishi-Kari T, Baltimore D. A transgenic immunoglobulin μ gene prevents rearrangement of endogenous genes. Cell. 1985;42:117–27. doi: 10.1016/s0092-8674(85)80107-0. [DOI] [PubMed] [Google Scholar]
- 119.Weaver D, Reis MH, Albanese C, Costantini F, Baltimore D, Imanishi-Kari T. Altered repertoire of endogenous immunoglobulin gene expression in transgenic mice containing a rearranged mu heavy chain gene. Cell. 1986;45:247–59. doi: 10.1016/0092-8674(86)90389-2. [DOI] [PubMed] [Google Scholar]
- 120.Westphal H, Overbeek PA, Khillan JS, Chepelinsky AB, Schmidt A, et al. Promoter sequences of murine αA crystallin, murine α2(I) collagen or avain sarcoma virus genes linked to the bacterial chloramphenicol acetyl transferase gene direct tissue-specific patterns of chloramphenicol acetyl transferase expression in transgenic mice. Cold Spring Harbor Symp Quant Biol. 1985;50:411–16. doi: 10.1101/sqb.1985.050.01.051. [DOI] [PubMed] [Google Scholar]
- 121.Wilkie TM, Brinster RL, Palmiter RD. Germline and somatic mosaicism in transgenic mice. Dev Biol. 1986 doi: 10.1016/0012-1606(86)90068-0. In press. [DOI] [PubMed] [Google Scholar]
- 122.Wirak DO, Chalifour LE, Wasserman PM, Muller WJ, Hassell JA, et al. Sequence-dependent DNA replication in preimplantation mouse embryos. Mol Cell Biol. 1985;5:2924–35. doi: 10.1128/mcb.5.11.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Woychik RP, Stewart TA, Davis LG, D'Eustachio P, Leder P. An inherited limb deformity created by insertional mutagenesis in a transgenic mouse. Nature. 1985;318:36–40. doi: 10.1038/318036a0. [DOI] [PubMed] [Google Scholar]
- 123a.Yamamoto KR. Steroid receptor regulated transcription of specific genes and gene networks. Ann Rev Genet. 1985;19:209–52. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- 124.Yamamura K, Kikutani H, Folsom V, Clayton LK, Kimoto M, et al. Functional expression of microinjected Eαd gene in C57BL/6 transgenicmice. Nature. 1985;316:67–69. doi: 10.1038/316067a0. [DOI] [PubMed] [Google Scholar]
- 125.Yamamura K, Kudo A, Ebihara T, Kamino K, Araki K, et al. Cell-type specific and regulated expression of a human γl immunoglobulin gene in transgenic mice. Proc Natl Acad Sci USA. 1986;83:2152–56. doi: 10.1073/pnas.83.7.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Note added in proof The following papers were published during the last few months. The titles reveal several exciting developments
- Bierberich C, Scangos G, Tanaka K, Jay G. Regulated expression of a murine class I gene in transgenic mice. Mol Cell Biol. 1986:61339–42. doi: 10.1128/mcb.6.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F, Chada K, Magram J. Correction of murine β-thalassemia by gene transfer into the germ line. Science. 1986;233:1192–94. doi: 10.1126/science.3461564. [DOI] [PubMed] [Google Scholar]
- Kollias G, Wrighton N, Hurst J, Grosveld F. Regulated expression of human Aγ-, β-, and hybrid γβ-globin genes in transgenic mice: manipulation of the developmental expression patterns. Cell. 1986;46:89–94. doi: 10.1016/0092-8674(86)90862-7. [DOI] [PubMed] [Google Scholar]
- Leder A, Pattengale PK, Kuo A, Stewart T, Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell. 1986;45:485–95. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- Morello D, Moore G, Salmon AM, Yaniv M, Babinet C. Studies on the expression of an H-2K/human growth hormone fusion gene in giant transgenic mice. EMBO J. 1986;5:1877–83. doi: 10.1002/j.1460-2075.1986.tb04439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M, Leopold P, Vailly J, Cuzin F. Germ line transmission of autonomous genetic elements in transgenic mouse strains. Cell. 1986;46:513–19. doi: 10.1016/0092-8674(86)90876-7. [DOI] [PubMed] [Google Scholar]
- Soriano P, Jaenisch R. Retroviruses as probes for mammalian development: allocation of cells to the somatic and germ cell lineages. Cell. 1986;46:19–29. doi: 10.1016/0092-8674(86)90856-1. [DOI] [PubMed] [Google Scholar]