Abstract
The myelodysplastic syndromes (MDS) are a group of clonal diseases characterized by inefficient haematopoiesis, increased apoptosis and risk of evolution to acute myeloid leukaemia. Alterations in epigenetic processes, including DNA methylation, histone modifications, miRNA and splicing machinery, are well known pathogenical events in MDS. Although many advances have been made in determining the mutational frequency, distribution and association affecting these epigenomic regulators, functional integration to better understand pathogenesis of the disease is a challenging and expanding area. Recent studies are shedding light on the molecular basis of myelodysplasia and how mutations and epimutations can induce and promote this neoplastic process through aberrant transcription factor function (RUNX1, ETV6, TP53), kinase signalling (FLT3, NRAS, KIT, CBL) and epigenetic deregulation (TET2, IDH1/2, DNMT3A, EZH2, ASXL1, SF3B1, U2AF1, SRSF2, ZRSR2). In this review we will try to focus on the description of these mutations, their impact on prognosis, the functional connections between the different epigenetic pathways, and the existing and future therapies targeting these processes.
Keywords: Genetics, Epigenetics, MDS
Introduction
The myelodysplastic syndromes (MDS) are a group of clonal diseases characterized by inefficient haematopoiesis, increased apoptosis and risk of evolution to acute myeloid leukaemia (AML). Disease complexity is not only entailed by morphological diversity, which translates in an array of entities encompassed under the term MDS, but by the increasing number of molecular pathways and hallmarks that participate in disease initiation, evolution and progression to AML. Approximately 10–20% patients with MDS will ultimately develop a secondary AML. Efforts have been made to try to identify patients at risk of this final event. Solid evidence of the molecular events related to disease progression still remains elusive, with frequent contradictory or scarce data on the field. Understanding of the different molecular mechanisms responsible for initiation and progression of the disease have been an area of active research in the last decades.
The main biological hallmarks in myelodysplasia have been well described and include both genomic and epigenomic alterations in transcription factors, epigenetic modulators, miRNA, microenvironment and innate immunity (Bejar et al, 2011). DNA methylation remains the single most important mechanism of epigenetic regulation through the activation or repression of gene expression both at CpG-enriched promoters (CpG islands), and gene enhancers present outside of these genomic areas (Bird, 2002; Issa 2013). Altered methylation patterns have been shown to exist in myelodysplasia, with global hypermethylation of CpG islands inducing silencing of target genes (many of them being cell cycle regulators), to appear in initial stages of the disease and to be associated with disease progression (Figueroa et al, 2009; Jiang et al, 2009; Issa, 2013). Histone modifications represent another of the essential cellular mechanisms implicated in tissue-specific, transcription factor-dependent regulation of gene expression. Through methylation, acetylation and ubiquitination of different histones, specific changes to transcriptional activation and repression participate in cell fate, differentiation and regulation of proliferation. Aberrant expression of these processes due to alterations in histone regulators is one of the key pathogenic features in MDS (Issa, 2013).
The goal of this review is to focus on the known epigenetic alterations described in MDS in order to try and integrate the current mutational landscapes in these modulators with functional modelling of the disease and existing and potential therapies.
Epigenetic modifications in MDS
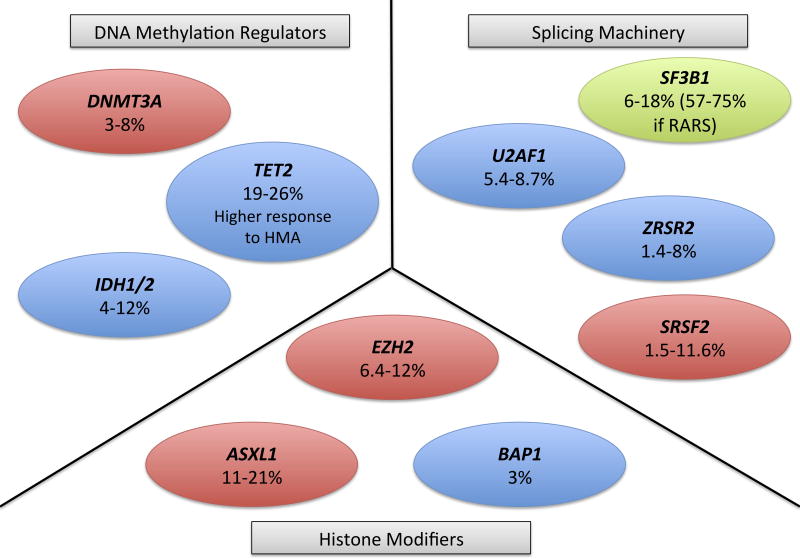
The epigenetics behind MDS remained a mystery until the advent of next-generation whole-genome and whole-exome sequencing techniques, allowing a more comprehensive evaluation of the epigenome in MDS. The majority of novel gene mutations that have been identified play a role in DNA methylation, histone modification and RNA splicing. Figure 1 summarizes the different epigenetic modifiers that are known to be mutated in MDS and their relation to overall prognosis or evolution to AML.
Figure 1.
Main mutated epigenetic regulators in myelodysplastic syndrome (MDS). Genes represented in red are known to be associated with worse prognosis. Genes in green are associated with better prognosis and those in blue have an unclear prognostic impact. Percentages represent the known prevalence of the mutation in MDS.
DNA methylation
In normal tissue, the CpG-rich promoters remain mainly unmethylated regardless of differentiation state (Bird 2002). In MDS, about 3–5% of these promoter-associated CpG islands become aberrantly hypermethylated (Figueroa et al, 2009; Jiang et al, 2009), often independent of cytogenetic changes. The hypermethylation may occur early in the disease and is found to be associated with more rapid progression to AML (Shen et al, 2010).
TET2
Loss-of-function mutations occur in about 19–26% of MDS patients (Guo et al 2011). Various studies have shown both hypermethylating and hypomethylating profiles related to TET2 mutations (Figueroa et al 2010, Ko et al 2010), with TET2 deletions leading to an increase in haematopoietic stem cell (HSC) compartment and self-renewal (Quivoron et al, 2011; Moran-Crusio et al, 2011; Ko et al, 2011; Li et al, 2011). The clinical implication of TET2 mutations remains unclear. A recent study demonstrated that patients with TET2 mutations had a higher overall response rate to azacytidinetreatment with no difference in OS (Itzykson et al, 2011).
DNMT3A
Mutations in DNMT3A, leading to a reduction in methyltransferase catalytic activity, are found in about 3–8% of MDS patients (Thol et al, 2011a; Walter et al, 2011), with nearly all bone marrow cells harbouring the mutation when present, therefore suggesting an early occurrence in the disease. It is associated with older age at diagnosis but not with other cytogenetic or clinical features (Walter et al, 2011). Some studies have demonstrated a worse clinical outcome with lower survival and rapid progression to AML (Walter et al, 2011); however this was not confirmed in further studies (Thol et al, 2011a; Bejar et al, 2012).
IDH1/IDH2
Isocitrate dehydrogenase 1 and 2 mutations lead to decreased α-ketoglutarate (α-KG), TET inhibition and widespread promoter hypermethylation of DNA (Xu et al, 2011). Other α-KG dependent enzymes have also been found to be inhibited by 2-hydroxyglutarate (2-HG) (Chowdhury et al, 2011; Lu et al, 2012). However, the impact on OS and evolution to AML remains unclear (Patnaik et al, 2012; Kosmider et al, 2010; Thol et al, 2010). About 4–12% of MDS cases show IDH1/2 gain-of-function mutations (Patnaik et al, 2012; Kosmider et al, 2010; Thol et al, 2010). Functional effects of IDH1 and IDH2 mutations on haematopoiesis are unclear, with early studies demonstrating impaired HSC differentiation (Figueroa et al, 2010).
Histone modification
Post-translational modifications of histones are also an important part of epigenetic regulation. These proteins can be acetylated, methylated, and ubiquinated by a group of histone-modifying enzymes.
EZH2
EZH2 is an important part of the Polycomb Repressive Complex 2 (PRC2), which trimethylates lysine 27 of histone 3 regulating stem cell differentiation and repression of gene transcription. Loss-of-function mutations are found in about 6.4–12% of patients (Bejar et al, 2012; Ernst et al, 2010; Bejar et al, 2011) and have been associated with poor overall survival (OS) independently of other prognostic factors and mutations.
ASXL1
Another gene that is a component of the PRC and plays a role in MDS is ASXL1. Loss-of-function mutation in this gene is the third most frequently mutated in MDS, occurring in about 11–21% of MDS patients (Rocquain et al, 2010; Gelsi-Boyer et al, 2009; Boultwood et al, 2010; Thol et al, 2011b) and in about 10–15% of patients with myeloproliferative neoplasm (MPN) syndromes (Rocquain et al, 2010; Gelsi-Boyer et al, 2009; Boultwood et al, 2010). Like the EZH2 mutation, it is associated with poor prognosis with worse OS independent of other clinical factors.
RNA splicing
RNA splicing is an essential part of pre-mRNA maturing into mRNA for translation into proteins. More than 90% of human genes undergo splicing and translate into various protein isoforms; therefore, splicing is an integral process of gene expression diversity (Wahl et al, 2009; Chen & Manley, 2009). RNA splicing is the most commonly mutated pathway in MDS, and appears to occur early in disease evolution (Papaemmanuil et al, 2013). These mutations play a major role in dictating clinical features of the disease.
SF3B1
SF3B1 is one of the most common mutations seen in MDS and is highly associated with the presence of ringed sideroblasts. It is seen in about 6–18% of non-ringed sideroblast MDS and in about 57–75% of ringed sideroblast MDS (Yoshida et al, 2011; Papaemmanuil et al, 2011). SF3B1 encodes a component of U2 snRNP that recognizes 3’ splice site at intron-exon junctions. Many studies have demonstrated improved OS, higher neutrophil and platelet counts, less bone marrow blasts, and low risk of leukemic evolution in patients with SF3B1 mutations (Bejar et al, 2012; Malcovati et al, 2011).
U2AF1
U2AF1 encodes the small subunit of U2 auxiliary factor complex, which is required for the binding of U2 snRNP to the pre-mRNA branch site. This mutation can be found in 5.4–8.7% of MDS patients (Graubert et al, 2012; Thol et al, 2012; Damm et al, 2012), especially in younger patients (Wu et al, 2013). Although it has been associated with ASXL1 and DNMT3A (Thol et al, 2012; Damm et al, 2012) data from other groups have shown no significant co-occurrence of these mutations (Wu et al, 2013). The clinical impact of this mutation remains unclear, with contradictory data regarding the risk of progression to secondary AML (Graubert et al, 2012; Thol et al, 2012; Damm et al, 2012; Makishima et al, 2012). Recent data suggest that this mutation appears to be stable during disease progression, implying it probably plays a role in the development of MDS but not in the progression to AML (Wu et al, 2013).
SRSF2
SRSF2 encodes a serine/arginine-rich splicing factor 2 that is important for selection of splice-site, assembly of spliceosome and both constitutive and alternative splicing (Long & Caceres, 2009; Wu et al, 2012). Mutation in this gene occurs in about 1.5–11.6% of patients (Bejar et al, 2012), is closely associated with the male gender and older age (Wu et al, 2012), poorer OS (Makishima et al, 2012; Wu et al, 2012) and other mutations such as RUNX1, IDH2 and ASXL1 (Wu et al, 2012; Nagata et al, 2011). Like U2AF1, it appears to be stable throughout disease progression, suggesting a role in disease initiation but not in the evolution to AML (Wu et al, 2012).
ZRSR2
Similar to UA2F1, ZRSR2 also encodes a subunit of U2 auxiliary unit and interacts with U2AF1 and SF3B1 to bind to U2snRNP (Yoshida, et al 2011). The mutation in this gene occurs in about 1.4–8% of MDS patients (Damm et al, 2012). The biological effect of this mutation remains unclear and needs to be further studied.
Epigenetic Modifications in AML
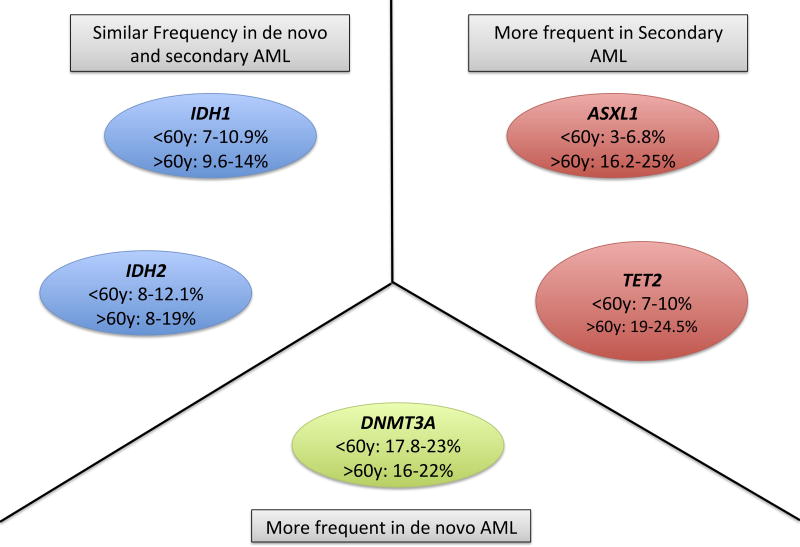
Although it is beyond the scope of this review to describe the epigenetics of AML in a detailed manner, it still remains important to note that many of the previously described mutations are also found in a substantial number of AML cases (Abdel-Wahab & Levine, 2013). This data is shown in Figure 2. Additionally, recent data suggest different frequencies in mutations in epigenetic modifiers may exist between de novo and secondary AML. One example of this is the elegant work done by Fernandez-Mercado et al (2012), analysing mutation patterns of 16 genes in 84 cases of primary and secondary AML. Similar frequencies of mutations in IDH1/2 were found in primary and secondary AML, with a higher frequency of TET2 mutations in AML evolving from chronic monomyelocytic leukaemia (CMML) and a higher frequency of ASXL1 mutations in secondary AML. Mutations in DNMT3A were found to be more frequent in primary AML. Further research analysing the frequency of these mutations in greater number of patients is nevertheless required to confirm and expand our knowledge in the field.
Figure 2.
Main mutated epigenetic regulators in acute myeloid leukaemia (AML). Frequencies of mutations include all AML cases (both de novo and secondary) in patients aged 60 years or younger (<60y) and older than 60 years (>60y).
Several studies have shown increased overall methylation in both de novo and secondary AML, with higher mean overall methylation in cases with an antecedent haematological disorder, such as MDS, compared to primary AML (Figueroa et al, 2009; Wilop et al, 2011; Deneberg et al, 2010). Moreover, current data points to the existence of methylation patterns that lead to distinct gene-promoter signatures in primary AML compared to secondary AML and MDS, and that these patterns seem to impact evolution and clinical outcome in both diseases (Figueroa et al, 2009; Deneberg et al, 2010; Galm et al, 2005; Toyota et al, 2001). Available evidence also indicates an impact of hypomethylating agents in global and gene-specific methylation levels (Figueroa et al, 2009; Negrotto et al, 2012), allowing us to speculate if the therapeutic potential of these drugs may be related to these methylation changes. Unfortunately, most of this data still remains anecdotal and we are lacking large confirmatory studies trying to further explore methylation patterns in both MDS and AML along with their evolution with epigenetic therapies. With the development of current technologies we may yet be able to better answer these uncertainties in the following years.
Integrating the Different Epigenetic Alterations in MDS and AML Evolution
Translational and basic research in the past decades has been of utmost importance in advancing our knowledge regarding the biological markers and hallmarks of MDS. Despite the development of new technologies, which have allowed for more sophisticated methods of study in molecular oncology, the scientific community still has to face the enigma posed by the integration and interpretation of the data these advances have offered. Our ability to determine the presence of cytogenetic and molecular alterations (both genomic and epigenomic) in different settings has, unfortunately, developed at a far quicker pace than our ability to understand in a profound manner the impact and relevance of these alterations in the initiation, progression and development of oncogenesis.
Many somatic mutations in different biological markers have been described in MDS. Apart from the previously described affecting regulators of DNA methylation (Abdel-Wahab & Figueroa, 2012; Issa, 2013) (TET2, IDH1/IDH2, DNMT3A), histone modifiers (ASXL1, EZH2) and splicesosome machinery (SF3B1, U2AF1, SRSF2, ZRSR2) there are others, such as transcription factors (RUNX1, TP53, ETV6) and signal transduction kinases (NRAS, FLT3, JAK2, KIT, CBL), whose detailed description is beyond the scope of this review.
In the current section of this review we will try to summarize the current evidence supporting the functional mechanisms by which these different mutations and epigenetic alterations interact in the development of MDS.
DNA methylation and its link to Histone Modification and Innate Immunity
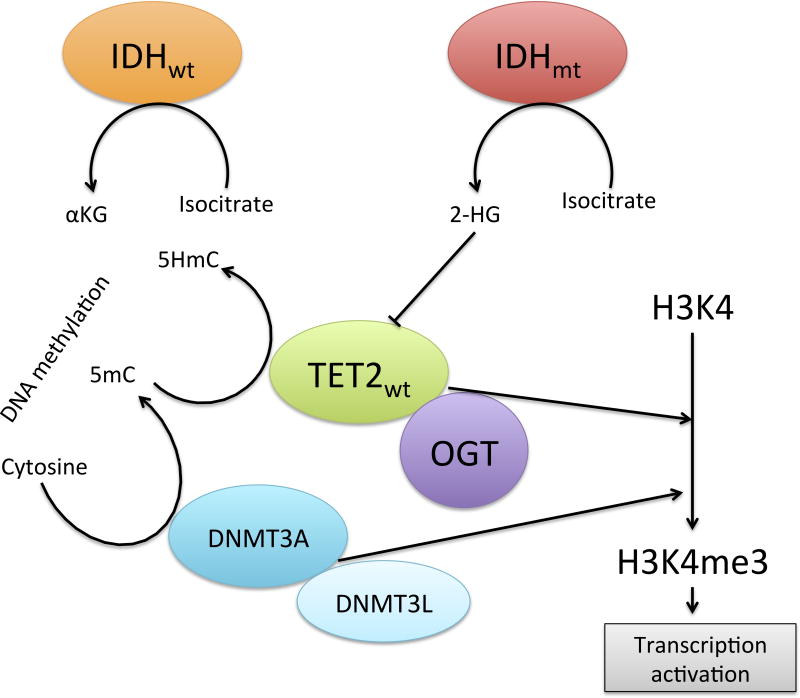
Current understanding of the biology and functional regulation of the different DNA methylation regulators has shed light in the progressive integration of epigenetic regulation. TET2 participates in the promotion of DNA demethylation through the conversion of 5-methylcytosine (5mC) into 5-Hydroxymethylytosine (5hmC), and has recently been shown to modulate other epigenetic processes beside methylation changes due to its hydroxylase activity.
Many groups have studied the implication of TET genes in myeloid malignancies, including MDS, with mutations in TET2 having been shown to induce a loss of function leading to a predominance of 5mC in DNA (Abdel-Wahab et al, 2009; Delhommeau et al, 2009; Mohamedali et al, 2009; Smith et al, 2010). This modification in the normal methylation status of DNA, due to a disruption in the normal methylation/demethylation balance, is partially responsible for the MDS phenotyp,e as has been elegantly shown using murine models by several groups (Quivoron et al, 2011; Moran-Crusio et al, 2011; Li et al, 2011; Ko et al, 2011). Data from these experiments suggests loss of function of Tet2 induces an increased self-renewal capacity and expansion with greater repopulating potential that could be linked to an impairment of the normal regulation of Hoxa genes in the affected haematopoietic compartment suggesting a tumour suppressor effect (through induction of differentiation and inhibition of proliferation/stem-cell expansion) of wild-type Tet2. Interestingly enough, Tet2 mutation was enough to induce myeloid expansion with a clear predominance of the monocytic compartment in a similar manner to what has been shown with other molecular markers, such as SETBP1 (Damm et al, 2013) and NRAS, in human samples. This functional finding is consistent with clinical observations regarding disease phenotype and mutational patterns, as is the case for CMML, a subtype of MDS exhibiting a high frequency of mutations in these three genes, and morphologically characterized by peripheral and bone marrow monocytic expansion (Cazzola et al, 2013). Although the exact mechanisms behind this specific monocyte differentiation still remain unclear, there is room for speculating a possible synergistic effect of different epigenetic mutations in the development of such a phenotype. In this sense, there is solid data supporting a clear relationship between CMML and the simultaneous occurrence of TET2 and SRSF2 mutations (Cazzola et al, 2013) possibly due to complex epigenetic and RNA-dependent deregulation of myeloid regulatory genes. Additional investigation is however required in order to determine mutational hierarchy, identify these candidate genes and elucidate the molecular mechanisms responsible for the specific phenotype.
TET2-mediated epigenetic gene activation is also regulated through the ability of TET to mediate histone modifications through the recruitment of OGT (O-linked β-D-N-Acetylglucosamine Transferase). Once OGT is recruited, it interacts with chromatin inducing O-GlcNAcylation of Histone 2B (Chen et al, 2013) and host cell factor 1 (HCF1), a key regulator of the H3K4 methyltransferase complex (SET1/COMPASS) (Solary et al, 2014). Histone 3 Lysine 4 trimethylation (H3K4me3) is a well-known histone marker with importance in transcriptional activation and lineage determination of haematopoietic stem cells and progenitors (Barski et al, 2007; Orford et al, 2008). Loss-of-function mutations in TET2 are responsible for a block in this histone regulation leading to a decrease in H3K4me3 and, therefore, a decrease in transcription of essential regulators of haematopoiesis (Deplus et al, 2013). These data clearly exemplify the complexity of functional consequences of mutations in epigenetic modifiers, and opens the possibility of speculation to possible functional links to other pathogenic events in MDS, such as innate immunity modifications. Both TET (through OGT) and JMJD3, a JmjC domain protein involved in histone methylation, act as regulators of H3K4 trimethylation whose inhibition has been shown to be related to MDS pathogenesis. A recent publication (Wei et al, 2013a) suggests innate immunity signals mediated through Toll-Like Receptor (TLR) signalling via nuclear factor (NF)-κB induce activation of JMJD3 which, in turn, leads to the transcriptional activation of genes promoting NF-κB expression and innate immunity signalling regulated by H3K4me3, therefore inducing an activation loop of this pathway.
An interesting observation in the original mutational studies in MDS was the mutually exclusiveness of TET2 and IDH1/2 mutations, which was later shown to be the result of a link between metabolism and epigenetics. TET enzymes require α-KG in order to be able to be active. Somatic mutations in the IDH genes lead to a disruption of their catalytic activity inducing a product shift from α-KG to its homolog, 2-HG (Ward et al, 2010). This phenomenon leads to an inhibition of TET not only through α-KG depletion, but through direct inhibition by 2-HG leading to increased HOXA activity (Xu et al, 2011), and therefore represents a biological explanation to the fact that TET and IDH1/2 mutations are not usually present in the same MDS clone. A similar α-KG dependency occurs with JmJC demethylases suggesting, again, new functional links between these epigenetic regulators (Chowdhury et al, 2011; Lu et al, 2012).
Consistent with the above epigenetic mechanisms, deregulation of DNMT3A, another DNA methylation regulator, has been described in MDS and AML (Walter et al, 2011; Bejar et al, 2011). The DNMTs (DNA methyltransferases) are a group of enzymes that catalyse the addition of a methyl group in carbon 5 of cytosine to produce 5mC, and can act as either de novo or as maintenance transferases regulating methylation, primarily in dinucleotides at CpG islands (Li et al, 2013). Constitute methylation is essential for HSCs self-renewal. Evidence of a fundamental impact on the normal homeostasis of these enzymes has not only been shown through the presence of point mutations in cases of MDS (as previously noted), but through functional essays using murine models, which prove the importance of DNMTs in haematopoiesis by modulation of tissue and context-specific gene expression (Challen et al, 2011). In this model, Dnmt3a–null mice HSCs were shown to exhibit an upregulation of multipotency genes (Runx1, Pbx1, Cdkn1a) and a downregulation of differentiation factors (Flt3, Ikzf1, Spi1, Mef2c) due to changes in the methylation signature (including substantial hypermethylation at CpG islands along with a global hypomethylation), further supporting the role of loss-of-function mutations in DNMT3A in leukaemia and MDS. A recent interesting observation by Jost et al (2013) is the characterization of epigenetic modifications in DNMT3A in AML due to aberrant hypermethylation at an internal promoter region of DNMT3A. Interestingly, this methylation pattern, with a silencing effect on the methyltransferase, was particularly observed in samples without mutations at the DNMT3A gene, and was correlated with changes in gene expression including upregulation of HOXA and HOXB clusters in a similar manner as the DNMT3mut cases. Comparable epigenetic modulation of essential key regulators of haematopoeisis and MDS pathogenesis may yet be described in the near future and may well be one of the reasons for mutational patterns in MDS, including co-occurrence and mutual exclusiveness of different mutations. Genomic alterations in an epigenetic modulator may lead to aberrant epigenetic silencing of a second gene (as the above mentioned DNMT3A epimutation), result in a proliferation disadvantage in the presence of other mutations (such as the previously described case of TET2 and IDH1/2) or induce a potentiation of its effect through cooperation of a mutation on another epigenetic regulator. An example of the latter is the cooperative effect of Tet2 mutations in Ezh2-deleted mice, which showed a more accelerated and advanced myelodysplasia than Ezh2−/− Tet2wt mice (Muto et al 2013).
Similarly to TET2, DNMT3A not only regulates transcription through DNA methylation, but can also bind to methylated and non-methylated H3K4 through its PWWP and ADD domains respectively (Li et al, 2013), therefore modulating histones and chromatin compaction by the formation of a DNA-DNMT3A–DNMT3L complex. In this complex, DNMT3L acts as a co-regulatory methyltransferase-like protein that modulates the activity of DNMT3A (Li et al 2013; Neri et al, 2013). As we will describe latter, DNMT recruitment to H3K4 participates in the regulation of transcription through the PRC1 and PRC2 complexes, some of whose components (such as ASXL1 and EZH2) are important players in MDS pathogenesis. It appears then that separation of different mutational events in epigenetic modulators in MDS will ultimately lead to deregulation of common hallmarks of gene regulation. Current molecular biology is slowly closing the gap between the different observational discoveries the scientific community has made in the last years, allowing us to understand the intricate mechanisms of homeostasis and disease evolution. A graphical representation of the main interactions and functions of these epigenetic modulators can be found in Figure 3.
Figure 3.
DNA methylation and histone modifications by TET2 and DNMT3A.
Histone Modifications in MDS: Polycomb Repressor Complexes and regulation of H3K27me3
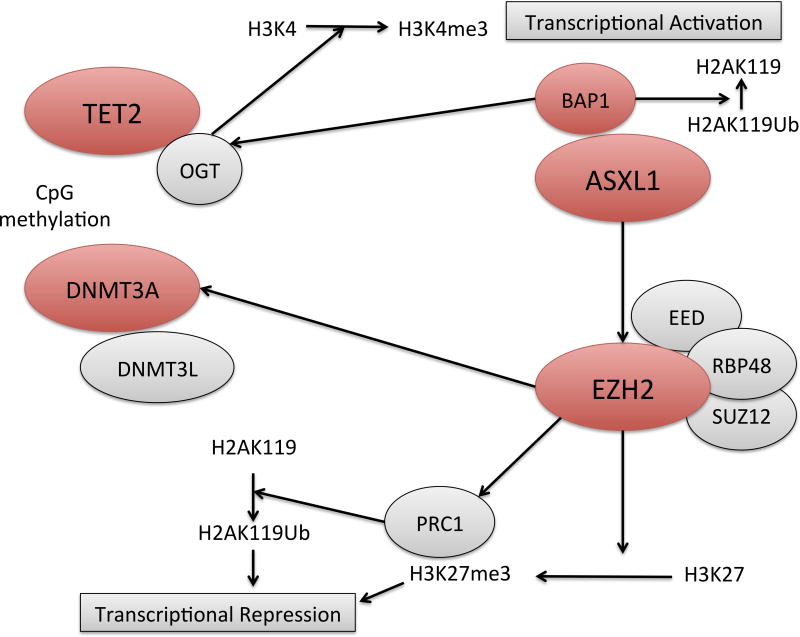
Modification of histones through different biochemical processes, including methylation and acetylation, represent one of the main epigenetic mechanisms in gene expression regulation in a transcription factor-dependent manner. The Polycomb pathway is a key regulator of cell-fate decisions and differentiation through H3K27 trymethylation, and is composed of two protein complexes (Polycomb group proteins or PcG), PRC1 and PRC2, which are functionally related and integrated (Morey & Helin, 2010).
PRC1 (Polycomb Repressor Complex 1) catalyses the ubiquitylation of lysine 119 in histone H2A (H2AK119Ub1), which is a repressor mark, and is dependent on PRC2 for recruitment to target genes and subsequent silencing (Müller & Verrijzer, 2009). PRC1 is composed of several proteins, including ASXL1. ASXL1 assembles various proteins to form several complexes with different functional effects, including ASXL1-BAP1, ASXL1-NHR and ASXL1-PRC2 (Katoh, 2013; Abdel-Wahab & Dey, 2013). Through its binding to PRC2, ASXL1 regulates H3K27, inducing its trimethylation through the SET domain in EZH2, inhibiting transcription of target genes (including the HOXA cluster of genes) by blocking transcription factors access to DNA (due to chromatin compaction), and further recruiting PRC1 for maintained gene repression. In vitro models assessing the effect of ASXL1 loss have shown a marked global loss of H3K27me3 independent of the expression levels of PRC2 members, which was reversible with re-expression of ASXL1 (Abdel-Wahab et al, 2012). Various murine models further support the driver effect of this mutation, with Asxl1+/− mice developing an MDS-like and MDS/MPN disease with increased apoptosis and proliferation in bone marrow (similarly to what is found in human MDS) (Wang et al, 2014). In this model, Asxl1-deficient mice showed upregulation of Hoxa genes that, again, represent a genomic hallmark of the disease common to other mutations such as TET2, IDH1/2, EZH2 and DNMT3A. Without ASXL1, there is an upregulation of microRNA 125a (MIR125A) and subsequent suppression of C-type lectin domain family 5, member a (CLEC5A), leading to impairment of myeloid differentiation and development of MDS (Inoue et al, 2013). Interestingly, despite the presence of ASXL1 mutations in different myeloid disorders, CMML shows a higher frequency of mutations than other MDS subtypes or AML (Cazzola et al, 2013) suggesting an essential role of this gene in granulomonocytic development. In fact, a recent study by Davies et al (2013) has demonstrated an impairment of granulomonocytic differentiation in ASXL1-deficient CD34+ cells due to downregulation of genes involved in myeloid and monocyte-macrophage development (MSR1, IL6, APOC1, CCL2, CCL and CD14), further supporting a possible connection between this gain of function-mutation and a monocytic phenotype, as in CMML.
BAP1 is a nuclear-localized deubiquitinating enzyme that binds to ASXL1 participating in H2AUb deubiquitination, thereby regulating myelopoiesis, mutated in 3% MDS cases. Interestingly, there is data to support the activity of BAP1 as a positive regulator of HCF-1 and OGT (Dey et al, 2012), the latter being related to TET2-mediated histone modulation as we previously described. All this data highlights the intertwined alterations in the epigenetic transcriptional regulation in MDS pathogenesis.
PRC2 catalyses the trimethylation of H3K27 in a direct manner, and indirectly regulates the ubiquitination of H2AK119 by recruitment of PRC1 (Morey & Helin, 2010; Müller & Verrijzer, 2009; Lund et al, 2014), inducing transcriptional repression. Components of PRC2 include EZH2, EED, SUZ12 and RBBP4. This assembled complex can bind to H3K27, inducing its trimethylation via the SET domain of EZH2 and, as a result, chromatin compaction and inhibition of transcription factor access to DNA. EZH2 can also induce DNA methylation by recruiting DNMT, further repressing transcription of target genes through gene promoter silencing (Vire et al, 2006). Through these mechanisms EZH2 is an important regulator of cell fate controlling the balance between self-renewal and differentiation, having both the ability to stabilize chromatin structure to maintain long-term self-renewal potential of HSCs (Kamminga et al, 2006), and to induce a shift toward proliferation by increased cell cycle genes (Bracken et al, 2003) via regulation of PI3K pathway, MYC expression and several miRNA. Deregulation of this balance and the functions of EZH2 are responsible for its contribution to oncogenesis through different mechanisms. EZH2 has been described to be altered in MDS through missense, frameshift and nonsense loss-of-function mutations in the SET, CXC and D2 domains (Ernst et al, 2010; Nikoloski et al, 2010), as well as in MDS cases with −7/del(7q) and MDS/MPN with uniparental disomy of 7q (which harboured concomitant inactivating mutations on EZH2 in 71% of cases) (Jerez et al, 2012). These findings suggest a tumour suppressor effect of EZH2 through loss of H3K27 trimethylation, in a similar manner as seen with ASXL1-inactivating mutations, leading to de-repression of proliferation and stem-cell expansion genes such as HOXA and HOXB.
Deregulation of histone modifications represents a key player in myelodysplasia initiation and progression to AML, with mutations and epigenetic alterations in different regulators being associated with different stages of the disease. Understanding of the molecular integration of these processes (including TET, DNMTs, PRC1 and PRC2) will allow us to develop new therapeutic strategies to target epigenetic deregulation in a much more effective manner. Representation of these molecular communications can be seen in Figure 4.
Figure 4.
Transcriptional regulation through PRC1, PRC2, TET2 and DNMT3A. Figures in red represent proteins known to harbour loss-of-function mutations in myelodysplastic syndrome.
Impact of Splicesosome mutations on MDS Pathogenesis and Evolution: Connection to other Epigenetic regulators, Transcription factors and Innate immunity
As described earlier in this review, mutations in the different components of the splicing machinery have become a common finding along the different stages of MDS, suggesting their role as founder mutations of the disease (Mian et al 2013). Recent data suggest most cases of MDS (up to 85%) (Yoshida et al, 2011) harbour a mutation in one of the members of the RNA-splicing machinery, and that these mutations tend to be mutually exclusive. These mutations can alter the normal function of splicesosome machinery inducing inappropriate inclusion of introns or exons into RNA, disrupting the normal function of target proteins and, therefore, the ability of the cell to differentiate, hence generating dysplasia (Abrahamsson et al, 2009). SF3B1 is common in RARS and RARS-T as described by several groups (Thol et al, 2010; Yoshida et al, 2011; Papaemmanuil et al, 2011; Mian et al, 2013) and recent data using murine models with Sf3b1+/− mice has shown its ability to induce a phenotype similar to RARS (Rogers et al, 2013). Interestingly enough, expression levels of Ezh2, as well as Npm1 and Tp53, were found to be significantly lower in Sf3b1-deficient mice, suggesting a regulatory effect over PRC2 and histone modification. This is consistent with other findings of the ability of Sf3b1 to physically interact with PRC1 proteins in Sf3b1+/− mice models.
Other murine models exploring the effects of Srsf2 in HSCs showed that both wild-type and P95H (the most common SRSF2 mutation in MDS) Srsf2 induced distinct changes in alternative splicing of more than 100 genes, with several changes in genes with known roles in haematopoietic malignancies being uniquely induced by the P95H mutant (Qiu et al, 2013). This finding suggests a possible gain of function effect on SRSF2 mutations, which may induce modifications in key regulators of MDS pathogenesis, further supporting the idea of splicesosome mutations as initiators of myelodysplasia by induction of additional mutations in epigenetic modulators, transcription factors and signal transductors as described by Mian et al (2013).
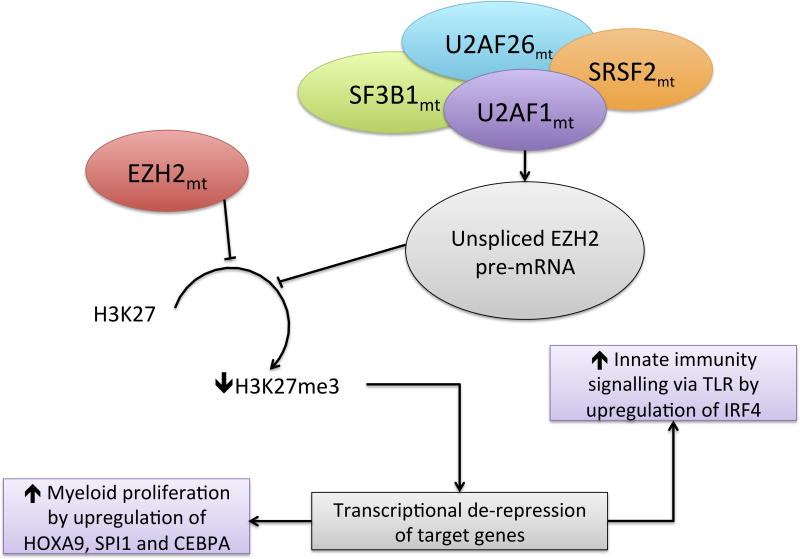
A recent publication (Khan et al 2013) details the effect of EZH2 mutations in H3k27 trimethylation and the expression of different known gene targets in 469 cases of different myeloid malignancies. Of special significance was the fact that cases with EZH2wt that harboured splicesosome mutations (including U2AF1, SRSF2 and U2AF26) were often related to a similar phenotype as those with EZH2 mutations. This was derived from a reduction of spliced compared to unspliced EZH2 pre-mRNA leading to decreased levels of the protein and, therefore, H3K27me3. Increased expression levels of IRF4 (IFN Regulatory Factor 4), HOXA9, SPI1 and CEBPA were found in these EZH2-deficient cases, suggesting there not only is an upregulation of genes that participate in haematopoiesis, stem-cell renewal and proliferation, but in innate immunity as well. This is represented in Figure 5. We can therefore hypothesize that these different hallmarks of MDS development (epigenetic regulation, splicing, proliferation deregulation and innate immunity) may well be connected in a more direct manner than could be originally anticipated. With the increasing complexity of MDS pathogenesis, further exploration and understanding of the extent of these molecular relationships is of paramount importance. Several innate immunity alterations in a variety of pathways are being discovered in MDS, including overexpression of TLR2 and TLR6 (Wei et al, 2013b), mutations in TLR2 (TLR2 F217S) (Wei et al, 2013b), upregulation of NF-κB (Wei et al, 2013a; Wei et al, 2013b), TNF signalling, PD-L1 (CD274)/ PD-L2 (PDCD1LG2)/ PD-1 (PDCD1)/CTLA4 (Yang et al, 2013) and others, with evidence of impact of these at an epigenetic level. If a stronger connection can be established between these microenvironment dependent factors and different epigenetic alterations, a new potential area of research and treatment development will arise.
Figure 5.
EZH2 downregulation effects and splicing mutations in MDS. This figure represents the connection between splicing mutations, EZH2 loss of function and deregulation of proliferataion and innate immunity signalling through IRF4. Adapted from Rogers et al (2013) and Khan et al (2013). mt, mutated.
Closing the gap with the Clinic: Therapeutic Implications
Several decades ago therapeutic options for patients with MDS were scarce. Fortunately, during recent years, research has enabled the development of several drugs with both proven and potential therapeutic activity in myelodysplasia. Although lower risk MDS are still mainly treated with growth factor support when needed, agents modulating epigenetic changes in the disease have become the standard of therapy in higher risk disease, with potential in lower risk MDS. DNA hypomethylating- and histone deacetylase-inhibiting agents have been largely studied in the last decade in patients with MDS and AML with promising results. Additional molecules directed at the epigenetic alterations of the disease are being developed to further enhance the therapeutic armamentarium of MDS and AML and are summarized in Table I.
Table I.
Approved and under-development drugs directed at epigenetic Regulation in MDS.
| Drug | Mechanism of Action | Development Stage | Clinical Outcomes |
|---|---|---|---|
| Azacytidine | DNMT1 inhibition | Approved by FDA Phase II Oral formulation trial Phase II Low-dose for low risk disease | Increased OS, TFS and ORR (including CR, CCyR, PR and HI) Reduced transfusion dependency |
| Decitabine | Approved by FDA Phase II Low-dose for low risk disease | Increased OS, PFS and ORR (including CR, CCyR, PR and HI) Reduced transfusion dependency | |
| SGI-110 | Phase I/II | 27% ORR in AML | |
| Vorinostat | HDAC inhibition | Phase II in combination with Azacytidine | Increased ORR and improved OS |
| Panobinostat | Phase I/II | ORR of 50% with median OS of 18 months | |
| Enzatiostat | Phase II in combination with Azacytidine | Increased ORR | |
| Pracinostat | Phase II in combination with Azacytidine | CCyR in patients with HMA failure |
DNMT1: DNA (cytosine-5-)-methyltransferase 1; FDA: US Food and Drug Administration; OS: Overall survival; TFS: transformation-free survival; ORR: Overall response rate; CR: complete response; PR: partial response; CCyR: Complete cytogenetic response; HI: Haematological improvement; AML, acute myeloid leukaemia; HMA, hypomethylating agent.
Hypomethylating agents (HMAs)
Azacytidine and decitabine are cytidine analogues that exert potent inhibition of DNA methylation both in vitro and in vivo by inducing a depletion of methyltransferase levels (Borthakur et al, 2008). Additionally, azacytidine also induces disruption of nucleic acid and protein metabolism by incorporation into RNA, and inhibits ribonucleotide reductase, an essential enzyme for DNA synthesis (Aimiuwu et al, 2012). However, the specific mechanism of action of these agents still remains controversial. Data suggests that DNMT1 depletion leading to hypomethylation may exert a transcriptional change, inducing cellular differentiation and subsequent TP53 independent apoptosis (Hollenbach et al, 2013). This may be the biological reasoning behind the responses observed to these therapies in patients with high-risk MDS with del(17p) and TP53 loss-of function mutations.
HMAs induce responses in patients with MDS and AML and reduce risk of transformation to AML compared to best supportive care (Fenaux et al, 2009), with azacytidine showing a slight advantage in survival compared with decitabine (Lee et al, 2013). Currently, both agents are approved for the treatment of MDS, and are considered the standard of care for patients with high risk MDS or AML not eligible for intensive chemotherapy or stem cell transplantation.
Some current studies are trying to elucidate if low doses of azacytidine or decitabine are clinically active and safe in low-risk disease patients with poor prognostic features.
Ultimate loss of response to therapy unfortunately remains a near constant in the history of the disease. Efficacy of one of these agents in the setting of failure to the other has been reported (Borthakur et al, 2008), however responses remain underwhelming with short OS (Jabbour et al, 2010). This has lead to increasing efforts in the development of additional therapies such as SGI-110, a new DNMT inhibitor with a longer half-life than decitabine due to decreased in vivo deamination. A phase I/II trial in patients with AML recently presented at the 2013 American Society of Hematology (ASH) annual meeting has shown promising results (Jabbour et al, 2013). Identification of the mechanisms behind failure to therapy still remains elusive, with increasing efforts having been made in order to determine the existence of possible predictive factors of response to therapy. Contradictory results have been obtained when analysing baseline methylation or reduction of methylation levels after therapy as predictors of response (Shen et al, 2010).
Efforts to identify molecular markers of response to therapy have also been made, with TET2 and DNMT3A mutations having been associated with better response to azacytidine, albeit with contradictory data regarding their impact on OS (Traina et al, 2014). Polycomb complex gene mutations have also been associated with longer survival after hypomethylating therapy (Kulasekararaj et al, 2010).
Despite these important advances, the identification of clear subgroups of patients at a potential risk of failure to HMAs remains a challenge.
Histone Deacetylase (HDAC) Inhibitors
Despite the revolution of HMA in MDS therapy, there is still much room for improvement in order to significantly reduce leukaemic transformation, increase OS and induce long-term haematological and cytogenetic responses in the majority of patients. To this end, development of additional therapeutic targets has been sought in recent decades. HDAC inhibitors represent the most promising and explored group of drugs in this context and have therefore been tested in MDS and AML both as single agents and in combination with HMA and other therapies, with promising results. Initial trials with valproic acid combined with azacytidine demonstrated a modest advantage in response rate compared to azacytidine alone (Garcia-Manero et al, 2006). Vorinostat has been extensively studied, with in vitro data suggesting its ability to promote cell cycle arrest and growth inhibition, and induce apoptosis and differentiation of bone marrow cells from patients with MDS and AML (Silva et al, 2013). Results from a Phase II trial in combination with azacytidine in patients with MDS have been recently presented at the 2013 ASH annual meeting, with very promising results (Verma et al, 2013).
Apart from vorinostat, other HDAC inhibitors have been developed. Entinostat showed increased rate of haematological responses when combined with azacytidine in patients with AML and MDS (Prebet et al, 2010). Pracinostat has shown extremely promising results in a preliminary Phase II study in combination with azacytidine in 9 patients with high-risk MDS (Quintas-Cardama et al, 2012). In this trial an overall response rate of 89% was observed, with 56% patients showing a complete cytogenetic response. Despite the small study population these encouraging results have led to further trials in different disease settings. Finally, Panobinostat is currently being evaluated in older patients with MDS and AML in combination with decitabine.
Future perspectives: Targeting EZH2, IDH and the splicing machinery
As has been described throughout this review, the progressive discovery of additional alterations on epigenetic modulators is opening the door to potential future therapies.
EZH2
Although EZH2 mutations in MDS are associated with loss of function and suggest a tumour suppressor function of the PRC2 component, there is solid biological data supporting the potential effect of EZH2 as an inducer of proliferation, stem-cell renewal and inhibitor of differentiation. Although in the early stages of development, several EZH2 inhibitors are currently being explored in lymphoma with promising preclinical data (Lund et al, 2014). 3-Deazaneplanocin, or DZNep, is a S-adenosylhomocysteine hydrolase inhibitor that seems to induce EZH2 depletion by degradation of PRC2 complex and upregulate MIR29, leading to decreased lymphoma growth. A more specific EZH2 inhibitor in development is EI1. This drug inhibits cell growth and induces apoptosis and differentiation in lymphoma cells by reduction of H3K27me3 levels. Further research is required in order to determine if a subset of patients with MDS in which EZH2 could be a potential pro-oncogenic player could be candidates to therapy with these new therapies.
IDH1/2
As has been previously stated, IDH mutations lead to an important metabolic and epigenetic deregulation. Recent studies in AML have elucidated the possible different mechanisms responsible for the metabolic chaos and epigenetic disruption. The presence of mutant IDH genes, especially in heterozygous cases, induces HOXA9 genes, promotes cell-cycle transition through CDKN2A and CDKN2B epigenetic silencing, and activates different signalling pathways, such as MAPK (Chaturvedi et al, 2013). This opens an area of potential therapies that could directly or indirectly target AML and, presumably, MDS cases harbouring IDH mutations. Targeting MAPK through drugs such as ARRY-614 (a dual p38/MAPK and Tie2 inhibitor) could be considered. Additionally, direct inhibition of mutant IDH is being explored in AML. Such is the case for HMS-101, a drug that has shown in vitro activity with reduction of 2-HG levels, induction of apoptosis and inhibition of cell division in mouse and human IDHmut AML cells (Chaturverdi et al, 2013). Evaluating the possible efficacy of this molecule in MDS could be an interesting future prospect with potential to impact the treatment of a subset of patients with both AML and MDS.
Splicesosome
Mutations in the splicesosome complex leading to alterations in normal splicing of important genes may be another potential target of therapy in MDS, however additional understanding of the molecular mechanisms leading to myelodysplasia is required in order to identify potential therapeutic strategies and to develop specific drugs that target this group of genes.
Concluding Remarks
Epigenetic regulation represents an essential biological process leading to cell fate and differentiation programmes that are paramount in normal tissue development and homeostasis. Aberrant modulation of the different epigenetic processes has been consistently demonstrated to be a primordial pathogenic event in MDS, and although increasing understanding has been attained during the last decades, there are still many unknown mechanisms leading to disease promotion and progression. New molecular techniques have allowed us to start subdividing MDS into different mutational patterns associated with distinct prognosis, response to therapy and phenotypes. Although available data is still lacking a thorough integration with the different disease processes and its correlation with the clinic, much of this epigenetic data may be the basis for developing new classifications and prognostic models of the disease that expand on the current World Health Organization classification and Revised International Prognostic Scoring System. The development of an array of epigenetic modulators, such as DNMT and HDAC inhibitors, has had an impact on the treatment of patients with MDS and may contribute to future improvements in OS, progression-free survival and a potential to change the course of the disease and the need of bone marrow transplantation. The field of epigenetic modulation certainly represents a fascinating area of research that may lead to important changes in the understanding of molecular oncology and molecular biology and revolutionize the field of clinical oncology.
Acknowledgments
Funding information: P30 CA016672
Footnotes
All authors were fully responsible for all content and involved in the different stages of the manuscript contributing equally to the development of this work.
Conflict of Interest
The authors declare no conflicts of interest.
References
- Abdel-Wahab O, Dey A. The ASXL-BAP1 axis: new factors in myelopoiesis, cancer and epigenetics. Leukemia. 2013;27:10–15. doi: 10.1038/leu.2012.288. [DOI] [PubMed] [Google Scholar]
- Abdel-Wahab O, Figueroa ME. Interpreting new molecular genetics in myelodysplastic syndromes. Hematology American Society of Hematology Education Program. 2012;2012:56–64. doi: 10.1182/asheducation-2012.1.56. [DOI] [PubMed] [Google Scholar]
- Abdel-Wahab O, Levine RL. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood. 2013;121:3563–3572. doi: 10.1182/blood-2013-01-451781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, Huberman K, Thomas S, Dolgalev I, Heguy A, Paietta E, Le Beau MM, Beran M, Tallman MS, Ebert BL, Kantarjian HM, Stone RM, Gilliland DG, Crispino JD, Levine RL. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, Pandey S, Patel JP, Chung YR, Koche R, Perna F, Zhao X, Taylor JE, Park CY, Carroll M, Melnick A, Nimer SD, Jaffe JD, Aifantis I, Bernstein BE, Levine RL. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamsson AE, Geron I, Gotlib J, Dao KH, Barroga CF, Newton IG, Giles FJ, Durocher J, Creusot RS, Karimi M, Jones C, Zehnder JL, Keating A, Negrin RS, Weissman IL, Jamieson CH. Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. Proceedings of the National Academy of Sciencies U S A. 2009;106:3925–3929. doi: 10.1073/pnas.0900189106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aimiuwu J, Wang H, Chen P, Xie Z, Wang J, Liu S, Klisovic R, Mims A, Blum W, Marcucci G, Chan KK. RNA-dependent inhibition of ribonucleotide reductase is a major pathway for 5-azacytidine activity in acute myeloid leukemia. Blood. 2012;119:5229–5238. doi: 10.1182/blood-2011-11-382226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, Kantarjian H, Raza A, Levine RL, Neuberg D, Ebert BL. Clinical effect of point mutations in myelodysplastic syndromes. New England Journal of Medicine. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Stevenson KE, Caughey BA, Abdel-Wahab O, Steensma DP, Galili N, Raza A, Kantarjian H, Levine RL, Neuberg D, Garcia-Manero G, Ebert BL. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. Journal of Clinical Oncology. 2012;30:3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Borthakur G, Ahdab SE, Ravandi F, Faderl S, Ferrajoli A, Newman B, Issa JP, Kantarjian H. Activity of decitabine in patients with myelodysplastic syndrome previously treated with azacitidine. Leukemia & Lymphoma. 2008;49:690–695. doi: 10.1080/10428190701882146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boultwood J, Perry J, Pellagatti A, Fernandez-Mercado M, Fernandez-Santamaria C, Calasanz MJ, Larrayoz MJ, Garcia-Delgado M, Giagounidis A, Malcovati L, Della Porta MG, Jadersten M, Killick S, Hellstrom-Lindberg E, Cazzola M, Wainscoat JS. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 2010;24:1062–1065. doi: 10.1038/leu.2010.20. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO Journal. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzola M, Della Porta MG, Malcovati L. The genetic basis of myelodysplasia and its clinical relevance. Blood. 2013;122:4021–4034. doi: 10.1182/blood-2013-09-381665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, Liang S, Lu Y, Darlington GJ, Meissner A, Issa JP, Godley LA, Li W, Goodell MA. Dnmt3a is essential for hematopoietic stem cell differentiation. Nature Genetics. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi A, Araujo Cruz MM, Jyotsana N, Sharma A, Yun H, Gorlich K, Wichmann M, Schwarzer A, Preller M, Thol F, Meyer J, Haemmerle R, Struys EA, Jansen EE, Modlich U, Li Z, Sly LM, Geffers R, Lindner R, Manstein DJ, Lehmann U, Krauter J, Ganser A, Heuser M. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood. 2013;122:2877–2887. doi: 10.1182/blood-2013-03-491571. [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nature Reviews Molecular Cell Biology. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, McDonough MA, King ON, Clifton IJ, Klose RJ, Claridge TD, Ratcliffe PJ, Schofield CJ, Kawamura A. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Reports. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm F, Kosmider O, Gelsi-Boyer V, Renneville A, Carbuccia N, Hidalgo-Curtis C, Della Valle V, Couronne L, Scourzic L, Chesnais V, Guerci-Bresler A, Slama B, Beyne-Rauzy O, Schmidt-Tanguy A, Stamatoullas-Bastard A, Dreyfus F, Prebet T, de Botton S, Vey N, Morgan MA, Cross NC, Preudhomme C, Birnbaum D, Bernard OA, Fontenay M, Groupe Francophone des M. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood. 2012;119:3211–3218. doi: 10.1182/blood-2011-12-400994. [DOI] [PubMed] [Google Scholar]
- Damm F, Itzykson R, Kosmider O, Droin N, Renneville A, Chesnais V, Gelsi-Boyer V, de Botton S, Vey N, Preudhomme C, Clavert A, Delabesse E, Park S, Birnbaum D, Fontenay M, Bernard OA, Solary E. SETBP1 mutations in 658 patients with myelodysplastic syndromes, chronic myelomonocytic leukemia and secondary acute myeloid leukemias. Leukemia. 2013;27:1401–1403. doi: 10.1038/leu.2013.35. [DOI] [PubMed] [Google Scholar]
- Davies C, Yip BH, Fernandez-Mercado M, Woll PS, Agirre X, Prosper F, Jacobsen SE, Wainscoat JS, Pellagatti A, Boultwood J. Silencing of ASXL1 impairs the granulomonocytic lineage potential of human CD34(+) progenitor cells. British Journal of Haematology. 2013;160:842–850. doi: 10.1111/bjh.12217. [DOI] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, Lecluse Y, Plo I, Dreyfus FJ, Marzac C, Casadevall N, Lacombe C, Romana SP, Dessen P, Soulier J, Viguie F, Fontenay M, Vainchenker W, Bernard OA. Mutation in TET2 in myeloid cancers. New England Journal of Medicine. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Deneberg S, Grovdal M, Karimi M, Jansson M, Nahi H, Corbacioglu A, Gaidzik V, Dohner K, Paul C, Ekstrom TJ, Hellstrom-Lindberg E, Lehmann S. Gene-specific and global methylation patterns predict outcome in patients with acute myeloid leukemia. Leukemia. 2010;24:932–941. doi: 10.1038/leu.2010.41. [DOI] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, Fuks F. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO Journal. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, Kirkpatrick DS, Pham VC, Lill JR, Bakalarski CE, Wu J, Phu L, Katavolos P, LaFave LM, Abdel-Wahab O, Modrusan Z, Seshagiri S, Dong K, Lin Z, Balazs M, Suriben R, Newton K, Hymowitz S, Garcia-Manero G, Martin F, Levine RL, Dixit VM. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, Hochhaus A, Drexler HG, Duncombe A, Cervantes F, Oscier D, Boultwood J, Grand FH, Cross NC. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature Genetics. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Fenaux P, Mufti GJ, Hellstrom-Lindberg E, Santini V, Finelli C, Giagounidis A, Schoch R, Gattermann N, Sanz G, List A, Gore SD, Seymour JF, Bennett JM, Byrd J, Backstrom J, Zimmerman L, McKenzie D, Beach C, Silverman LR, International Vidaza High-Risk MDS Survival Study Group Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncology. 2009;10:223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Mercado M, Yip BH, Pellagatti A, Davies C, Larrayoz MJ, Kondo T, Perez C, Killick S, McDonald EJ, Odero MD, Agirre X, Prosper F, Calasanz MJ, Wainscoat JS, Boultwood J. Mutation patterns of 16 genes in primary and secondary acute myeloid leukemia (AML) with normal cytogenetics. PLoS One. 2012;7:e42334. doi: 10.1371/journal.pone.0042334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Skrabanek L, Li Y, Jiemjit A, Fandy TE, Paietta E, Fernandez H, Tallman MS, Greally JM, Carraway H, Licht JD, Gore SD, Melnick A. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009;114:3448–3458. doi: 10.1182/blood-2009-01-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Lowenberg B, Licht JD, Godley LA, Delwel R, Valk PJ, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galm O, Wilop S, Luders C, Jost E, Gehbauer G, Herman JG, Osieka R. Clinical implications of aberrant DNA methylation patterns in acute myelogenous leukemia. Annals of Hematology. 2005;84(Suppl 1):39–46. doi: 10.1007/s00277-005-0005-0. [DOI] [PubMed] [Google Scholar]
- Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, Yang H, Rosner G, Verstovsek S, Rytting M, Wierda WG, Ravandi F, Koller C, Xiao L, Faderl S, Estrov Z, Cortes J, O'Brien S, Estey E, Bueso-Ramos C, Fiorentino J, Jabbour E, Issa JP. Phase 1/2 study of the combination of 5-aza-2'-deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108:3271–3279. doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelsi-Boyer V, Trouplin V, Adelaide J, Bonansea J, Cervera N, Carbuccia N, Lagarde A, Prebet T, Nezri M, Sainty D, Olschwang S, Xerri L, Chaffanet M, Mozziconacci MJ, Vey N, Birnbaum D. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. British Journal of Haematology. 2009;145:788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, Krysiak K, Harris CC, Koboldt DC, Larson DE, McLellan MD, Dooling DJ, Abbott RM, Fulton RS, Schmidt H, Kalicki-Veizer J, O'Laughlin M, Grillot M, Baty J, Heath S, Frater JL, Nasim T, Link DC, Tomasson MH, Westervelt P, DiPersio JF, Mardis ER, Ley TJ, Wilson RK, Walter MJ. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nature Genetics. 2012;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbach PW, Nguyen AN, Brady H, Williams M, Ning Y, Richard N, Krushel L, Aukerman SL, Heise C, MacBeth KJ. A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS One. 2010;5:e9001. doi: 10.1371/journal.pone.0009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D, Kitaura J, Togami K, Nishimura K, Enomoto Y, Uchida T, Kagiyama Y, Kawabata KC, Nakahara F, Izawa K, Oki T, Maehara A, Isobe M, Tsuchiya A, Harada Y, Harada H, Ochiya T, Aburatani H, Kimura H, Thol F, Heuser M, Levine RL, Abdel-Wahab O, Kitamura T. Myelodysplastic syndromes are induced by histone methylation-altering ASXL1 mutations. Journal of Clinical Investigation. 2013;123:4627–4640. doi: 10.1172/JCI70739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP. The myelodysplastic syndrome as a prototypical epigenetic disease. Blood. 2013;121:3811–3817. doi: 10.1182/blood-2013-02-451757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzykson R, Kosmider O, Cluzeau T, Mansat-De Mas V, Dreyfus F, Beyne-Rauzy O, Quesnel B, Vey N, Gelsi-Boyer V, Raynaud S, Preudhomme C, Ades L, Fenaux P, Fontenay M, Groupe Francophone des M. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia. 2011;25:1147–1152. doi: 10.1038/leu.2011.71. [DOI] [PubMed] [Google Scholar]
- Jabbour E, Garcia-Manero G, Batty N, Shan J, O'Brien S, Cortes J, Ravandi F, Issa JP, Kantarjian H. Outcome of patients with myelodysplastic syndrome after failure of decitabine therapy. Cancer. 2010;116:3830–3834. doi: 10.1002/cncr.25247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour E, Yee K, Kropf P, O'Connell C, Stock W, Tibes R, Rizzieri D, Walsh K, Griffiths EA, Roboz GJ, Savona M, Ervin T, Podoltsev NA, Pemmaraju N, Daver N, Garcia-Manero G, Borthakur G, Wierda WG, Ravandi F, Cortes JE, Brandwein JM, Odenike O, Feldman EJ, Chung W, Naim S, Choy G, Taverna P, Hao Y, Dimitrov G, Azab M, Issa J-P. First Clinical Results Of a Randomized Phase 2 Study Of SGI-110, a Novel Subcutaneous (SQ) Hypomethylating Agent (HMA), In Adult Patients With Acute Myeloid Leukemia (AML) Blood. 2013;122:497. [Google Scholar]
- Jerez A, Sugimoto Y, Makishima H, Verma A, Jankowska AM, Przychodzen B, Visconte V, Tiu RV, O'Keefe CL, Mohamedali AM, Kulasekararaj AG, Pellagatti A, McGraw K, Muramatsu H, Moliterno AR, Sekeres MA, McDevitt MA, Kojima S, List A, Boultwood J, Mufti GJ, Maciejewski JP. Loss of heterozygosity in 7q myeloid disorders: clinical associations and genomic pathogenesis. Blood. 2012;119:6109–6117. doi: 10.1182/blood-2011-12-397620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Dunbar A, Gondek LP, Mohan S, Rataul M, O'Keefe C, Sekeres M, Saunthararajah Y, Maciejewski JP. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2009;113:1315–1325. doi: 10.1182/blood-2008-06-163246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost E, Lin Q, Weidner CI, Wilop S, Hoffmann M, Walenda T, Schemionek M, Herrmann O, Zenke M, Brummendorf TH, Koschmieder S, Wagner W. Epimutations mimic genomic mutations of DNMT3A in acute myeloid leukemia. Leukemia. 2013 doi: 10.1038/leu.2013.362. Epub ahead of print 27 November 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, Weersing E, Dontje B, de Haan G. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107:2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Functional and cancer genomics of ASXL family members. British Journal of Cancer. 2013;109:299–306. doi: 10.1038/bjc.2013.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SN, Jankowska AM, Mahfouz R, Dunbar AJ, Sugimoto Y, Hosono N, Hu Z, Cheriyath V, Vatolin S, Przychodzen B, Reu FJ, Saunthararajah Y, O'Keefe C, Sekeres MA, List AF, Moliterno AR, McDevitt MA, Maciejewski JP, Makishima H. Multiple mechanisms deregulate EZH2 and histone H3 lysine 27 epigenetic changes in myeloid malignancies. Leukemia. 2013;27:1301–1309. doi: 10.1038/leu.2013.80. [DOI] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proceedings of the National Academy of Sciences U S A. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmider O, Gelsi-Boyer V, Slama L, Dreyfus F, Beyne-Rauzy O, Quesnel B, Hunault-Berger M, Slama B, Vey N, Lacombe C, Solary E, Birnbaum D, Bernard OA, Fontenay M. Mutations of IDH1 and IDH2 genes in early and accelerated phases of myelodysplastic syndromes and MDS/myeloproliferative neoplasms. Leukemia. 2010;24:1094–1096. doi: 10.1038/leu.2010.52. [DOI] [PubMed] [Google Scholar]
- Kulasekararaj AG, Mohamedali AM, Smith AE, Lea NC, Kizilors A, Abdallah A, Nasser EE, Mian SA, Yiu R, Gaken J, Pomplun S, Jiang J, Gaymes TJ, Pasipanodya P, Hayden J, Ireland RM, Lim Z, Ho AY, Marsh JCW, Mufti GJ. Polycomb Complex Group Gene Mutations and Their Prognostic Relevance In 5-Azacitidine Treated Myelodysplastic Syndrome Patients. Blood (ASH Annual Meeting Abstracts) 2010;116:125. [Google Scholar]
- Lee YG, Kim I, Yoon SS, Park S, Cheong JW, Min YH, Lee JO, Bang SM, Yi HG, Kim CS, Park Y, Kim BS, Mun YC, Seong CM, Park J, Lee JH, Kim SY, Lee HG, Kim YK, Kim HJ, Korean Society of Haematology AML/MDS working party Comparative analysis between azacitidine and decitabine for the treatment of myelodysplastic syndromes. British Journal of Haematology. 2013;161:339–347. doi: 10.1111/bjh.12256. [DOI] [PubMed] [Google Scholar]
- Li KK, Luo LF, Shen Y, Xu J, Chen Z, Chen SJ. DNA methyltransferases in hematologic malignancies. Seminars in Hematology. 2013;50:48–60. doi: 10.1053/j.seminhematol.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochemical Journal. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O'Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund K, Adams PD, Copland M. EZH2 in normal and malignant hematopoiesis. Leukemia. 2014;28:44–49. doi: 10.1038/leu.2013.288. [DOI] [PubMed] [Google Scholar]
- Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, Jerez A, Przychodzen B, Bupathi M, Guinta K, Afable MG, Sekeres MA, Padgett RA, Tiu RV, Maciejewski JP. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119:3203–3210. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcovati L, Papaemmanuil E, Bowen DT, Boultwood J, Della Porta MG, Pascutto C, Travaglino E, Groves MJ, Godfrey AL, Ambaglio I, Galli A, Da Via MC, Conte S, Tauro S, Keenan N, Hyslop A, Hinton J, Mudie LJ, Wainscoat JS, Futreal PA, Stratton MR, Campbell PJ, Hellstrom-Lindberg E, Cazzola M, Chronic Myeloid Disorders Working Group of the International Cancer Genome, C. & of the Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie, M Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118:6239–6246. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian SA, Smith AE, Kulasekararaj AG, Kizilors A, Mohamedali AM, Lea NC, Mitsopoulos K, Ford K, Nasser E, Seidl T, Mufti GJ. Spliceosome mutations exhibit specific associations with epigenetic modifiers and proto-oncogenes mutated in myelodysplastic syndrome. Haematologica. 2013;98:1058–1066. doi: 10.3324/haematol.2012.075325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamedali AM, Smith AE, Gaken J, Lea NC, Mian SA, Westwood NB, Strupp C, Gattermann N, Germing U, Mufti GJ. Novel TET2 mutations associated with UPD4q24 in myelodysplastic syndrome. Journal of Clinical Oncology. 2009;27:4002–4006. doi: 10.1200/JCO.2009.22.6985. [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD, Melnick A, Godley LA, Aifantis I, Levine RL. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends in Biochemical Sciences. 2010;35:323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Muller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Current Opinion in Genetics & Development. 2009;19:150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y, Sanada M, Miyagi S, Saraya A, Kamio A, Nagae G, Nakaseko C, Yokote K, Shimoda K, Koseki H, Suzuki Y, Sugano S, Aburatani H, Ogawa S, Iwama A. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. Journal of Experimental Medicine. 2013;210:2627–2639. doi: 10.1084/jem.20131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y, Sanada M, Kon A, Yoshida K, Shiraishi Y, Sato-Otsubo A, Mori H, Ishiyama K, Sakata-Yanagimoto M, Obara N, Nagasaki M, Miyawaki S, Chiba S, Miyano S, Yung SL, Koeffler HP, Ogawa S. Mutational Spectrum Analysis of Interesting Correlation and Interrelationship Between RNA Splicing Pathway and Commonly Targeted Genes in Myelodysplastic Syndrome. Blood (ASH Annual Meeting Abstracts) 2011;118:273. [Google Scholar]
- Negrotto S, Ng KP, Jankowska AM, Bodo J, Gopalan B, Guinta K, Mulloy JC, Hsi E, Maciejewski J, Saunthararajah Y. CpG methylation patterns and decitabine treatment response in acute myeloid leukemia cells and normal hematopoietic precursors. Leukemia. 2012;26:244–254. doi: 10.1038/leu.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F, Krepelova A, Incarnato D, Maldotti M, Parlato C, Galvagni F, Matarese F, Stunnenberg HG, Oliviero S. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell. 2013;155:121–134. doi: 10.1016/j.cell.2013.08.056. [DOI] [PubMed] [Google Scholar]
- Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nature Genetics. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- Orford K, Kharchenko P, Lai W, Dao MC, Worhunsky DJ, Ferro A, Janzen V, Park PJ, Scadden DT. Differential H3K4 methylation identifies developmentally poised hematopoietic genes. Developmental Cell. 2008;14:798–809. doi: 10.1016/j.devcel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, Godfrey AL, Rapado I, Cvejic A, Rance R, McGee C, Ellis P, Mudie LJ, Stephens PJ, McLaren S, Massie CE, Tarpey PS, Varela I, Nik-Zainal S, Davies HR, Shlien A, Jones D, Raine K, Hinton J, Butler AP, Teague JW, Baxter EJ, Score J, Galli A, Della Porta MG, Travaglino E, Groves M, Tauro S, Munshi NC, Anderson KC, El-Naggar A, Fischer A, Mustonen V, Warren AJ, Cross NC, Green AR, Futreal PA, Stratton MR, Campbell PJ, Chronic Myeloid Disorders Working Group of the International Cancer Genome, C Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. New England Journal of Medicine. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A, Shlien A, Groves MJ, Forbes SA, Raine K, Hinton J, Mudie LJ, McLaren S, Hardy C, Latimer C, Della Porta MG, O'Meara S, Ambaglio I, Galli A, Butler AP, Walldin G, Teague JW, Quek L, Sternberg A, Gambacorti-Passerini C, Cross NC, Green AR, Boultwood J, Vyas P, Hellstrom-Lindberg E, Bowen D, Cazzola M, Stratton MR, Campbell PJ, Chronic Myeloid Disorders Working Group of the International Cancer Genome, C Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik MM, Hanson CA, Hodnefield JM, Lasho TL, Finke CM, Knudson RA, Ketterling RP, Pardanani A, Tefferi A. Differential prognostic effect of IDH1 versus IDH2 mutations in myelodysplastic syndromes: a Mayo Clinic study of 277 patients. Leukemia. 2012;26:101–105. doi: 10.1038/leu.2011.298. [DOI] [PubMed] [Google Scholar]
- Prebet T, Gore SD, Sun Z, Greenberg PL, Juckett M, Malick L, Smith MR, Paietta E, Czader M, Gabrilove J, Erba HP, Tallman MS. Prolonged Administration of Azacitidine with or without Entinostat Increases Rate of Hematologic Normalization for Myelodysplastic Syndrome and Acute Myeloid Leukemia with Myelodysplasia-Related Changes: Results of the US Leukemia Intergroup Trial E1905. Blood (ASH Annual Meeting Abstracts) 2010;116:601. [Google Scholar]
- Qiu J, Lin L, Xu Y, Thol F, Monterroza DD, Dekelver R, Chen L, Heuser M, Fu X-D, Zhang D-E. SRSF2 Is Essential For Hematopoiesis and Its Mutations Dysregulate Alternative RNA Splicing In MDS. Blood. 2013;122:261. [Google Scholar]
- Quintas-Cardama A, Kantarjian HM, Ravandi F, Foudray C, Pemmaraju N, Kadia TM, Borthakur G, Daver NG, Faderl S, Jabbour E, Cortes JE, Garcia-Manero G. Very High Rates of Clinical and Cytogenetic Response with the Combination of the Histone Deacetylase Inhibitor Pracinostat (SB939) and 5-Azacitidine in High-Risk Myelodysplastic Syndrome. Blood (ASH Annual Meeting Abstracts) 2012;120:3821. [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, Godley L, Opolon P, Tilly H, Solary E, Duffourd Y, Dessen P, Merle-Beral H, Nguyen-Khac F, Fontenay M, Vainchenker W, Bastard C, Mercher T, Bernard OA. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Rocquain J, Carbuccia N, Trouplin V, Raynaud S, Murati A, Nezri M, Tadrist Z, Olschwang S, Vey N, Birnbaum D, Gelsi-Boyer V, Mozziconacci MJ. Combined mutations of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias. BMC Cancer. 2010;10:401. doi: 10.1186/1471-2407-10-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HJ, Tabarroki A, Zhang L, Parker Y, Hasrouni E, Mahfouz RZ, Ebrahem Q, Isono K, Koseki H, Advani AS, Saunthararajah Y, Sekeres MA, Lindner D, Barnard J, Tiu RV. Splicing Factor 3b Subunit 1 (SF3B1) Heterozygous Mice Manifest a Hematologic Phenotype Similar To Low Risk Myelodysplastic Syndromes With Ring Sideroblasts. Blood (ASH Annual Meeting Abstracts) 2013;122:259. [Google Scholar]
- Shen L, Kantarjian H, Guo Y, Lin E, Shan J, Huang X, Berry D, Ahmed S, Zhu W, Pierce S, Kondo Y, Oki Y, Jelinek J, Saba H, Estey E, Issa JP. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. Journal of Clinical Oncology. 2010;28:605–613. doi: 10.1200/JCO.2009.23.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva G, Cardoso BA, Belo H, Almeida AM. Vorinostat induces apoptosis and differentiation in myeloid malignancies: genetic and molecular mechanisms. PLoS One. 2013;8:e53766. doi: 10.1371/journal.pone.0053766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AE, Mohamedali AM, Kulasekararaj A, Lim Z, Gaken J, Lea NC, Przychodzen B, Mian SA, Nasser EE, Shooter C, Westwood NB, Strupp C, Gattermann N, Maciejewski JP, Germing U, Mufti GJ. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116:3923–3932. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- Solary E, Bernard OA, Tefferi A, Fuks F, Vainchenker W. The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia. 2014;28:485–496. doi: 10.1038/leu.2013.337. [DOI] [PubMed] [Google Scholar]
- Thol F, Weissinger EM, Krauter J, Wagner K, Damm F, Wichmann M, Gohring G, Schumann C, Bug G, Ottmann O, Hofmann WK, Schlegelberger B, Ganser A, Heuser M. IDH1 mutations in patients with myelodysplastic syndromes are associated with an unfavorable prognosis. Haematologica. 2010;95:1668–1674. doi: 10.3324/haematol.2010.025494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thol F, Friesen I, Damm F, Yun H, Weissinger EM, Krauter J, Wagner K, Chaturvedi A, Sharma A, Wichmann M, Gohring G, Schumann C, Bug G, Ottmann O, Hofmann WK, Schlegelberger B, Heuser M, Ganser A. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. Journal of Clinical Oncology. 2011a;29:2499–2506. doi: 10.1200/JCO.2010.33.4938. [DOI] [PubMed] [Google Scholar]
- Thol F, Winschel C, Ludeking A, Yun H, Friesen I, Damm F, Wagner K, Krauter J, Heuser M, Ganser A. Rare occurrence of DNMT3A mutations in myelodysplastic syndromes. Haematologica. 2011b;96:1870–1873. doi: 10.3324/haematol.2011.045559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thol F, Kade S, Schlarmann C, Loffeld P, Morgan M, Krauter J, Wlodarski MW, Kolking B, Wichmann M, Gorlich K, Gohring G, Bug G, Ottmann O, Niemeyer CM, Hofmann WK, Schlegelberger B, Ganser A, Heuser M. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119:3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- Toyota M, Kopecky KJ, Toyota MO, Jair KW, Willman CL, Issa JP. Methylation profiling in acute myeloid leukemia. Blood. 2001;97:2823–2829. doi: 10.1182/blood.v97.9.2823. [DOI] [PubMed] [Google Scholar]
- Traina F, Visconte V, Elson P, Tabarroki A, Jankowska AM, Hasrouni E, Sugimoto Y, Szpurka H, Makishima H, O'Keefe CL, Sekeres MA, Advani AS, Kalaycio M, Copelan EA, Saunthararajah Y, Olalla Saad ST, Maciejewski JP, Tiu RV. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia. 2014;28:78–87. doi: 10.1038/leu.2013.269. [DOI] [PubMed] [Google Scholar]
- Verma A, Odchimar-Reissig R, Feldman EJ, Navada SC, Demakos EP, Baer MR, Najfeld V, Sparano JA, Piekarz R. A Phase II Trial Of Epigenetic Modulators Vorinostat In Combination With Azacitidine (azaC) In Patients With The Myelodysplastic Syndrome (MDS): Initial Results Of Study 6898 Of The New York Cancer Consortium. Blood. 2013;122:386. [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Walter MJ, Ding L, Shen D, Shao J, Grillot M, McLellan M, Fulton R, Schmidt H, Kalicki-Veizer J, O'Laughlin M, Kandoth C, Baty J, Westervelt P, DiPersio JF, Mardis ER, Wilson RK, Ley TJ, Graubert TA. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li Z, He Y, Pan F, Chen S, Rhodes S, Nguyen L, Yuan J, Jiang L, Yang X, Weeks O, Liu Z, Zhou J, Ni H, Cai CL, Xu M, Yang FC. Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood. 2014;123:541–553. doi: 10.1182/blood-2013-05-500272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, Thompson CB. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chen R, Dimicoli S, Bueso-Ramos C, Neuberg D, Pierce S, Wang H, Yang H, Jia Y, Zheng H, Fang Z, Nguyen M, Ganan-Gomez I, Ebert B, Levine R, Kantarjian H, Garcia-Manero G. Global H3K4me3 genome mapping reveals alterations of innate immunity signaling and overexpression of JMJD3 in human myelodysplastic syndrome CD34+ cells. Leukemia. 2013a;27:2177–2186. doi: 10.1038/leu.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Dimicoli S, Bueso-Ramos C, Chen R, Yang H, Neuberg D, Pierce S, Jia Y, Zheng H, Wang H, Wang X, Nguyen M, Wang SA, Ebert B, Bejar R, Levine R, Abdel-Wahab O, Kleppe M, Ganan-Gomez I, Kantarjian H, Garcia-Manero G. Toll-like receptor alterations in myelodysplastic syndrome. Leukemia. 2013b;27:1832–1840. doi: 10.1038/leu.2013.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilop S, Fernandez AF, Jost E, Herman JG, Brummendorf TH, Esteller M, Galm O. Array-based DNA methylation profiling in acute myeloid leukaemia. British Journal of Haematology. 2011;155:65–72. doi: 10.1111/j.1365-2141.2011.08801.x. [DOI] [PubMed] [Google Scholar]
- Wu SJ, Kuo YY, Hou HA, Li LY, Tseng MH, Huang CF, Lee FY, Liu MC, Liu CW, Lin CT, Chen CY, Chou WC, Yao M, Huang SY, Ko BS, Tang JL, Tsay W, Tien HF. The clinical implication of SRSF2 mutation in patients with myelodysplastic syndrome and its stability during disease evolution. Blood. 2012;120:3106–3111. doi: 10.1182/blood-2012-02-412296. [DOI] [PubMed] [Google Scholar]
- Wu SJ, Tang JL, Lin CT, Kuo YY, Li LY, Tseng MH, Huang CF, Lai YJ, Lee FY, Liu MC, Liu CW, Hou HA, Chen CY, Chou WC, Yao M, Huang SY, Ko BS, Tsay W, Tien HF. Clinical implications of U2AF1 mutation in patients with myelodysplastic syndrome and its stability during disease progression. American Journal of Hematology. 2013;88:E277–282. doi: 10.1002/ajh.23541. [DOI] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Bueso-Ramos C, Dinardo C, Estecio MR, Davanlou M, Geng QR, Fang Z, Nguyen M, Pierce S, Wei Y, Parmar S, Cortes J, Kantarjian H, Garcia-Manero G. Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic syndromes is enhanced by treatment with hypomethylating agents. Leukemia. 2013 doi: 10.1038/leu.2013.355. Epub ahead of print 25 November 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, Nolte F, Hofmann WK, Miyawaki S, Sugano S, Haferlach C, Koeffler HP, Shih LY, Haferlach T, Chiba S, Nakauchi H, Miyano S, Ogawa S. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]