Abstract
Verbal memory is typically studied using immediate recall (IR) and delayed recall (DR) scores, although DR is dependent on IR capability. Separating these components may be useful for deciphering the genetic variation in age-related memory abilities. This study was conducted to (a) construct individual trajectories in IR and independent aspects of delayed recall, or residualized-DR (rDR), across older adulthood; and (b) identify genetic markers that contribute to four estimated phenotypes: IR and rDR levels and changes after age 60. A cognitively intact sample (N = 20,650 with 125,164 observations) was drawn from the U.S. Health and Retirement Study, a nationally representative study of adults aged 50 and older. Mixed effects regression models were constructed using repeated measures from data collected every two years (1996–2012) to estimate level at age 60 and change in memory post-60 in IR and rDR. Genome-wide association scans (GWAS) were conducted in the genotypic subsample (N = 7,486) using ~1.2 million single nucleotide polymorphisms (SNPs). One SNP (rs2075650) in TOMM40 associated with rDR level at the genome-wide level (p = 5.0x10-08), an effect that replicated in an independent sample from the English Longitudinal Study on Ageing (N = 6,898 with 41,328 observations). Meta-analysis of rDR level confirmed the association (p = 5.0x10-11) and identified two others in TOMM40 (rs71352238 p = 1.0x10-10; rs157582 p = 7.0x10-09), and one in APOE (rs769449 p = 3.1 x10-12). Meta-analysis of IR change identified associations with three of the same SNPs in TOMM40 (rs157582 p = 8.3x10-10; rs71352238 p = 1.9x10-09) and APOE (rs769449 p = 2.2x10-08). Conditional analyses indicate GWAS signals on rDR level were driven by APOE, whereas signals on IR change were driven by TOMM40. Additionally, we found that TOMM40 had effects independent of APOE e4 on both phenotypes. Findings from this first U.S. population-based GWAS study conducted on both age-related immediate and delayed verbal memory merit continued examination in other samples and additional measures of verbal memory.
Introduction
Memory impairment is the hallmark characteristic of mild cognitive impairment (MCI), Alzheimer disease (AD), and other types of dementia. It is estimated that currently (in 2016), ~5.2 million, or 11.1% of the 47 million persons aged 65 or older in the United States will develop AD in their lifetimes [1]. With an expanding population of older adults, these figures are expected to increase by 2050 to ~13.7 million, or 15.6% of the 87.7 million persons aged 65+ [1]. Verbal memory ability has particular relevance to aging research in that there is normative yet profound decline in older age [2–4]. The molecular basis of such decline is increasingly being disentangled as is the intricate relationship between decline and memory-related deficits [5–10]. Identifying the extent to which genetic variants underlie individual differences in specific aspects of memory trajectories are useful for understanding disease-related mechanisms that contribute to memory impairments as well as age-related memory changes.
Distinguishing immediate and delayed recall
Verbal memory is one aspect of episodic memory that requires verbal processing for encoding or retrieval of words, sentences, or prose [11]. It is typically assessed using both immediate recall and delayed recall tasks, which represent distinct cognitive processes [12, 13]. Immediate recall involves use of working memory, by which recently-presented information is rehearsed until recollection. Delayed recall requires intact immediate recall as it assesses retrieval of the information learned during the initial processing involved in immediate recall. This conceptual distinction between immediate and delayed recall is supported by functional brain imaging studies suggesting different localization of these processes [14–18]. Moreover, each component has distinct clinical relevance and implications when studying the underlying genetics.
Studies involving candidate genes have found learning and retrieval are partially distinct. For example, a study of community-dwelling older adults (ages 59 to 95) evaluated change in cognitive performance, comparing individuals with the Apolipoprotein E (APOE) e3/e3 and e3/e4 genotypes to assess the effect of the e4 risk allele. With regard to verbal memory, e3/e4 individuals exhibited significantly greater decline in delayed recall but not immediate recall [19]. In another study among healthy older adults (ages 60 to 100), the catechol-O-methyltransferase (COMT) enzyme was evaluated for effects on prefrontal cognitive functions. While there were no associations found between COMT genotypes and verbal memory overall, upon testing differences in the association by gender, men who were homozygous for the Val allele (vs. Val/Met and Met/Met) performed better on delayed recall only [20]. There were no differences in either recall phase by genotype that were detected for women. These studies provide preliminary indication that there is a distinct molecular basis for immediate and delayed recall.
From the clinical perspective, mild impairments in recall of verbal information over time are commonly experienced in aging and not considered pathological, whereas greater impairments particularly in delayed recall is a key early sign of Alzheimer disease and other dementias. Delayed recall performance has been shown to be a better predictor than immediate recall of AD severity, conversion to AD in the dementia process, or in differentiating normative changes from MCI and AD [21–23]. Despite the clinical importance, decline in delayed recall independent of decline in immediate memory has not been well-characterized. Generally, longitudinal studies report steady decline in overall verbal memory performance begins around 60 years of age [2, 24–26]. Distinguishing patterns in decline for immediate and delayed verbal recall is critical because of the potential differences in age-related trajectories as well as differences in implications for disease identification and prognosis.
Distinguishing memory level and change
A related issue for studying trajectories of memory over age is whether the factors underlying level of memory performance in early life and midlife differ from the processes that produce memory decline in later life. To use an analogy from physical growth, it seems unlikely that the (behavioral and genetic) factors contributing to loss of height in later life are the same as those contributing to stature in early adulthood. There has been increasing support of this age effect with episodic memory, and other cognitive aging phenotypes, as more studies have found that the effect of genes in older age (e.g., APOE, KIBRA, BDNF, DRD2, GRM3, COMT) tends to be stronger [27–30]. In parallel, evidence from longitudinal studies of aging twins indicates that inter-individual variation in level is highly influenced by genetic factors, but the genetic factors contributing to memory level are partially distinct from those underlying memory decline [28, 31–41]. Taken together, these suggest that distinguishing between level and change in verbal memory abilities would be useful for detecting underlying genetic relationships.
Study aims
This study addresses three important limitations common in genetic research on memory decline in aging: (a) confounding of measurement of delayed memory with immediate memory and (b) reliance on candidate genes. We are unaware of any prior study that has used a genome-wide approach to identify unique genetic associations with immediate and delayed verbal recall.
The first aim of this study was to characterize trajectories of four discrete components of verbal memory across older adulthood: level and decline in immediate recall, and level and change in independent aspects of delayed recall. This research employs data from a large population representative sample (N = 20,650) of older individuals from the U.S. Health and Retirement Study (HRS), with repeated measures (up to eight assessments per person) of verbal memory.
Our second aim was to identify genetic variants that associate with individual differences in the four components and to evaluate whether the associations are specific to each component. We employed genome wide association scans (GWAS) that had broad coverage of ~1.2 million SNP markers with the HRS genetic subsample (N = 7,486), followed by GWAS in an independent replication sample from the English Longitudinal Study of Ageing (ELSA; N = 6,898).
Material and methods
Participants
Discovery cohort. The Health and Retirement Study (HRS) is a nationally representative longitudinal study of U.S. households containing adults aged 50 years and older, with interviews conducted every two years, starting in 1992. Analyses of memory trajectories are based on data from a phenotypic sample comprising HRS participants who had completed at least two assessments of verbal recall between 1996 and 2012 and were at least 50 years of age during the study. Scores for some individuals were excluded if recorded prior to their turning age 50. We excluded scores of individuals who had a likely diagnosis of dementia, based on a question to the respondent on whether they had been diagnosed by a healthcare provider (with Alzheimer disease, dementia, senility, or serious memory-related problem) or the respondent requiring a proxy to complete their interview. Memory scores for these individuals were excluded for the wave in which the diagnosis was reported, the wave prior to reporting the diagnosis, and all subsequent waves when the individual was interviewed. These criteria yielded a sample of N = 20,650, with individuals assessed up to 8 times over the 16 years (125,164 observations total), with an average of 6.1 assessments per person. This phenotypic sample is 57.9% female, and the self-reported racial/ethnic composition is 82.2% Non-Hispanic White (NHW), 14.1% African-American/Black, and 3.8% Hispanic/Latino.
To reduce potential bias from population stratification bias and increase comparability with the ELSA replication sample, the GWAS was limited to individuals who identified as NHW (i.e., having primarily European ancestry). Other selection criteria were having verbal memory data on at least two occasions and providing a DNA sample, resulting in N = 7,486 (57.6% female) with 65,937 observations.
Replication cohort. The English Longitudinal Study of Ageing (ELSA) is a companion study of HRS, with a nationally representative cohort of individuals living in England aged 50 and older. The study began in 2002 with 11,391 participants and has since included interviews every two years, with measures collected for the purposes of harmonizing with HRS, including assessment of verbal memory (see Steptoe et al., 2013 [42]). Inclusion criteria for the present study were the same as for the HRS discovery sample. Memory scores and genetic information were available for a sample of N = 6,898 unrelated individuals (53.8% female) assessed up to 6 times (41,328 observations, collected 2002–2012).
Measures
Memory recall scores
In HRS and ELSA, a verbal memory task was administered as part of a series of cognitive tests, based on the Modified Telephone Interview for Cognitive Status (TICS-m [43]). This assessment was designed to screen for serious impairment and to aid in characterizing cognitive decline and mild impairment in large-scale community based studies of older adults where it is impractical to conduct more detailed assessments. This format is administered in many large-scale population-based studies conducted in the U.S. and internationally [42, 44–47].
In both HRS and ELSA, immediate recall (IR) and delayed recall (DR) were assessed as follows: Participants were alerted to pay attention and listen carefully, then asked to confirm readiness prior to being read a list of 10 nouns (administered by interviewer or taped recording) at the rate of one per second; then they were asked to recall the words immediately and again after a 5-minute delay [48] which was filled by asking non-cognitive survey items. The possible range of scores for each test is 0 to 10, for number of words recalled correctly, in any order. In HRS, the first assessment was typically in-person whereas most subsequent assessments were by telephone. For respondents age 80 or older, follow-ups were conducted face-to-face. Between 1996 and 2012, HRS used alternating lists across waves to reduce practice effects [44]. McArdle and colleagues studied construct equivalence of the cognitive measures in HRS and confirmed measurement invariance across waves of testing and identified only small effects from modality of testing [44, 49]. IR and DR scores were used as dependent variables as was a residual delayed recall (rDR) score. The rDR score represents the component of DR that is independent of IR (See pages 1–3 in S1 File).
Genetic marker data
For HRS, genotype data were accessed from the National Center for Biotechnology Information Genotypes and Phenotypes Database (dbGaP [50]). DNA samples from HRS participants were collected in two waves. In 2006, the first wave was collected from buccal swabs using the Qiagen Autopure method (Qiagen, Valencia, CA). In 2008, the second wave was collected using Oragene saliva kits and extraction method. Both waves were genotyped by the NIH Center for Inherited Disease Research (CIDR; Johns Hopkins University) using the HumanOmni2.5-4v1 array from Illumina (San Diego, CA). Raw data from both phases were clustered and called together. HRS followed standard quality control recommendations to exclude samples and markers that obtained questionable data, including CIDR technical filters [51], removing SNPs that were duplicates, had missing call rates ≥ 2%, > 4 discordant calls, > 1 Mendelian error, deviations from Hardy-Weinberg equilibrium (at P-value < 10−4 in European samples), and sex differences in allelic frequency ≥ 0.2). Further detail is provided in HRS documentation [52]. To reduce generating inflated P-values from inclusion of rare SNPs, we excluded SNPs with a MAF < 5%. Applying these criteria resulted in available data on 1,198,956 SNPs for the 9,532 NHW individuals, of whom 7,486 had phenotypic data and form our analytic sample.
For ELSA, genotype data were accessed from the European Genome-phenome Archive (EGA; http://www.elsa-project.ac.uk/). DNA was extracted from blood samples collected in 2004 and 2008, and samples were genotyped by University College London (UCL) Genomics Institute on the HumanOmni2.5–8 array from Illumina (San Diego, CA). Because the two studies were designed to be similar (with 2,368,902 markers in common across the studies), the same quality control procedures used with HRS were applied to the ELSA data, except that UCL excluded SNPs with missing call rates >5% (rather than 2%), yielding available data on 1,204,603 SNPs for 7,404 individuals with European ancestry, of whom 6,898 had phenotypic data also.
Statistical analysis
As detailed below, we followed five analytical steps: (1) Phenotype construction. Mixed effects regression models were used to evaluate the age-based trajectories and between-subject variation in trajectories of IR and rDR (analyzed separately). Four phenotypes were estimated for each individual: IR level, IR change, rDR level, and rDR change. (2) GWAS. Separate GWASs were run, first in the discovery cohort, to identify genetic loci associated with each of the four phenotypes. Replication GWASs were run in the independent cohort to evaluate replication. (3) SNP Comparison. We compared GWAS results across the phenotypes, and between the discovery and replication cohorts. (4) Meta-analysis. We conducted a meta-analysis of the discovery and replication cohorts to assess the robustness of the combined effects. (5) Functional genomic and pathway analysis. We evaluated the plausible biological pathways in which implicated genes play a role in memory outcomes.
Phenotype construction
To isolate the component of delayed recall that is independent of immediate recall, we formed a residualized-delayed recall (rDR) score. Mixed effects regression models were used to regress DR on IR using data from all participants and observations. rDR is the residual value (difference between the observed DR score and predicted DR score based on IR). IR and rDR scores were used as dependent variables in spline modeling to obtain four estimates for each participant to be used as phenotypes in genetic analyses: immediate recall level (IR-L), immediate recall change (IR-C), residual delayed recall level (rDR-L), and residual delayed recall change (rDR-C).
Splines are piecewise segments that fit linear trends, but each spline has its own intercept and slope to reflect the changing trends of the score over age. We conducted preliminary analyses to identify the number of splines needed to summarize the overall trajectories in the data and to identify the knot points (the dividing point between splines). The two-spline model was selected as the most parsimonious for characterizing overall trajectories for both IR and rDR. Based on prior literature [2, 24–26] and preliminary analyses, age 60 was selected as the spline knot point, i.e., allowing individuals to have different slopes before and after age 60.
All spline models were conducted as mixed effects analyses using PROC MIXED in SAS 9.4 [53] to estimate subject-specific intercepts and linear slopes, using empirical Bayes methods [54]. This technique allowed us to create comparable scores for participants, regardless of their age and duration of participation (as long as they had two observations after age 60) and is similar to the approach used to estimate level and decline of other cognitive measures in HRS [55, 56].
Genome-wide association scans
Separate GWASs, run as linear regressions under an additive model, were conducted for each of the four phenotypes (IR-L, IR-C, rDR-L, and rDR-C), adjusting for sex and population stratification (the first three principal components). As with any statistical analysis of association, if the correlation between dependent and independent variables differs for subpopulations, spurious associations may result in GWAS [57]. To reduce such type 1 error, we conducted GWAS adjusting for population structure as indicated by latent factors from principal components analysis (PCA) [58, 59]. Detailed descriptions of the processes employed for running PCA, including SNP selection, are provided by HRS [52], and follow methods outlined by Patterson and colleagues [60]. Two PCAs were run. The first PCA included 1,230 HapMap anchors from various ancestries, and were used to test against self-reported race and ethnic classifications. Several corrections to the dataset were made based on this analysis. The second PCA was run on the corrected dataset, on unrelated individuals and excluding HapMap anchors, to create eigenvectors to serve as covariates and adjust for population stratification in association tests. From the second PCA, the first two eigenvalues with the highest values accounted for less than 4.5% of the overall genetic variance, with additional components (3–8) increasing this minimally, by a total of ~1.0% [52]. We conducted correlational analysis for IR and DR level and change scores and found the first eight components had small and non-significant associations (all r’s<0.02) with each of the four phenotypes. As a check for non-linear associations, we examined graphs in which level of immediate recall was divided into quartiles and plotted against the first three principal components (PCs) (Supporting Information, Fig A-C in S1 Fig). The pattern did not suggest population substructures consistent with false associations in GWAS. Based on all this, we opted for a strategy that does not ignore population substructure, but also does not over-correct, and adjusted for the first three PCs in all analyses. This is a similar approach used in another recent GWAS conducted using HRS data (e.g., Ramanan et al., 2014). When coupling this approach of adjusting for PCs with all quality control procedures performed, excluding any related individuals and limiting the dataset for ancestral homogeneity, we reduce the likelihood of false associations resulting from population stratification [59–65]. All GWASs were completed using PLINK 1.9 [66, 67].
To estimate the fraction of phenotypic variation explained by common SNPs (with minor allele frequency ≥ 5%) for each phenotype, we implemented genomic-relationship-matrix restricted maximum likelihood (GREML) in unrelated individuals from all genotyped SNPs in the data set. We included only individuals falling within 1 SD of all self-identified non-Hispanic Whites for eigenvectors 1 and 2 in the principal components analysis of all unrelated participants. More information on the process for screening and selection of the sample by ancestral homogeneity and relatedness is provided in HRS documentation [52]. SNP heritability estimates were run adjusted for sex and three PCs. GREML was implemented in the software Genome-Wide Complex Trait Analysis (GCTA [68]) version 1.26.0.
SNP comparison
We evaluated SNP associations in the GWASs by p-value, using the genome-wide significance level of p ≤ 5.0x10-08 [69]. We also discuss additional findings at the “suggestive association” level of p ≤ 5.0x10-06 because we prefer not to miss potential new findings (Type II error) at the cost of an overly stringent Type I cutoff. To compare SNP associations from discovery and replication data sets we: 1) assessed the independent contribution of SNPs meeting genome-wide significance in the discovery scan; and 2) compared effect sizes and p-values to those estimated for these SNPs in the replication scans. For visualization of the SNP plots, we used R (CRAN; https://www.r-project.org) and LocusZoom v1.1 [70].
Meta-analysis
We conducted a meta-analysis of the discovery and replication cohorts using the inverse variance weighted model approach implemented in the METAL package (http://www.sph.umich.edu/csg/abecasis/Metal [71]).
Functional genomic and pathway analysis
Genes and flanking genes were identified using dbSNP and the “SNP and CNV Annotation Database” (SCAN) (http://www.scandb.org/newinterface/index.html). Gene functions were identified using the literature search feature of the “Gene Expression Analysis” (GEA) tool (http://chrisarm.com/gea/lsearch/). Gene network analysis was conducted using the Ingenuity Knowledge Base and Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA; http://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/). Additional interactions were added sourced from the Qinsight Biomedical Search Engine (Quertle LLC; Henderson, NV; https://www.quetzal-search.info/) and the BaseSpace Correlation Engine (Illumina, San Diego, CA; https://basespace.illumina.com/home/index), which summarizes evidence from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/).
Results
Sample characteristics and outcome variables
Table 1 provides selected characteristics for HRS participants in the full phenotypic sample (N = 20,650) and the NHW genotypic sample used for the GWAS (N = 7,486). The mean IR and DR scores are based on the first assessment occasion for each individual. In the phenotypic sample, the mean score (out of 10 words) was 5.7 for IR and 4.5 for DR. The full HRS sample and the NHW phenotypic sub-sample are similar in score distributions and gender ratio, but the genotypic sample was slightly younger and had memory scores that averaged 0.2 SD for both IR and DR. After adjusting for a linear effect of age, GWAS sample membership was not associated with IR or DR scores (ß = 0.02).
Table 1. Characteristics of HRS participants at the first assessment point, tested for immediate and delayed recall between 1996 and 2012, in the HRS full and Non-Hispanic White (NHW) genotypic samples.
| Full HRS Sample (N = 20,650) |
NHW Genotypic Sample (N = 7,486) |
|||
|---|---|---|---|---|
| Mean (SD) | % | Mean (SD) | % | |
| Age (range 50–103) | 64.4 (9.8) | 61.7 (8.2) | — | |
| Female | — | 57.9 | — | 57.9 |
| Male | — | 42.1 | — | 42.1 |
| Race/Ethnicity | ||||
| Non-Hispanic white | — | 82.2 | — | 100.0 |
| Hispanic/Latino | — | 14.1 | — | — |
| African-American | — | 3.8 | — | — |
| Immediate recall scorea | 5.7 (1.8) | — | 6.1 (1.6) | — |
| Delayed recall scorea | 4.5 (2.2) | — | 5.0 (2.0) | — |
a Immediate and delayed recall scores ranged from 0 to 10.
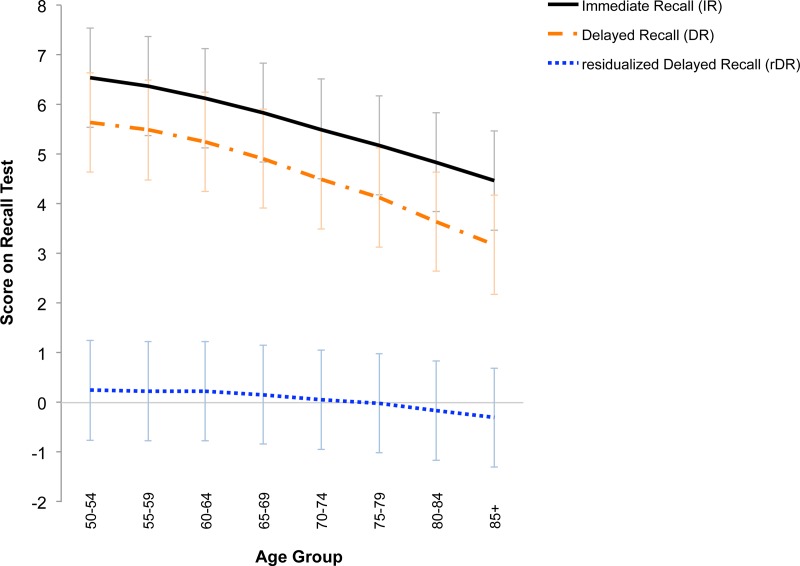
In the full phenotypic HRS sample, IR was strongly correlated with DR (r = .77), but (as expected) not with residualized delayed recall (rDR; r = -.01), reflecting our goal of isolating the immediate and delayed components of verbal recall. Fig 1 shows recall scores, including the overlap of IR and DR, as well as rDR, or the independent component of delayed scores after partialling out immediate scores.
Fig 1. Scores (as words recalled) for immediate, delayed, and residualized delayed recall among 20,650 participants in the HRS.
Aim 1 of the study was to characterize the longitudinal trajectories of the phenotypes: IR level (IR-L), IR change (IR-C), residual DR level (rDR-L) and residual DR change (rDR-C). To achieve this, we calculated and compared the estimated parameters from spline models for the four phenotypes for the HRS full phenotypic sample. The first slope component (s1) is a linear function of the scores obtained prior to age 60 and a second slope component (s2) is a linear function of scores obtained after age 60. There turned out to be negligible variation in slope prior to age 60 (i.e., memory performance was stable for the majority of participants; IR-C variance = 0.028, DR-C variance = 0.006, rDR-C = 0.004). Because there was more between-person variation after age 60 for IR and DR (IR-C variance = 0.094, DR-C variance = 0.113, rDR-C = 0.004), the key model parameters are subject-specific intercepts (random effects) for memory level at age 60 (i.e., representing IR-L or rDR-L) and subject-specific estimates of slope (change) after age 60, representing IR-C or rDR-C. However, variance in rDR-C post-60 was considerably smaller than for IR-C and DR-C.
As shown in Table 2, higher recall level negatively correlated with decline for both IR (r = -.62) and DR (r = -.64), indicating that individuals with initially better recall had more rapid decline, whereas those who began lower did not decline as much across subsequent measurements. In contrast, rDR level was only weakly associated with decline (r = -.18), suggesting that change on recall is independent of ability.
Table 2. Age-adjusted values for estimated immediate recall (IR), delayed recall (DR), and residual delayed recall (rDR) in HRS.
| Genotypic Sample (N = 7,486) | |||||||
|---|---|---|---|---|---|---|---|
| Mean | SD | 1 | 2 | 3 | 4 | 5 | |
| 1. IR level | 6.17 | 0.89 | |||||
| 2. IR change | -0.50 | 0.30 | -0.62 | ||||
| 3. DR level | 5.23 | 1.11 | 0.85 | -0.49 | |||
| 4. DR change | -0.67 | 0.36 | -0.55 | 0.76 | -0.64 | ||
| 5. rDR level | 0.22 | 0.34 | 0.15 | -0.08 | 0.62 | -0.31 | |
| 6. rDR change | -0.20 | 0.10 | -0.03 | -0.01 | -0.19 | 0.55 | -0.18 |
SD = standard deviation. Immediate and delayed recall scores represent the number of words recalled correctly, between 0 and 10. Correlations of .03 or greater had p-values < 0.01. rDR scores are IR-adjusted. IR and rDR level are estimated scores for age 60. IR and rDR change are estimated linear slopes per decade after age 60.
Evaluation of GWAS results
Quantile-quantile (QQ) plots for the HRS sample were constructed to show the ratio of observed to expected p-values for each SNP in the four GWASs, each adjusted for three PCs (see Figs A-D in S2 Fig). The majority of SNPs fell within the 95% confidence interval of the expectation; thus, it is reasonable to assume that there is little concern for inflation (type 1 error) of the observed p-values. The genomic inflation factor (lambda) for each scan was close to 1 (range λ’s = 1.001 to 1.010), and thus, not suggestive of problems with population stratification [72]. Notably, comparison scans using eight PCs showed nearly identical effect sizes and a similar pattern of p-values, and hence the three PCs controlled for in the analyses appear sufficient. Similarly, QQ plots for the ELSA sample did not indicate problems with population stratification (see Figs A-D in S3 Fig).
GWAS findings
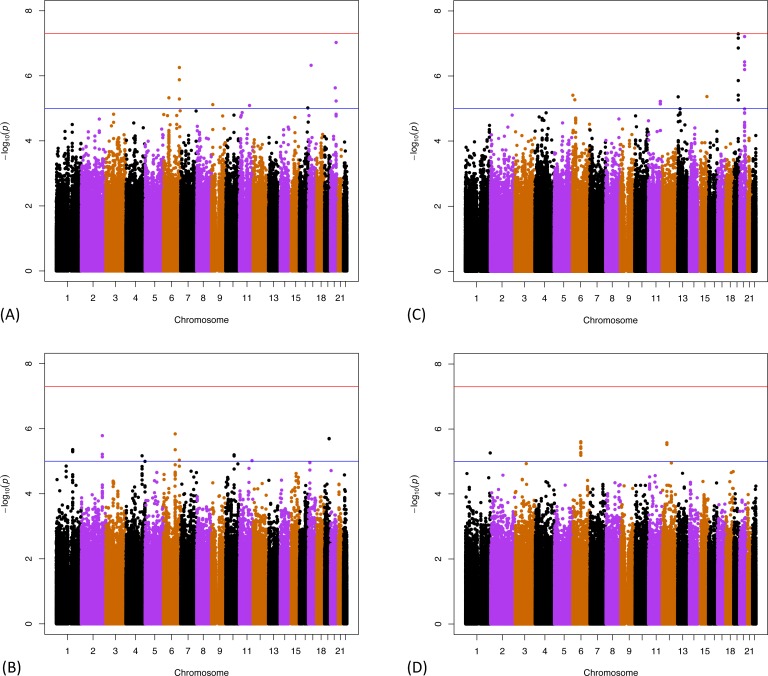
Fig 2 shows Manhattan plots for the discovery GWAS in HRS, for IR-L, IR-C, rDR-L, and rDR-C, respectively. The x-axis shows each of the 1.2M SNPs arranged in order of chromosomes and map location. The y-axis shows the log of the p-value for the association between the SNP and the phenotype. One SNP in the scan for rDR-L (Fig 2C) surpassed the genome-wide significance threshold (p ≤ 5.0x10-08), and several other SNPs in all four scans met our criterion for suggestive association (p ≤ 5.0x10-06). Corresponding Manhattan plots for the ELSA sample are provided in the Supporting Information (Figs A-D in S4 Fig).
Fig 2. P-values for all 1,198,956 SNP associations from GWAS on the HRS genetic sample.
(A) Level of Immediate Recall (IR-L); (B) Change in Immediate Recall (IR-C); (C) Level of Residualized Delayed Recall (rDR-L); (D) Change in Residualized Delayed Recall (rDR-C). For these figures, the upper (red) horizontal line demarcates the threshold of p = -log(5.0x10-08) and the lower (blue) horizontal line demarcates p = -log(1x10-05). SNPs are arranged by their chromosomal position (x-axis).
Overall, there were 29 SNPs that exceeded the p≤5.0x10-06 cutoff for a suggestive association with one or more phenotypes. These are listed in Table 3, ordered by chromosome, and chromosomal location, and presented with the associated phenotype, corresponding gene location, minor allele, effect size, and p-value from the HRS GWAS. Also shown are the effect sizes and p-values from the ELSA association tests. Additional findings found in the ELSA cohort only are provided in Supporting Information (Table C in S1 File).
Table 3. Strongest associated SNPs detected by GWASs in the HRS discovery sample and comparisons in the ELSA replication sample, for immediate recall level (IR-L) and change (IR-C) and residual delayed recall level (rDR-L) and change (rDR-C).
| Pheno. | Chr. | SNP | Position | Ref. allele | Minor Allele | Gene(s) | HRS | ELSA | ||
|---|---|---|---|---|---|---|---|---|---|---|
| (freq.) | Beta | P | Beta | P | ||||||
| IR-C | 1 | rs11804244 | 163212633 | G | G (0.150) | LOC100113374 or NUF2 | -0.029 | 4.E-06 | 0.004 | 1.E-01 |
| IR-C | 1 | rs6704030 | 163218278 | G | C (0.149) | LOC100113374 or NUF2 | -0.029 | 5.E-06 | 0.004 | 2.E-01 |
| IR-C | 2 | rs12463410 | 207715608 | A | T (0.021) | FASTKD2 a or CPO a | -0.054 | 2.E-06 | 0.001 | 8.E-01 |
| rDR-L | 6 | rs9504162 | 4599442 | A | T (0.187) | LOC100129052 or KU-MEL-3 | -0.028 | 4.E-06 | 0.002 | 8.E-02 |
| IR-L | 6 | rs7766458 | 54140128 | C | C (0.335) | C6orf142 or TINAG | -0.095 | 5.E-06 | 0.003 | 9.E-01 |
| rDR-C | 6 | rs12195716 | 79535412 | G | T (0.497) | LOC100128162 or IRAK1BP1 | -0.005 | 4.E-06 | 0.000 | 9.E-01 |
| rDR-C | 6 | rs2174743 | 79591805 | A | G (0.488) | IRAK1BP1 | -0.005 | 4.E-06 | 0.000 | 9.E-01 |
| rDR-C | 6 | rs7756858 | 79619968 | G | A (0.460) | IRAK1BP1 or PHIP a | -0.005 | 2.E-06 | 0.000 | 9.E-01 |
| rDR-C | 6 | rs1572585 | 79690576 | A | C (0.494) | PHIP a | -0.005 | 3.E-06 | 0.000 | 1.E+00 |
| IR-C | 6 | rs9372456 | 116592956 | G | G (0.384) | RPS5P1 or TSPYL1 | 0.028 | 1.E-06 | -0.003 | 4.E-01 |
| IR-C | 6 | rs10581090 | 116658967 | A | C (0.482) | DSE a | 0.026 | 4.E-06 | -0.004 | 2.E-01 |
| IR-L | 6 | rs147391086 | 157984507 | A | A (0.015) | — | -0.153 | 1.E-06 | 0.038 | 2.E-01 |
| IR-L | 6 | rs150510877 | 157994175 | A | T (0.019) | — | -0.159 | 6.E-07 | 0.032 | 2.E-01 |
| rDR-C | 12 | rs10880835 | 46079245 | C | G (0.498) | LOC100131290 or LOC400027 | 0.005 | 3.E-06 | na | na |
| rDR-C | 12 | rs35162469 | 46081065 | A | A (0.485) | LOC100131290 or LOC400027 | 0.005 | 3.E-06 | 0.000 | 8.E-01 |
| rDR-L | 13 | rs9510784 | 24188270 | A | C (0.450) | TNFRSF19 a | -0.025 | 4.E-06 | 0.005 | 5.E-01 |
| rDR-L | 15 | rs4076414 | 86441452 | C | G (0.418) | KLHL25 or AGBL1 | 0.025 | 4.E-06 | 0.013 | 9.E-02 |
| IR-L | 17 | rs8072199 | 26116848 | A | T (0.227) | NOS2A | 0.074 | 5.E-07 | -0.002 | 9.E-01 |
| IR-C | 19 | rs283815 | 45390333 | G | G (0.304) | PVRL2 a | -0.032 | 2.E-06 | -0.012 | 2.E-04 |
| rDR-L | -0.031 | 4.E-06 | -0.027 | 3.E-03 | ||||||
| rDR-L | 19 | rs71352238 | 45394336 | G | C (0.085) | — | -0.043 | 7.E-08 | -0.039 | 2.E-04 |
| rDR-L | 19 | rs2075650 | 45395619 | G | G (0.119) | TOMM40 a | -0.043b | 5.E-08 | -0.041 | 1.E-04 |
| IR-C | 19 | rs157582 | 45396219 | A | T (0.294) | TOMM40 a | -0.032 | 2.E-06 | -0.013 | 9.E-05 |
| rDR-L | -0.033 | 1.E-06 | -0.030 | 9.E-04 | ||||||
| rDR-L | 19 | rs769449 | 45410002 | A | A (0.065) | APOE a | -0.045 | 1.E-07 | -0.053 | 5.E-06 |
| IR-L | 20 | rs11906369 | 46829233 | A | A (0.146) | LOC100130372 or LOC284749 | -0.123 | 2.E-06 | na | na |
| rDR-L | 20 | rs6512614 | 48792663 | C | C (0.483) | TMEM189 a or CEBPB a | -0.028 | 5.E-07 | -0.005 | 5.E-01 |
| rDR-L | 20 | rs6012871 | 48792992 | G | C (0.486) | TMEM189 a or CEBPB a | -0.028 | 6.E-07 | -0.006 | 4.E-01 |
| rDR-L | 20 | rs6125931 | 48795413 | G | G (0.484) | TMEM189 a or CEBPB a | -0.028 | 4.E-07 | -0.004 | 6.E-01 |
| rDR-L | 20 | rs6125934 | 48803937 | A | T (0.396) | TMEM189 a or CEBPB a | -0.030 | 6.E-08 | -0.004 | 6.E-01 |
| IR-L | 20 | rs6025261 | 55503259 | A | A (0.286) | PTMAP6 or LOC728902 | 0.079 | 9.E-08 | 0.008 | 5.E-01 |
Genome build GRCh37. Pheno = phenotype; Chr = chromosome; Ref. allele = reference allele; freq = frequency.—Indicates that a gene name in which the SNP resides has not been identified. na = not available because when quality control filters were applied to the genotyped data, some SNPs were excluded from the analyses with ELSA. SNPs in the table are those with associations of p≤5x10-06 in the discovery cohort, HRS. When two gene names are provided, they represent the closest left and right flanking genes, respectively.
a Indicates there are prior reported associations with the gene and memory phenotypes (source: PubMed).
b Indicates the SNP association surpassed the genome-wide significance threshold of 5E-08 in the HRS discovery cohort and replicated in the independent cohort at p < .05.
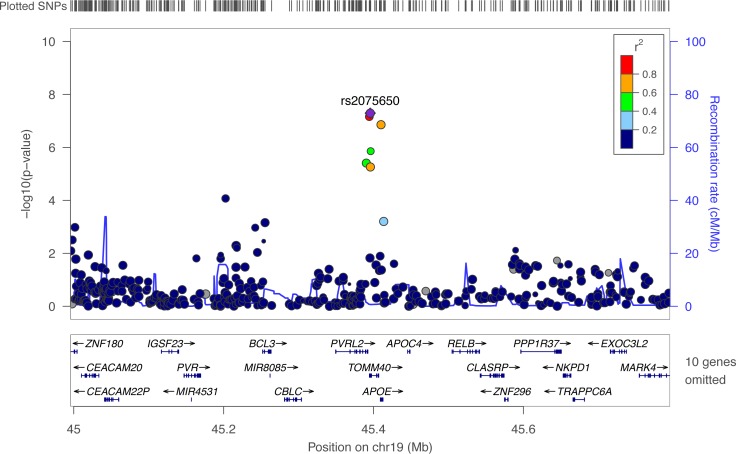
Of these 29 SNPs, the strongest associations across the scans were found for rDR-L, with rs2075650 (b = -0.043, p = 5.0x10-08) and rs71352238 (b = -0.043, p = 7.0x10-08) on chromosome 19 (Fig 3C). These two SNPs passed replication criteria in the ELSA cohort, with effect sizes similar to those found in HRS (rs2075650: b = -0.041, p = 1.0x10-04 and rs71352238: b = -0.039, p = 2.0x10-04) and the. Two SNPs on chromosome 19 in the PVRL2 (rs283815, b’s = -.03, p’s<4.0x10-06) and TOMM40 (rs157582, b’s = -.03, p’s<2.0x10-06) genes had suggestive association with both IR-C and rDR-L. Effect sizes for these two SNPs replicated in ELSA for rDR-L (b’s = -.03, p’s<3.0x10-03), but not IR-C (b’s = -.01, p’s<2.0x10-04). Interpretation of the size of genetic effects is provided in the Supporting Information (page 4 of S3 File).
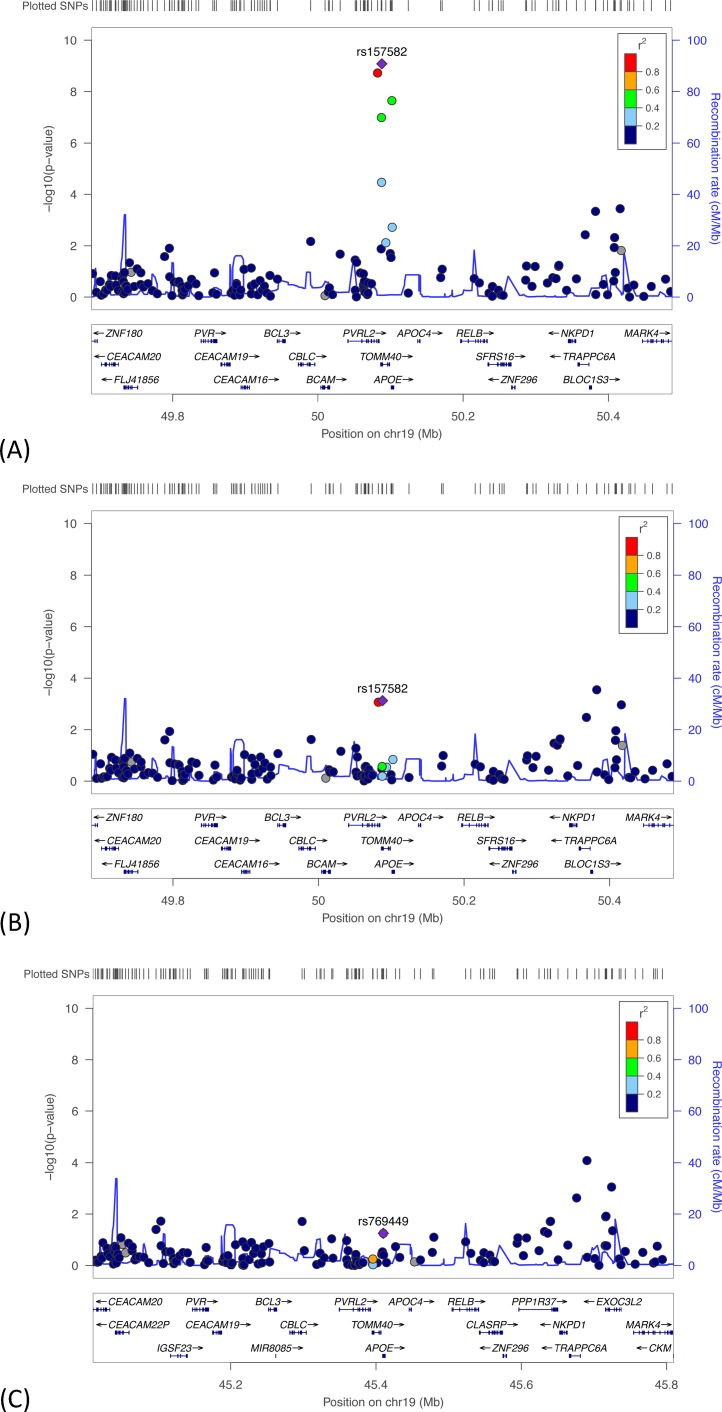
Fig 3. Regional plot showing the results for the association between rDR-L in HRS, with the top SNP in TOMM40 (rs2075650) identified.
The y-axis shows -log10 P-values; x-axis shows position of genes on chromosome 19. The diamond (purple) represents the top genome-wide significant SNP. The circles represent each genotyped SNP in the region 400-kb in both directions from rs2075650; the circle color indicates pairwise linkage disequilibrium (LD) in relation to the top SNP (calculated from hg19/1000 Genomes Nov 2014 EUR). The solid (blue) line indicates the recombination rate.
One SNP on chromosome 20, rs6125934 (b = -0.030, p = 6.0x10-08) approached the genome-wide cutoff for an association with rDR-L in HRS but that effect size did not replicate in ELSA (b = 0.004, p = 6.0x10-01). Similarly, a SNP on chromosome 20, rs6025261 (b = 0.079, p = 9.0x10-08) exceeded the suggestive level for an association with IR-L, but the effects did not replicate in ELSA (b = 0.008, p = 5.0x10-01). Several other SNPs met the criterion for suggestive association with IR-C (7 SNPs) or rDR-C (6 SNPs), but effects did not replicate in ELSA.
For the results presented in Table 3, SNPs were not pruned for linkage disequilibrium (LD, an index of the correlation between loci) because it is unknown which of the available SNPs may be the active variant for a specific gene versus serving as a marker for another variant. Fig 3 plots the surrounding SNPs and provides the pairwise LD values to the SNP (rs2075650) in the TOMM40 region associated at the genome-wide significance level with rDR-L. As evident from the plot, the genotyping array provided enough coverage of SNPs in the region to detect three nearby SNPs in LD with rs2075650 at a level of r2 ≥ 0.6.
SNP heritability (SNP h2) in the HRS cohort was estimated for each phenotype based on the combined genetic effect across all SNPs included in the GWAS, adjusted for sex and three PCs. Estimates were significantly greater than 0 for IR-L (h2 = 22.4%), IR-C (h2 = 12.0%) and DR-L (h2 = 13.4%), but not for rDR-L (h2 = 3.4%), rDR-C (h2 = 6.7%) or DR-C (h2 = 8.6%).
Meta-analysis results
Meta-analyses of the HRS and ELSA cohorts identified four SNPs that surpassed the genome-wide significance cut-off for rDR-L, three of which were also significantly associated with IR-C, and none of which significantly associated with rDR-C or IR-L (Table 4). All SNPs were on chromosome 19, with the TOMM40 SNPs within 6000 base pairs of each other, and the APOE SNP 19,700 base pairs away. Multiple SNPs from these meta-analyses reached the suggestive threshold of 5x10-06 and are reported in Supporting Information (Tables A–D in S2 File). QQ and Manhattan Plots from meta-analyses for the four phenotypes are shown in Supporting Information, S5 (Figs A-D in S5 Fig) and S6 Fig (Figs A-D in S6 Fig), respectively. For characterizing the effects of the top SNPs in the TOMM40 and APOE regions (reported below), we focus on SNPs that surpassed genome-wide significance and use the single top SNP to represent each of the TOMM40 and APOE effects.
Table 4. Meta-analyses results for SNPs reaching genome-wide significance for all four phenotypes: Immediate recall level (IR-L), immediate recall change (IR-C), residualized delayed recall level (rDR-L), and residualized delayed recall change (rDR-C).
| Meta-analysis p-value | ||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | BP | Effect allele | Gene | Direction | IR-L | IR-C | rDR-L | rDR-C |
| rs283815 | 45390333 | G | TOMM40 | ++ | 5.1E-02 | 1.9E-09 | 8.6E-07 | 9.0E-03 |
| rs71352238 | 45394336 | G | TOMM40 | — | 4.2E-03 | 1.2E-06 | 1.0E-10 | 2.2E-03 |
| rs2075650 | 45395619 | G | TOMM40 | — | 4.7E-03 | 1.0E-07 | 5.0E-11 | 2.5E-03 |
| rs157582 | 45396219 | A | TOMM40 | — | 3.3E-02 | 8.3E-10 | 7.0E-09 | 5.6E-03 |
| rs769449 | 45410002 | A | APOE | — | 1.9E-04 | 2.2E-08 | 3.0E-12 | 1.9E-02 |
Evaluation of the meta-analysis SNPs associated with rDR-L and IR-C
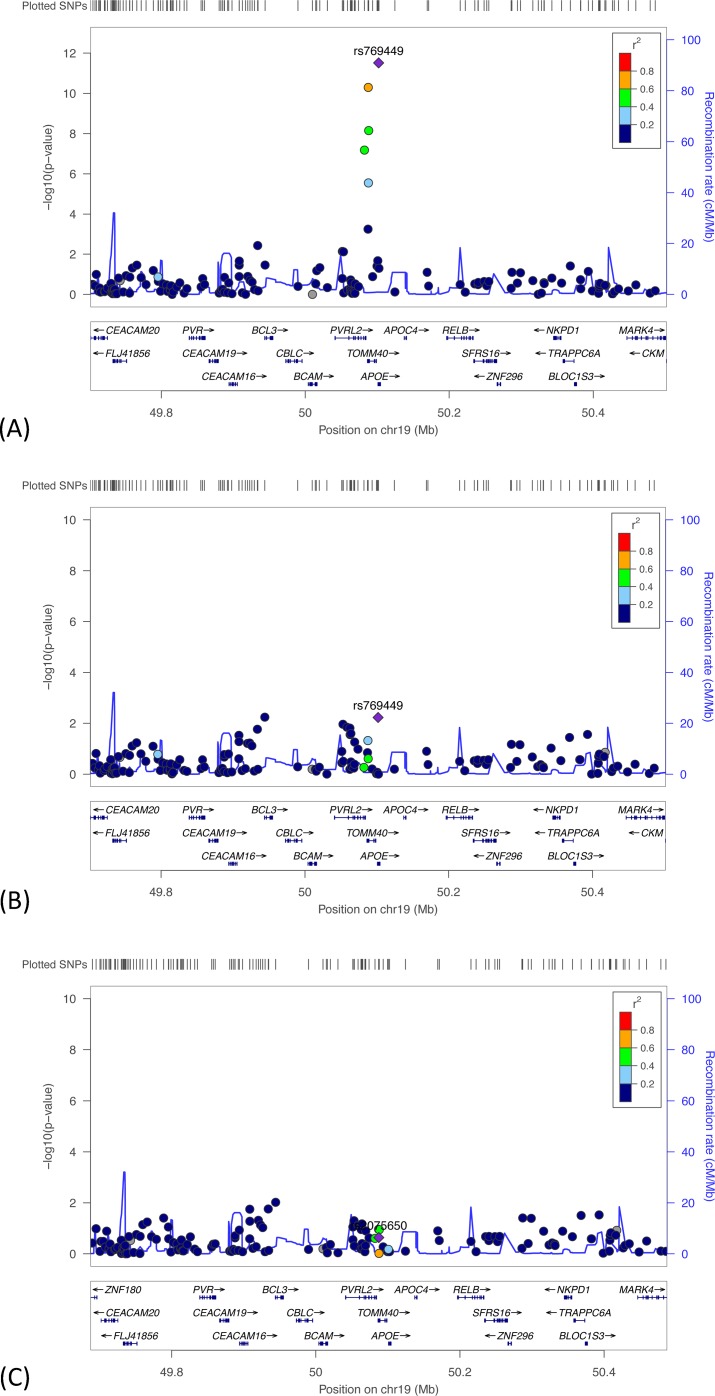
Evaluation of meta-analysis results for rDR-L. In the meta-analysis of rDR-L, the top two SNPs in the APOE (rs769449) and TOMM40 (rs2075650) are in LD with at r2 ≥ 0.6. These are represented by the top diamond and circle in Fig 4A. Because of evidence from the meta-analysis and prior findings that suggest that the effect of TOMM40 on nonpathological cognitive decline is not independent of APOE effects [6], we performed a series of analyses to test this. First, we performed a meta-analysis of the GWASs that were run conditioned on the top genome-wide significant SNP in TOMM40, rs2075650 (Fig 4B), and found the effect of the top APOE SNP diminished (rs769449 p = 5.9x10-03). Second, we performed a meta-analysis of the GWASs that were run conditioned on the top genome-wide significant SNP in APOE, rs769449 (Fig 4C), and found the effect of the top TOMM40 SNP diminished (rs2075650 p = 0.23) to a non-significant effect.
Fig 4. Regional plots showing meta-analyses results for rDR-L.
(A) Results with the top associated SNP in the APOE region labeled. Two conditional meta-analyses were performed for rDR-L, (B) estimating the association with the APOE SNP shown (rs769449), conditioning on the top TOMM40 SNP (rs2075650); and (C) estimating the association with the TOMM40 SNP shown (rs2075650), conditioning on the top APOE SNP (rs769449). The y-axis shows -log10 P-values; x-axis shows position of genes on chromosome 19 with SNPs 400-kb in both directions of the SNP of interest. The diamond represents the top SNP of interest. The circles represent each genotyped SNP in this region; the circle color indicates pairwise linkage disequilibrium (LD) in relation to the top SNP (calculated from hg19/1000 Genomes Nov 2014 EUR). The solid (blue) line indicates the recombination rate.
Third, we analyzed rDR-L, conditioned on APOE e4. SNP rs769449 resides in APOE, and is in high pairwise LD with the SNPs that determines the APOE e4 isoform, rs429358 (r2 = 0.779), as calculated using the Phase 3 of the 1000 Genomes Project and CEU reference panel [73]. The presence of APOE e4, and thus APOE genotype, was not determined from directly genotyped SNPs in HRS or ELSA, and thus it was not included in the primary analyses of this study. However, because the two non-synonymous SNPs that characterize the isoforms (rs429358 and rs7412) were inferred through imputation with estimated certainties ≥.997 in HRS [74], and have demonstrated high agreement with directly genotyped SNPs in other studies [75, 76], we were able to conduct additional evaluation of our finding using the isoforms in the HRS cohort. Details of the imputation process are provided by HRS [77, 78] and summarized in the Supporting Information (see page 1, in S3 File). When performing linear regression for the effects of TOMM40 rs2075650 on rDR-L, with APOE e4 in the model, and covariates (sex, 3 PCs), we found that the effect of TOMM40 remained (b = -0.0259, SE = 0. 0108, p = .017) with effect of e4 also significant (b = -0.0295, SE = 0.0124, p = .017).
Fourth, we tested the effects of TOMM40 on rDR-L in other APOE genotypes to examine effects beyond e4. Results of each model predicting rDR-L are provided in the Supporting Information (Table A in S3 File). Most notably, there was an effect of the rs2075650 G allele on rDR-L within e3/e3 individuals (b = -0.0390, SE = 0.0180, p = .031).
Evaluation of meta-analysis results for IR-C. From the meta-analysis of IR-C, the top two identified SNPs were in TOMM40 (rs157582) and APOE (rs769449), and in LD at r2 ≥ 0.8. Fig 5A shows the results of this meta-analysis, with the top associated SNP labeled. We examined whether the effects of the top two SNPs on this phenotype were independent by running a series of analyses. First, we performed a meta-analysis of the GWASs, conditioned on the top genome-wide significant APOE SNP (rs769449), to examine the change in effect of TOMM40 SNP rs157582 (Fig 5B). Compared to the joint analysis (Fig 5A), the conditional analysis found the association between IR-C and rs157582 diminished (p = 7.4x10-04), as did the effect of another TOMM40 SNP (rs71352238 p = 8.5x10-04). Second, we performed a meta-analysis of the GWASs, conditioned on the top genome-wide significant TOMM40 SNP (rs157582), to examine the change in effect of APOE SNP rs769449 (Fig 5C). The association between IR-C and APOE was no longer significant (rs769449, p = 0.06).
Fig 5. Regional plots showing results of the meta-analyses of IR-C.
(A) Meta-analysis results with the top SNP in TOMM40 (rs157582) shown; (B) results when estimating the association with the TOMM40 SNP shown (rs157582), conditioning on the top APOE SNP (rs769449); and (C) results when estimating the association with the APOE SNP (rs769449), conditioning on the top TOMM40 SNP (rs157582). The y-axis shows -log10 P-values; x-axis shows position of genes on chromosome 19 with SNPs 400-kb in both directions of the SNP of interest. The diamond represents the top SNP of interest. The circles represent each genotyped SNP in this region; the circle color indicates pairwise linkage disequilibrium (LD) in relation to the top SNP (calculated from hg19/1000 Genomes Nov 2014 EUR). The solid (blue) line indicates the recombination rate.
Third, we constructed linear regression models using the HRS cohort, predicting IR-C from the e4 allele, adjusted for covariates, and found a significant but small effect of e4 (b = -0.032, SE = 0.009, p = .001). Next, in a model predicting IR-C from both the TOMM40 SNP, rs157582 (b = -0.031, SE = 0.013, p < .0001) and APOE e4 (b = -0.008, SE = 0.012, p = .53), the effect of TOMM40 remained while the effect of APOE was reduced and no longer significant.
Lastly for IR-C, we tested the effects of the TOMM40 rs157582 A (risk) allele within each of the APOE genotypes. The models showed that there were significant effects of the rs157582 A allele on IR-C within the e3/e3 (b = -0.0310, SE = 0.016, p = .047) and e2/e3 strata (b = -0.0477, SE = 0.022, p = .032). Results of each model predicting IR-C are provided in the Supporting Information (Table B in S3 File).
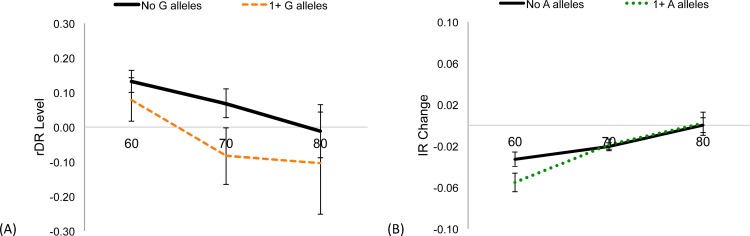
SNP effects on rDR-L and IR-C across age decades
Because our phenotypes reflected age-related trajectories, and due to evidence that genes have stronger effects on cognitive performance in older ages [27, 29], we evaluated whether the effect of TOMM40 on rDR-L and IR-C independent of APOE e4 differs at older age decades. Using the HRS cohort and mixed effects modeling, additional splines were fit with the knot point at age 70 and age 80, adjusted for APOE e4 carrier status, sex, and 3 PCs. When plotting the age-related rDR trajectories were stratified by presence of the rs2075650 ‘G’ allele, we find that in older ages, there were larger differences in the effects of the TOMM40 allele that were independent of APOE e4 (Fig 6A). In contrast, for IR change, the effect of the TOMM40 rs157582 ‘A’ allele that was independent of APOE e4 was evident only at age 60, when the A allele associated with a larger, negative change score (i.e., steeper decline); in older age decades, there were no differences in effects of the A allele on IR change (Fig 6B).
Fig 6. Plot of effect sizes of top TOMM40 risk alleles against age, adjusted for APOE e4 presence.
(A) Effect of the G allele in rs2075650 on rDR-L and (B) Effect of the A allele in rs157582 on IR-C. The lines represent age-related trajectories of the phenotype for individuals who have no risk alleles (solid line) or 1 or more risk allele (dashed line).
Gene network analysis
We evaluated the networks and molecular pathways including the relationships between TOMM40, APOE, and memory phenotypes from the IPA library of canonical pathways, disease processes, and biological processes, as well as customized pathways that were within the network. IPA analyses was seeded with established risk genes for AD (listed via AlzGene: APOE, BIN1, CLU, ABCA7, CR1, PICALM, MS4A6A, CD33, MS4A4E, CD2AP) and from the current study, as associated with rDR-L and IR-C, in order to elucidate relationships between genes and the variants involved in AD-related processes. The network (S7 Fig), shows that TOMM40 has possible interactions or functions in conjunction with several genes (i.e., APOE, BAX, INSR, APP, VDAC4, FOXB1, SNCA, HSD17B10, HSPA4) in the pathway of influence on verbal memory, episodic memory, AD, cognitive impairment, memory deficits, and reduced hippocampal volume. Some of these pathways likely involve other genes (e.g., CLU, PICALM, BDNF, CD33, CEBPB, TNFRSF19, ABCA7, BIN1, PPARGC1A) with upstream influence from F2. The network also shows interactions between APOE and other genes in nearly every pathway to a memory and cognition-related outcome.
Discussion
In this study, we used a population-based sample repeatedly assessed on verbal memory to characterize individual trajectories of change over age, giving greater precision in assessing level and decline of memory ability than available in most other studies. Trajectories were constructed to reflect immediate and delayed recall after age 60, an age at which other longitudinal studies suggest consistent decline in memory ability occurs. One SNP (rs2075650) on chromosome 19, in the TOMM40 region, associated at the genome-wide significant level with the phenotype of delayed recall level that is independent of immediate recall. This finding replicated in a completely independent sample and remained in our subsequent meta-analysis. The meta-analysis on delayed recall level also identified two other TOMM40 SNPs (rs71352238, rs157582) and an APOE SNP (rs769449) with genome-wide significant associations. Additional evaluation of the TOMM40 and APOE SNPs indicated that the GWAS signal for delayed recall level was mostly attributed to APOE; however, there were effects of the TOMM40 variant independent of e4. The meta-analysis on the separate phenotype of immediate recall change identified genome-wide significant SNPs in both genes (TOMM40 rs157582 and rs71352238; APOE rs769449). Additional evaluation indicated that TOMM40 had not only stronger effects than APOE on immediate recall change, but also effects independent of e4. Several other SNPs that reached the suggestive association level in the present study reside within genes associated with memory phenotypes in other studies. More critically, our results underscore the importance for specificity in phenotypes when assessing the genetic influence on components of memory abilities.
Discrete components of verbal memory
An innovation of this study was to separate the components involved in different aspects of recall. Whereas immediate recall reflects initial encoding and rehearsal, our residualized-delayed recall score, created by partialling the immediate score from the delayed score, is a distinct phenotype reflecting the “pure” retrieval component of memory (depicted in Fig 1). A similar approach has been used previously to create a pure measure of delayed memory function [79]. In the current study, separating the components provided greater insight into the basis for changes in verbal memory over age. About half of the variation in decline in DR after age 60 can be attributed to decline in IR (See Supporting Information, Tables A-B in S1 File). Also, after adjusting for what is initially encoded, there are small individual differences in what people can retrieve and even smaller individual differences in how people decline in retrieval.
Genetic specificity for aging-related changes in verbal memory components
The overarching goal of this study was to examine the complexity of verbal memory abilities over age, and whether genetic variants associate uniquely with specific components of verbal memory, represented by four phenotypes. Our finding with regard to rDR level supports work from prior cognitive aging studies. Taken together, although rs2075650, in the TOMM40 region, associates with rDR level in our study, and with episodic memory (p<5.0x10-08) and late-onset Alzheimer disease (LOAD) in other studies, that effect on verbal information retrieval detected through GWAS is primarily driven by linked APOE variants [6]. With regard to IR change, our findings indicate that TOMM40 plays a larger role, specifically, in decline of verbal learning after age 60. Further, our analysis showed that there are unique effects of TOMM40 beyond APOE e4 effects on both level of delayed recall prior to age 60 and decline in immediate recall after 60.
With the IR-C finding, additional evaluation of the effect of the TOMM40 SNP (rs157582) indicates that it has significant effects outside of e4. Upon cursory assessment with TOMM40 being 16 kb upstream of e4, conclusions could be drawn that the TOMM40 SNP simply serves as a marker for the e4 effect; however, several critical pieces of information suggest otherwise. First, the APOE SNP rs769449 resides 2 kb from e4 and is in high LD with it (r2 = .799) such that if the effect of TOMM40 on IR-C were attributable solely to APOE, we would have observed the effect of TOMM40 disappear when it was included in a model with rs769449 predicting IR-C. Yet, the effect of TOMM40 remained statistically significant whereas the effect of rs769449 did not. Second, if the signal were attributable solely to APOE, when the effect of the TOMM40 SNP was conditioned on the e4 allele (inferred from rs429358), the effect size of TOMM40 would have diminished. Conversely, the effect size of TOMM40 remained the same while the effect of e4 diminished to non-significance. Several additional observations point towards an independent TOMM40 effect. One relates to linkage disequilibrium (LD) and that the TOMM40 SNP (rs157582) is in imperfect LD with the APOE SNPs (r2 = .456 with rs769449; r2 = .590 with rs429358 [73]). Therefore, these SNPs cannot serve as interchangeable proxies for one another. Additional support comes from the observed associations with late onset Alzheimer disease (LOAD). When both rs157581 (residing 500 bp from rs157582 on TOMM40) and rs429358 were directly genotyped and tested in separate additive logit models for associations with LOAD, they showed different degrees of effect (ORs of 2.9 and 4.1, respectively [80]). Third, and perhaps most surprising, is the finding that rs157582 showed statistically significant effects among e3/e3 individuals, but not those who carry e4. Taken together, these findings suggest there is some degree of effect of TOMM40 that occurs independently of APOE e4.
Our findings are in line with several recent studies that have implemented newer methods to evaluate APOE-TOMM40 haplotype associations [81] and found TOMM40 variants influence memory performance and dementia risk beyond that conferred by the APOE e4 variant [82–94]. For example, individuals with an APOE e3-TOMM40 VL haplotype developed LOAD an average of seven years earlier than those with the APOE e3-TOMM40 S haplotype (70.5±1.2 years vs. 77.6±2.1 years, p = 0.02 [95]). This indicates that the TOMM40 polymorphism significantly influences age of disease onset for those with an e3 allele, which had been considered a low risk allele prior to these haplotype studies. Other studies have found that within APOE e3/e3 individuals, the VL variant of the TOMM40 polymorphism is associated with lower performance on episodic memory [84, 94]. Our study showed there were differences in effects of TOMM40 on delayed verbal memory and immediate recall change depending on APOE genotype, with notable effects among e3/e3 individuals. Given these findings, devising a study in which the TOMM40 poly-T length polymorphisms (also known as the ‘523’ repeat) have been characterized to test the association between APOE e3/e3-TOMM40 variant haplotypes with delayed verbal recall level and immediate recall change could provide further understanding of these findings.
Second, and more relevant to genetic specificity for verbal memory, prior GWAS studies use phenotypes (such as a TICS total score) that aggregate multiple cognitive domains, including crystallized ability, fluid ability, speed, and episodic memory. Since these processes differ in their phenotypic trajectories over age [96, 97] and genetic structure [98, 99], such aggregation may obscure genetic associations specific to memory. Our findings suggest even greater specificity, that variation in the TOMM40/APOE region is associated with level of ability in the retrieval component of verbal memory and change in initial encoding and rehearsal. Given the key importance of delayed recall in discriminating degrees of cognitive impairment, partialling out the phenotype to reflect true retrieval provides a more refined measure with which to study and potentially link underlying genetic associations with impairment. The interpretation of our findings would be clarified further by following up these results in a sample that has genotyping data and assessment of memory in non-verbal domains and probing of encoding by recognition as well as recall.
Biological implications and specificity
Our findings that TOMM40 and APOE associate with level of delayed recall and change in immediate recall have plausible biological mechanisms. An examination of gene interactions and protein expression through gene network analyses identified possible pathways from both genes to AD and memory-related conditions. For example, cognitively intact, older APOE e4 carriers have been shown to exhibit greater age-related reductions in hippocampal volume [100] and reduced functional activity in the right hippocampus [101]. In another study of healthy older adults (ages 60 to 87), lower hippocampal volume was associated with lower recall abilities among APOE e4 carriers in combination with the TOMM40 rs11556505 ‘T’ (r = 0.28, p<0.01, R2 = 0.08) and rs2075650 (the SNP in our study) ‘G’ (r = 0.28, p<0.01, R2 = 0.08) “risk” alleles [89]. This suggests that the additional effects of TOMM40 “risk” alleles on recall abilities, beyond effects of APOE, may be mediated via brain morphology.
Although verbal memory components may share some genetic underpinnings and biologic pathways, prior research and our findings support the notion that phenotypic specificity remains critically important. In a prior study, left hippocampal body volume was shown to be associated with delayed verbal memory (r = -0.17, p < .05), but not immediate memory among healthy older adults, ages 55 to 83 [16]. In another study, in which a residualized DR score was also used to remove the influences of IR, hippocampal size was associated with rDR performance, but not with IR [79]. Numerous findings have correlated verbal memory with other regions of the medial temporal lobes, outside of the hippocampus, although more recent evidence from functional neuroimaging studies of AD patients indicates that this is driven by learning rather than retrieval processes [15]. These findings suggest the need for further investigation of the relationship between hippocampal size, other regions of the medial temporal lobe, specific component of verbal memory, and the associated genetic profile to provide greater insight into biological mechanisms for age-related differences in memory abilities.
Repeated measures and other study strengths
This study addressed several limitations of prior research aimed at identifying genetic loci associated with memory decline, including the use of cross-sectional data to estimate age-related change and using an index of delayed memory that is confounded with immediate. We harnessed repeated measures from a large population-based study and created distinct phenotypes that better reflect the complexity of verbal memory ability and change over time. Results from longitudinal studies of episodic memory can be limited by practice effects [26], but this is of less concern in HRS because assessments were given every two years using alternate forms. We reduced the potential impact of selective attrition [102] by including individuals who had participated in HRS at least two occasions within 16 assessment years (1996 to 2012).
Limitations
This study had several limitations. First, our measure of verbal memory was based on a single presentation of a word list administered by a lay-interviewer and likely contains greater measurement error than other approaches using repeated presentations of a word list or multiple memory measures. However, this would be expected to introduce noise rather than false positive results. This illustrates a significant challenge for GWAS study designs: achieving samples sufficiently large to detect the expected effect sizes often means a trade-off in measurement intensity. By using a repeated measures design in a large sample we were able to characterize individual trajectories of age-related change and increased power compared to cross-sectional designs [103], but we still had limited power to detect additional SNP associations unique to the components of verbal memory. Third, variance in rDR change over time was particularly small. This partially explains the difficulty in detecting associations between genetic loci and change. Fourth, there are a variety of covariates that could have been included that may mediate the association between genetic effects and later life memory–including indices of health and socioeconomic status. We intentionally did not incorporate these in the analyses because it is arguable that these are outcomes rather than causes of memory ability and decline (for more discussion, see page 5 of S3 File). Further, for the GWAS in particular, there may be genetic pleiotropy between memory ability and these variables, especially educational attainment. Fifth, in examining the joint effect of TOMM40 and APOE on rDR-L and IR-C, we were not able to directly account for the effects of APOE e4, nor were we able to examine haplotype effects. Thus, our analyses cannot confirm causality between TOMM40 and phenotypes, nor can we confirm the independence of the effect of TOMM40 given the proximity to other susceptibility genes. For further confirmation, the finding warrants replication with a sample in which the genotyped APOE alleles are available, as well as the TOMM40 poly-T variant.
Conclusion
The current study provides evidence of genetic underpinnings for specific components of aging-related verbal memory performance, delayed recall at age 60 that is unconfounded with attention or memory span, and decline in immediate recall after age 60. The results underscore the importance of studying phenotypes that represent distinct components of memory, or other aspects of cognition, when assessing genetic influences. One identified genetic variant, a SNP in the TOMM40 region, has biological plausibility for its effect on delayed recall beyond its association with APOE e4 risk because of findings linking TOMM40, hippocampal formation and volume, and delayed recall. With regard to change in immediate recall ability, there is evidence for the genetic effects of both TOMM40 and APOE, as well as independent effects of TOMM40 among individuals who do not carry the APOE e4 allele. These findings need confirmation in other samples, but support the existence of differential effects of TOMM40 and APOE on specific components of verbal memory.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We wish to thank all participants of the Health and Retirement Study and the ELSA Longitudinal Study of Ageing. We would also like to thank Dr. Eileen Crimmins for her contributions in applying for ELSA data access.
Data Availability
The phenotypic data for the Health and Retirement Study (HRS) is public release data to registered users and researchers can become registered users through the University of Michigan (UM) data portal (http://hrsonline.isr.umich.edu/index.php?p=reg&_ga=2.214793619.503473076.1498851443-177726954.1424977713); however, registered users are not permitted to redistribute the data to third parties per the data use agreement (http://hrsonline.isr.umich.edu/index.php?p=regcou). The HRS genotype data is available to approved users through the NCBI Database of Genotypes and Phenotypes (dbGaP). A description of the available data is available through UM (https://hrs.isr.umich.edu/data-products/genetic-data/products#gdv1) and a description for gaining authorized access to the genotype data is available through dbGaP (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login). The phenotypic datasets for the English Longitudinal Study of Ageing (ELSA) are available to registered users to the U.K. Data Service (https://discover.ukdataservice.ac.uk/catalogue/?sn=5050&type=Data%20catalogue); registered users are not permitted to redistribute the data to third parties. The genotypic data for ELSA is available to approved users through the ELSA DNA Repository (EDNAR) and cannot be passed on to third parties per the user agreement. Conditions for use and instructions for the application to gain access through EDNAR are available online (http://doc.ukdataservice.ac.uk/doc/5050/mrdoc/pdf/5050_ELSA_Genetics_Data_Access_v3.pdf).
Funding Statement
This research was supported by the National Institute on Aging, of the National Institutes of Health, under award numbers F32AG048681 (PI: TEA) and P30AG017265 (Sub-contract: CAP). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The Health and Retirement Study [genetic] data is sponsored by the National Institute on Aging (grant numbers U01AG009740, [RC2AG036495, and RC4AG039029]) and was conducted by the University of Michigan. Funding for the English Longitudinal Study of Ageing is provided by the National Institute of Aging [grants 2RO1AG7644-01A1 and 2RO1AG017644] and a consortium of UK government departments coordinated by the Office for National Statistics. Funding for the English Longitudinal Study of Ageing genotyping data was provided by the Economic and Social Research Council (ESRC) and by the National Institute of Aging grants. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hebert LE, Weuve J, Scherr PA, Evans DA. Alzheimer disease in the United States (2010–2050) estimated using the 2010 census. Neurology. 2013;80(19):1778–83. doi: 10.1212/WNL.0b013e31828726f5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nilsson LG. Memory function in normal aging. Acta Neurologica Scandinavica. 2003;107(s179):7–13. [DOI] [PubMed] [Google Scholar]
- 3.Wilson RS, Beckett LA, Barnes LL, Schneider JA, Bach J, Evans DA, et al. Individual differences in rates of change in cognitive abilities of older persons. Psychol Aging. 2002;17(2):179 [PubMed] [Google Scholar]
- 4.Salthouse TA. Memory aging from 18 to 80. Alzheimer Disease & Associated Disorders. 2003;17(3):162–7. [DOI] [PubMed] [Google Scholar]
- 5.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43(5):429–35. Epub 2011/04/05. doi: 10.1038/ng.803 ; PubMed Central PMCID: PMC3084173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies G, Harris S, Reynolds C, Payton A, Knight H, Liewald D, et al. A genome-wide association study implicates the APOE locus in nonpathological cognitive ageing. Molecular Psychiatry. 2014;19(1):76–87. doi: 10.1038/mp.2012.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kao PF, Banigan MG, Vanderburg CR, McKee AC, Polgar PR, Seshadri S, et al. Increased expression of TrkB and Capzb2 accompanies preserved cognitive status in early Alzheimer disease pathology. Journal of neuropathology and experimental neurology. 2012;71(7):654–64. Epub 2012/06/20. doi: 10.1097/NEN.0b013e31825d06b7 ; PubMed Central PMCID: PMC3398703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsson LG, Nyberg L, Backman L. Genetic variation in memory functioning. Neuroscience and biobehavioral reviews. 2002;26(7):841–8. Epub 2002/12/10. . [DOI] [PubMed] [Google Scholar]
- 9.Luciano M, Wright M, Smith G, Geffen G, Geffen L, Martin N. Genetic covariance among measures of information processing speed, working memory, and IQ. Behavior genetics. 2001;31(6):581–92. [DOI] [PubMed] [Google Scholar]
- 10.Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, et al. Common Kibra alleles are associated with human memory performance. Science. 2006;314(5798):475–8. doi: 10.1126/science.1129837 [DOI] [PubMed] [Google Scholar]
- 11.Tulving E. Episodic memory: from mind to brain. Annual review of psychology. 2002;53(1):1–25. [DOI] [PubMed] [Google Scholar]
- 12.Loftus GR. Evaluating forgetting curves. Journal of Experimental Psychology: Learning, Memory, and Cognition. 1985;11(2):397. [Google Scholar]
- 13.Slamecka NJ, McElree B. Normal forgetting of verbal lists as a function of their degree of learning. Journal of Experimental Psychology: Learning, Memory, and Cognition. 1983;9(3):384. [Google Scholar]
- 14.Elger CE, Grunwald T, Lehnertz K, Kutas M, Helmstaedter C, Brockhaus A, et al. Human temporal lobe potentials in verbal learning and memory processes. Neuropsychologia. 1997;35(5):657–67. [DOI] [PubMed] [Google Scholar]
- 15.Wolk DA, Dickerson BC, Initiative AsDN. Fractionating verbal episodic memory in Alzheimer's disease. Neuroimage. 2011;54(2):1530–9. doi: 10.1016/j.neuroimage.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen KH, Chuah LY, Sim SK, Chee MW. Hippocampal region-specific contributions to memory performance in normal elderly. Brain and cognition. 2010;72(3):400–7. doi: 10.1016/j.bandc.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 17.Helmstaedter C, Grunwald T, Lehnertz K, Gleissner U, Elger C. Differential involvement of left temporolateral and temporomesial structures in verbal declarative learning and memory: evidence from temporal lobe epilepsy. Brain and cognition. 1997;35(1):110–31. doi: 10.1006/brcg.1997.0930 [DOI] [PubMed] [Google Scholar]
- 18.Rosen AC, Prull MW, Gabrieli JD, Stoub T, O'Hara R, Friedman L, et al. Differential associations between entorhinal and hippocampal volumes and memory performance in older adults. Behavioral neuroscience. 2003;117(6):1150 doi: 10.1037/0735-7044.117.6.1150 [DOI] [PubMed] [Google Scholar]
- 19.O'Hara R, Yesavage JA, Kraemer HC, Mauricio M, Friedman LF, Murphy GM. The APOE∍ 4 allele Is Associated with Decline on Delayed Recall Performance in Community‐Dwelling Older Adults. Journal of the American Geriatrics Society. 1998;46(12):1493–8. [DOI] [PubMed] [Google Scholar]
- 20.O’Hara R, Miller E, Liao C-P, Way N, Lin X, Hallmayer J. COMT genotype, gender and cognition in community-dwelling, older adults. Neuroscience letters. 2006;409(3):205–9. doi: 10.1016/j.neulet.2006.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomar JJ, Bobes-Bascaran MT, Conejero-Goldberg C, Davies P, Goldberg TE, Initiative AsDN. Utility of combinations of biomarkers, cognitive markers, and risk factors to predict conversion from mild cognitive impairment to Alzheimer disease in patients in the Alzheimer's disease neuroimaging initiative. Arch Gen Psychiatry. 2011;68(9):961–9. doi: 10.1001/archgenpsychiatry.2011.96 [DOI] [PubMed] [Google Scholar]
- 22.Greenaway MC, Lacritz LH, Binegar D, Weiner MF, Lipton A, Cullum CM. Patterns of verbal memory performance in mild cognitive impairment, Alzheimer disease, and normal aging. Cognitive and Behavioral Neurology. 2006;19(2):79–84. doi: 10.1097/01.wnn.0000208290.57370.a3 [DOI] [PubMed] [Google Scholar]
- 23.Welsh K, Butters N, Hughes J, Mohs R, Heyman A. Detection of abnormal memory decline in mild cases of Alzheimer's disease using CERAD neuropsychological measures. Archives of Neurology. 1991;48(3):278–81. [DOI] [PubMed] [Google Scholar]
- 24.Rönnlund M, Bäckman L, Lövdén M, Nilsson L, Nyberg L. Five-year changes in recall across levels of support: parallel age-related deficits. Int J Psychol. 2000;35:366. [Google Scholar]
- 25.Schaie KW. Intellectual development in adulthood: The Seattle longitudinal study: Cambridge University Press; 1996. [Google Scholar]
- 26.Rönnlund M, Nyberg L, Bäckman L, Nilsson L-G. Stability, growth, and decline in adult life span development of declarative memory: cross-sectional and longitudinal data from a population-based study. Psychol Aging. 2005;20(1):3 doi: 10.1037/0882-7974.20.1.3 [DOI] [PubMed] [Google Scholar]
- 27.Lindenberger U, Nagel IE, Chicherio C, Li S-C, Heekeren HR, Bäckman L. Age-related decline in brain resources modulates genetic effects on cognitive functioning. Front Neurosci. 2008;2:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson B, Whitfield K, Pedersen NL, Hofer SM, Ahern F, McClearn GE. Origins of individual differences in episodic memory in the oldest-old: A population-based study of identical and same-sex fraternal twins aged 80 and older. The Journals of Gerontology Series B: Psychological Sciences and Social Sciences. 1999;54(3):P173–P9. [DOI] [PubMed] [Google Scholar]
- 29.Papenberg G, Lindenberger U, Bäckman L. Aging-related magnification of genetic effects on cognitive and brain integrity. Trends in cognitive sciences. 2015;19(9):506–14. doi: 10.1016/j.tics.2015.06.008 [DOI] [PubMed] [Google Scholar]
- 30.Papenberg G, Salami A, Persson J, Lindenberger U, Bäckman L. Genetics and functional imaging: effects of APOE, BDNF, COMT, and KIBRA in aging. Neuropsychology review. 2015;25(1):47–62. doi: 10.1007/s11065-015-9279-8 [DOI] [PubMed] [Google Scholar]
- 31.Alarcón M, Plomin R, Fulker D, Corley R, DeFries JC. Multivariate path analysis of specific cognitive abilities data at 12 years of age in the Colorado Adoption Project. Behavior Genetics. 1998;28(4):255–64. [DOI] [PubMed] [Google Scholar]
- 32.Ando J, Ono Y, Wright MJ. Genetic structure of spatial and verbal working memory. Behavior genetics. 2001;31(6):615–24. [DOI] [PubMed] [Google Scholar]
- 33.Bouchard TJ Jr., Segal NL, Lykken DT. Genetic and environmental influences on special mental abilities in a sample of twins reared apart. Acta geneticae medicae et gemellologiae. 1990;39(2):193–206. Epub 1990/01/01. . [DOI] [PubMed] [Google Scholar]
- 34.Finkel D, Pedersen N, McGue M. Genetic influences on memory performance in adulthood: Comparison of Minnesota and Swedish twin data. Psychol Aging. 1995;10(3):437 [DOI] [PubMed] [Google Scholar]
- 35.Swan GE, Reed T, Jack LM, Miller BL, Markee T, Wolf PA, et al. Differential genetic influence for components of memory in aging adult twins. Arch Neurol. 1999;56(9):1127–32. Epub 1999/09/17. . [DOI] [PubMed] [Google Scholar]
- 36.Volk HE, McDermott KB, Roediger HL 3rd, Todd RD. Genetic influences on free and cued recall in long-term memory tasks. Twin Res Hum Genet. 2006;9(5):623–31. Epub 2006/10/13. doi: 10.1375/183242706778553462 . [DOI] [PubMed] [Google Scholar]
- 37.Panizzon MS, Lyons MJ, Jacobson KC, Franz CE, Grant MD, Eisen SA, et al. Genetic architecture of learning and delayed recall: A twin study of episodic memory. Neuropsychology. 2011;25(4):488 doi: 10.1037/a0022569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kremen WS, Panizzon MS, Franz CE, Spoon KM, Vuoksimaa E, Jacobson KC, et al. Genetic complexity of episodic memory: A twin approach to studies of aging. Psychol Aging. 2014;29(2):404 doi: 10.1037/a0035962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynolds CA, Finkel D, Gatz M, Pedersen NL. Sources of influence on rate of cognitive change over time in Swedish twins: An application of latent growth models. Experimental Aging Research. 2002;28(4):407–33. doi: 10.1080/03610730290103104 [DOI] [PubMed] [Google Scholar]
- 40.Reynolds CA, Finkel D, McArdle JJ, Gatz M, Berg S, Pedersen NL. Quantitative genetic analysis of latent growth curve models of cognitive abilities in adulthood. Dev Psychol. 2005;41(1):3 doi: 10.1037/0012-1649.41.1.3 [DOI] [PubMed] [Google Scholar]
- 41.Finkel D, Reynolds CA, McArdle JJ, Pedersen NL. The longitudinal relationship between processing speed and cognitive ability: Genetic and environmental influences. Behavior genetics. 2005;35(5):535–49. doi: 10.1007/s10519-005-3281-5 [DOI] [PubMed] [Google Scholar]
- 42.Steptoe A, Breeze E, Banks J, Nazroo J. Cohort profile: the English longitudinal study of ageing. Int J Epidemiol. 2013;42(6):1640–8. Epub 2012/11/13. doi: 10.1093/ije/dys168 ; PubMed Central PMCID: PMC3900867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welsh KA, Breitner JC, Magruder-Habib KM. Detection of dementia in the elderly using telephone screening of cognitive status. Cognitive and Behavioral Neurology. 1993;6(2):103–10. [Google Scholar]
- 44.McArdle JJ, Fisher GG, Kadlec KM. Latent variable analyses of age trends of cognition in the Health and Retirement Study, 1992–2004. Psychol Aging. 2007;22(3):525–45. Epub 2007/09/19. doi: 10.1037/0882-7974.22.3.525 . [DOI] [PubMed] [Google Scholar]
- 45.Stephan Y, Caudroit J, Jaconelli A, Terracciano A. Subjective age and cognitive functioning: A 10-year prospective study. The American Journal of Geriatric Psychiatry. 2014;22(11):1180–7. doi: 10.1016/j.jagp.2013.03.007 [DOI] [PubMed] [Google Scholar]
- 46.Shega JW, Sunkara PD, Kotwal A, Kern DW, Henning SL, McClintock MK, et al. Measuring Cognition: The Chicago Cognitive Function Measure in the National Social Life, Health and Aging Project, Wave 2. The Journals of Gerontology Series B: Psychological Sciences and Social Sciences. 2014;69(Suppl 2):S166–S76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crimmins EM, Kim JK, Langa KM, Weir DR. Assessment of cognition using surveys and neuropsychological assessment: the Health and Retirement Study and the Aging, Demographics, and Memory Study. The Journals of Gerontology Series B: Psychological Sciences and Social Sciences. 2011;66(suppl 1):i162–i71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ofstedal MB, Fisher GG, Herzog AR. Documentation of cognitive functioning measures in the Health and Retirement Study. Ann Arbor, MI: University of Michigan; 2005. [Google Scholar]
- 49.McArdle JJ. Longitudinal dynamic analyses of cognition in the health and retirement study panel. AStA Advances in Statistical Analysis. 2011;95(4):453–80. doi: 10.1007/s10182-011-0168-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Health and Retirement Study [Internet]. National Center for Biotechnology Information. 2012 [cited September 16, 2014]. Available from: http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000428.v1.p1.
- 51.Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, et al. Quality control and quality assurance in genotypic data for genome‐wide association studies. Genet Epidemiol. 2010;34(6):591–602. doi: 10.1002/gepi.20516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.HRS. Quality control report for genotypic data. St. Louis, MO: University of Washington, 2012. [Google Scholar]
- 53.SAS. SAS Institute version 9.4. Cary NC; 2013.
- 54.Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982:963–74. [PubMed] [Google Scholar]
- 55.McArdle JJ, Ferrer-Caja E, Hamagami F, Woodcock RW. Comparative longitudinal structural analyses of the growth and decline of multiple intellectual abilities over the life span. Dev Psychol. 2002;38(1):115 [PubMed] [Google Scholar]
- 56.McArdle JJ, Plassman BL. A biometric latent curve analysis of memory decline in older men of the NAS-NRC twin registry. Behavior genetics. 2009;39(5):472–95. doi: 10.1007/s10519-009-9272-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, et al. Genes mirror geography within Europe. Nature. 2008;456(7218):98–101. doi: 10.1038/nature07331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Human molecular genetics. 2008;17(R2):R143–R50. doi: 10.1093/hmg/ddn268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. doi: 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 60.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190 doi: 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Q, Yu K. Improved correction for population stratification in genome‐wide association studies by identifying hidden population structures. Genet Epidemiol. 2008;32(3):215–26. doi: 10.1002/gepi.20296 [DOI] [PubMed] [Google Scholar]
- 62.Serre D, Montpetit A, Paré G, Engert JC, Yusuf S, Keavney B, et al. Correction of population stratification in large multi-ethnic association studies. PLoS One. 2008;3(1):e1382 doi: 10.1371/journal.pone.0001382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang S, Zhu X, Zhao H. On a semiparametric test to detect associations between quantitative traits and candidate genes using unrelated individuals. Genet Epidemiol. 2003;24(1):44–56. doi: 10.1002/gepi.10196 [DOI] [PubMed] [Google Scholar]
- 64.Zhu X, Zhang S, Zhao H, Cooper RS. Association mapping, using a mixture model for complex traits. Genet Epidemiol. 2002;23(2):181–96. doi: 10.1002/gepi.210 [DOI] [PubMed] [Google Scholar]
- 65.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nature Reviews Genetics. 2010;11(7):459–63. doi: 10.1038/nrg2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. arXiv preprint arXiv:14104803. 2014. [DOI] [PMC free article] [PubMed]
- 67.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. Epub 2007/08/19. doi: 10.1086/519795 ; PubMed Central PMCID: PMC1950838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. Epub 2010/12/21. doi: 10.1016/j.ajhg.2010.11.011 ; PubMed Central PMCID: PMC3014363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science-AAAS-Weekly Paper Edition. 1996;273(5281):1516–7. [DOI] [PubMed] [Google Scholar]
- 70.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. doi: 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bacanu SA, Devlin B, Roeder K. Association studies for quantitative traits in structured populations. Genet Epidemiol. 2002;22(1):78–93. Epub 2002/01/05. doi: 10.1002/gepi.1045 . [DOI] [PubMed] [Google Scholar]
- 73.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–7. doi: 10.1093/bioinformatics/btv402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.HRS. Cognitive and Behavioral Phenotypes: SNPs of Interest. Ann Arbor, MI: University of Michigan, 2014. [Google Scholar]
- 75.Oldmeadow C, Holliday EG, McEvoy M, Scott R, Kwok JB, Mather K, et al. Concordance between direct and imputed APOE genotypes using 1000 Genomes data. Journal of Alzheimer's Disease. 2014;42(2):391–3. doi: 10.3233/JAD-140846 [DOI] [PubMed] [Google Scholar]
- 76.Radmanesh F, Devan WJ, Anderson CD, Rosand J, Falcone GJ. Accuracy of imputation to infer unobserved APOE epsilon alleles in genome-wide genotyping data. European Journal of Human Genetics. 2014;22(10):1239–42. doi: 10.1038/ejhg.2013.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.CIDR. CIDR Health and Retirement Study: Imputation Report—1000 Genomes Project reference panel. Seattle, WA: University of Washington, 2012. [Google Scholar]
- 78.Faul J, Smith J, Zhao W. Health and Retirement Study: Candidate Genes for Cognition/Behavior. Ann Arbor, MI: University of Michigan, 2014. [Google Scholar]
- 79.Golomb J, Kluger A, de Leon MJ, Ferris SH, Convit A, Mittelman MS, et al. Hippocampal formation size in normal human aging: a correlate of delayed secondary memory performance. Learning & Memory. 1994;1(1):45–54. [PubMed] [Google Scholar]
- 80.Yu CE, Seltman H, Peskind ER, Galloway N, Zhou PX, Rosenthal E, et al. Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer's disease: patterns of linkage disequilibrium and disease/marker association. Genomics. 2007;89(6):655–65. Epub 2007/04/17. doi: 10.1016/j.ygeno.2007.02.002 ; PubMed Central PMCID: PMC1978251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martin ER, Lai EH, Gilbert JR, Rogala AR, Afshari A, Riley J, et al. SNPing away at complex diseases: analysis of single-nucleotide polymorphisms around APOE in Alzheimer disease. The American Journal of Human Genetics. 2000;67(2):383–94. doi: 10.1086/303003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roses AD, Lutz MW, Crenshaw DG, Grossman I, Saunders AM, Gottschalk WK. TOMM40 and APOE: Requirements for replication studies of association with age of disease onset and enrichment of a clinical trial. Alzheimer's & dementia: the journal of the Alzheimer's Association. 2013;9(2):132–6. Epub 2013/01/22. doi: 10.1016/j.jalz.2012.10.009 . [DOI] [PubMed] [Google Scholar]
- 83.Ma X-Y, Yu J-T, Wang W, Wang H-F, Liu Q-Y, Zhang W, et al. Association of TOMM40 polymorphisms with late-onset Alzheimer’s disease in a Northern Han Chinese population. Neuromolecular medicine. 2013;15(2):279–87. doi: 10.1007/s12017-012-8217-7 [DOI] [PubMed] [Google Scholar]
- 84.Johnson SC, La Rue A, Hermann BP, Xu G, Koscik RL, Jonaitis EM, et al. The effect of TOMM40 poly-T length on gray matter volume and cognition in middle-aged persons with APOEɛ3/ɛ3 genotype. Alzheimer's & Dementia. 2011;7(4):456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Caselli RJ, Dueck AC, Huentelman MJ, Lutz MW, Saunders AM, Reiman EM, et al. Longitudinal modeling of cognitive aging and the TOMM40 effect. Alzheimer's & Dementia. 2012;8(6):490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cacabelos R, Goldgaber D, Vostrov A, Matsuki H, Torrellas C, Corzo D, et al. APOE-TOMM40 in the Pharmacogenomics of Dementia. Journal of Pharmacogenomics & Pharmacoproteomics. 2014;2014. [Google Scholar]
- 87.Laczó J, Andel R, Vyhnalek M, Matoska V, Kaplan V, Nedelska Z, et al. The effect of TOMM40 on spatial navigation in amnestic mild cognitive impairment. Neurobiology of aging. 2015;36(6):2024–33. doi: 10.1016/j.neurobiolaging.2015.03.004 [DOI] [PubMed] [Google Scholar]
- 88.Ferencz B, Gerritsen L. Genetics and underlying pathology of dementia. Neuropsychol Rev. 2015;25(1):113–24. Epub 2015/01/09. doi: 10.1007/s11065-014-9276-3 . [DOI] [PubMed] [Google Scholar]
- 89.Ferencz B, Laukka EJ, Lövdén M, Kalpouzos G, Keller L, Graff C, et al. The influence of APOE and TOMM40 polymorphisms on hippocampal volume and episodic memory in old age. Frontiers in human neuroscience. 2013;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lyall DM, Harris SE, Bastin ME, Maniega SM, Murray C, Lutz MW, et al. Alzheimer's disease susceptibility genes APOE and TOMM40, and brain white matter integrity in the Lothian Birth Cohort 1936. Neurobiology of aging. 2014;35(6):1513. e25-. e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Seripa D, Bizzarro A, Pilotto A, Palmieri O, Panza F, D'Onofrio G, et al. TOMM40, APOE, and APOC1 in primary progressive aphasia and frontotemporal dementia. Journal of Alzheimer's Disease. 2012;31(4):731–40. doi: 10.3233/JAD-2012-120403 [DOI] [PubMed] [Google Scholar]
- 92.Payton A, Sindrewicz P, Pessoa V, Platt H, Horan M, Ollier W, et al. A TOMM40 poly-T variant modulates gene expression and is associated with vocabulary ability and decline in nonpathologic aging. Neurobiology of aging. 2016;39:217. e1-. e7. [DOI] [PubMed] [Google Scholar]
- 93.Greenbaum L, Springer RR, Lutz MW, Heymann A, Lubitz I, Cooper I, et al. The TOMM40 poly-T rs10524523 variant is associated with cognitive performance among non-demented elderly with type 2 diabetes. European Neuropsychopharmacology. 2014;24(9):1492–9. doi: 10.1016/j.euroneuro.2014.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hayden KM, McEvoy JM, Linnertz C, Attix D, Kuchibhatla M, Saunders AM, et al. A homopolymer polymorphism in the TOMM40 gene contributes to cognitive performance in aging. Alzheimer's & Dementia. 2012;8(5):381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, Sundseth SS, et al. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease. Pharmacogenomics J. 2010;10(5):375–84. Epub 2009/12/24. doi: 10.1038/tpj.2009.69 ; PubMed Central PMCID: PMC2946560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Horn JL, Cattell RB. Age differences in fluid and crystallized intelligence. Acta psychologica. 1967;26:107–29. [DOI] [PubMed] [Google Scholar]
- 97.Horn JL, McArdle JJ. A practical and theoretical guide to measurement invariance in aging research. Experimental aging research. 1992;18(3):117–44. [DOI] [PubMed] [Google Scholar]
- 98.Deary IJ, Johnson W, Houlihan LM. Genetic foundations of human intelligence. Hum Genet. 2009;126(1):215–32. doi: 10.1007/s00439-009-0655-4 [DOI] [PubMed] [Google Scholar]
- 99.Papassotiropoulos A, Dominique J-F. Genetics of human episodic memory: dealing with complexity. Trends in cognitive sciences. 2011;15(9):381–7. doi: 10.1016/j.tics.2011.07.005 [DOI] [PubMed] [Google Scholar]
- 100.Cohen R, Small C, Lalonde F, Friz J, Sunderland T. Effect of apolipoprotein E genotype on hippocampal volume loss in aging healthy women. Neurology. 2001;57(12):2223–8. [DOI] [PubMed] [Google Scholar]
- 101.Lind J, Persson J, Ingvar M, Larsson A, Cruts M, Van Broeckhoven C, et al. Reduced functional brain activity response in cognitively intact apolipoprotein E ε4 carriers. Brain. 2006;129(5):1240–8. [DOI] [PubMed] [Google Scholar]
- 102.Hedden T, Gabrieli JD. Insights into the ageing mind: a view from cognitive neuroscience. Nature Reviews Neuroscience. 2004;5(2):87–96. doi: 10.1038/nrn1323 [DOI] [PubMed] [Google Scholar]
- 103.Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6(2):109–18. Epub 2005/02/18. doi: 10.1038/nrg1522 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
The phenotypic data for the Health and Retirement Study (HRS) is public release data to registered users and researchers can become registered users through the University of Michigan (UM) data portal (http://hrsonline.isr.umich.edu/index.php?p=reg&_ga=2.214793619.503473076.1498851443-177726954.1424977713); however, registered users are not permitted to redistribute the data to third parties per the data use agreement (http://hrsonline.isr.umich.edu/index.php?p=regcou). The HRS genotype data is available to approved users through the NCBI Database of Genotypes and Phenotypes (dbGaP). A description of the available data is available through UM (https://hrs.isr.umich.edu/data-products/genetic-data/products#gdv1) and a description for gaining authorized access to the genotype data is available through dbGaP (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login). The phenotypic datasets for the English Longitudinal Study of Ageing (ELSA) are available to registered users to the U.K. Data Service (https://discover.ukdataservice.ac.uk/catalogue/?sn=5050&type=Data%20catalogue); registered users are not permitted to redistribute the data to third parties. The genotypic data for ELSA is available to approved users through the ELSA DNA Repository (EDNAR) and cannot be passed on to third parties per the user agreement. Conditions for use and instructions for the application to gain access through EDNAR are available online (http://doc.ukdataservice.ac.uk/doc/5050/mrdoc/pdf/5050_ELSA_Genetics_Data_Access_v3.pdf).