Abstract
Hepatitis C virus (HCV) infection is a rapidly increasing global health problem with an estimated 170 million people infected worldwide. HCV is a hepatotropic, positive-sense RNA virus of the family Flaviviridae. As a positive-sense RNA virus, the HCV genome itself must serve as a template for translation, replication and packaging. The viral RNA must therefore be a dynamic structure that is able to readily accommodate structural changes to expose different regions of the genome to viral and cellular proteins to carry out the HCV life cycle. The ∼9600 nucleotide viral genome contains a single long open reading frame flanked by 5′ and 3′ non-coding regions that contain cis-acting RNA elements important for viral translation, replication and stability. Additional cis-acting RNA elements have also been identified in the coding sequences as well as in the 3′ end of the negative-strand replicative intermediate. Herein, we provide an overview of the importance of these cis-acting RNA elements in the HCV life cycle.
Keywords: Hepatitis C virus, cis-acting RNA element, Internal Ribosome Entry Site, miR-122
1. Introduction
Hepatitis C virus (HCV) is a positive-sense RNA virus of the family Flaviviridae (genus hepacivirus). It has been estimated that approximately 170 million people are infected worldwide, eventually suffering from liver cirrhosis and hepatocellular carcinoma [1, 2]. Until recently, the only therapy included a regimen of pegylated interferon α and ribavirin that was poorly effective in most patients [3]. With the recent development of direct-acting antiviral combination therapies that produce sustained virological response (cure) rates of over 95%, there is great enthusiasm that HCV infection can be effectively controlled in the near future.
The viral genome is approximately ∼9600 nucleotides (nt) in length. The HCV genome has a single open reading frame (ORF) that encodes a ∼3000 amino acid polyprotein that is subsequently cleaved by virus-encoded and cellular proteinases into the ten mature viral proteins (Fig. 1A). The ORF is flanked by highly structured 5′ and 3′ non-coding regions (NCRs) that contain RNA secondary and tertiary structures important for viral translation, replication and stability (Fig. 1B-C). Herein, we provide a comprehensive overview of the importance of these cis-acting RNA elements (CRE) in the HCV life cycle.
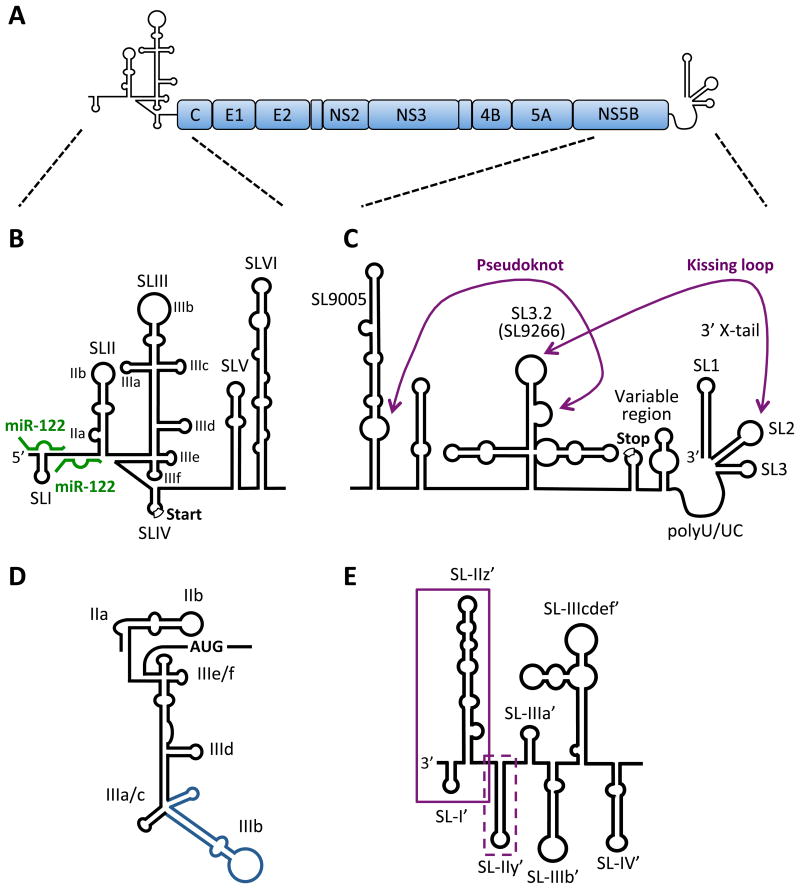
Figure 1. RNA secondary structures in the HCV genome.
A) HCV genome organization. The HCV genome is a positive-sense, single-stranded RNA encoding a single large open reading frame (∼3000 amino acids), flanked by highly structured 5′ and 3′ NCRs. B) Schematic diagram of the 5′ NCR and RNA structures of the core-coding region. The 5′ NCR consists of four stem-loop structures (SLI to SLIV) and the core coding region contains two stem-loop structures (SLV and SLVI). SLII through SLIV comprise the HCV IRES element required for cap-independent translation (start codon is indicated) and SLI through SLII are required for viral RNA replication. The miR-122 sites are indicated (green). C) The 3′ NCR has a tripartite structure containing a variable region (with the polyprotein stop codon), poly-U/UC tract and 3′ × region (containing 3′ SL1, SL2 and SL3). The kissing-loop interaction between 3′ SL2 and SL3.2 (SL9266) of the NS5B-coding region (part of a larger cruciform CRE) as well as a pseudoknot between the 3′ sub-terminal bulge of SL3.2 and residues in the bulge of the extended stem-loop SL9005 (around nt 9110) are indicated. SL nomenclature used is based on the H77 complete genome sequence (Genbank Accession #AF011753). D) The secondary structure of the IRES reflecting its orientation in complex with the 40S subunit and eIF3. SLIIa induces a bent structure in SLII to direct SLIIb to the ribosomal E-site in the head region of the 40S subunits. The SLIIIad junction binds to the surface of the 40S whereas the SLIIIbc motif (blue) associates with eIF3. SLIIIef and the SLIV pseudoknot (not shown) position the AUG codon in the P-site of the 40S subunit. E) The 3′ end of the negative-strand replicative intermediate. Boxed sequences indicate required (solid) and contributing (dashed) sequences to initiation of positivestrand synthesis.
2. CREs in HCV translation
The start-site AUG codon for translation initiation in the 5′ NCR of the HCV RNA genome is located at nucleotide 342. Thus, it was unclear how the viral mRNA would be efficiently translated by a conventional cap-dependent scanning mechanism where the 40S ribosomal subunits would have to traverse several higher-ordered RNA structures and bypass several upstream AUG codons. It was known from studies with picornaviruses that long structured RNA elements, which were located in the 5′ NCRs, could mediate translation initiation by an internal ribosome entry site mechanism. Specifically, these so-called internal ribosome entry site (IRES) sequences could, aided by canonical and non-canonical translation initiation factors, bind 40S subunits to internal sequences and independently of the 5′ end of the viral RNA. Thus, IRES elements function like Shine-Dalgarno sequences in bacterial mRNAs, where 30S subunits are recruited via base-pair interactions with the rRNA and small sequences upstream of the initiation codon. However, unlike a Shine-Dalgarno sequence, IRES elements are long and structured. Indeed, IRES activity was detected in the HCV RNA genome when investigators found that the HCV 5′ NCR, when located in the intergenic spacer of an artifical bicistronic mRNA, mediated translation of the second cistron independently of the first cistron [4, 5]. Furthermore, insertion of an AUG codon upstream of the AUG start codon at position 342 was not recognized by the 40S subunit [6], suggesting that the HCV IRES binds the 40S subunit directly at the start-site AUG. Over the past decade, numerous single-particle cryoelectron microscopy, NMR and X-ray structures of the HCV IRES have been reported (discussed in [7]). These structures and the recent solution structure of the entire HCV IRES [8] have uncovered the molecular details of how the HCV IRES assembles translation-competent 80S ribosomes.
2.1 The Internal Ribosomal Entry Site (IRES)
The HCV 5′ NCR contains four major stem loop (SL) structures (Fig. 1B). SLI is important to augment viral replication (see below), while SLII-IV comprises the HCV IRES. Thus, the IRES functions as a three SL-domain structure, in which the individual SLs are folded independently, and are separated by small linker regions [9](Fig. 1B). First, we will summarize the function of each loop in IRES activity and then describe the steps at which these SL structures assemble functional 80S ribosomes at the start-site AUG codon.
SLIIa induces an ion-dependent bent structure in SLII to direct SLIIb to the ribosomal E-site in the head region of the 40S subunits [10] (Fig. 1D). Deletion of SLIIa eliminates the bent structure of SLII. While SLIIa IRES deletions still allow the formation of 80S-IRES complexes, these complexes do not elongate because GTP hydrolysis in Met-tRNAi-eIF2 complexes and eIF2 release from the 40S is inhibited. It is thought that the bent topology of SLII in the E-site needs to undergo a conformational switch to allow its removal and the subsequent occupancy of the E-site with deacylated tRNA, resulting in translation elongation [11, 12]. SLII also reaches into the mRNA binding cleft in the 40S subunit where it causes a conformational change in the 40S subunits, resulting in rotation of the head relative to the body of the 40S, clamping down of the attached HCV mRNA [12].
SLIII contains several domains [13], including a SLIIIabc four-way junction [14] and a SLIIIef/SLIV pseudoknot structure [15, 16] (Fig. 1D). Upon formation of IRES-40S complexes the SLIIIad junction binds to the surface of the 40S, whereas the SLIIIbc RNA motif associates with translation factor eIF3 [9]. It is thought that binding of eIF3 to 40S subunits prevents the premature association of 60S subunits [17]. Furthermore, reconstitution IRES-40S experiments have shown that eIF3 enhances the formation of 40S-IRES complexes likely by stabilizing the Met-tRNAi-eIF2 complex [18]. SLIIIef interacts with SLIV to form a complex double pseudoknot structure that is important in positioning the start-site AUG codon in the viral mRNA into the P-site of the 40S subunit [15, 16].
The assembly of IRES-40S complexes is governed by dynamic changes in the IRES that occur after interaction of the IRES with its cellular binding partners. First, the basal domain of SLIII (SLIIIad) binds to the backside of the 40S and subsequently the apical domain of SLIII (SLIIIbc) recruits factor eIF3 (Fig. 1D). In the second step, domain SLII reorients and interacts with the decoding center of the 40S subunit, resulting in a head rotation in the 40S and clamping down of the mRNA in the 40S mRNA binding cleft. Finally, pseudoknots in SLIIIef aid in positioning of the initiator tRNAmet in the ribosomal P-site of the 40S subunit. Remarkably, the IRES sequence and structure is highly conserved among all HCV genotypes, but is very different from IRES elements found, for example, in picornaviral RNA genomes. Picornaviral IRES elements also require more initiation factors than the HCV IRES, suggesting that diversification likely occurred as a result of tissue tropism and pathogenic signatures of these viruses.
2.2 Modulation of the HCV IRES by host factors
Several host proteins are recruited to the HCV IRES to mediate IRES activity. These factors are collectively referred to as IRES trans-acting factors (ITAFs, reviewed in [19]). ITAFs contain one or more RNA-binding domains and are involved in a variety of post-transcriptional mechanisms of cellular RNAs
One of the most studied ITAFs is the La protein that is normally involved in the processing of precursor tRNAs in eukaryotic cells [20]. La protein binds to SLIV and the region around the AUG start codon in the HCV IRES [21, 22]. It has been shown that La contacts SLIV and ribosomal protein S5 in the 40S ribosomal subunit [23]), resulting in enhanced IRES activity by an unknown mechanism. Several heterogeneous nuclear RNA proteins (hnRNPs) have also been shown to modulate HCV IRES activity. For example, SYNCRIP (synaptotagmin-binding, cytoplasmic RNA-interacting protein)/hnRNP Q/NSAP1 [24, 25] has diverse functions in RNA metabolism including splicing and translational regulation, and binds to the HCV IRES immediately downstream of the start codon to promote the correct positioning of the 40S subunit [26]. Similarly, hnRNP L is normally involved in regulation of alternative splicing and binds to the HCV IRES downstream of the AUG start codon to stimulate translation by an unknown mechanism [27, 28]. Curiously, hnRNP D, which is typically involved in cellular mRNA destabilization, stimulates the HCV IRES after binding to SLII of the IRES [29]. IMP-1 (insulin-like growth factor 2 mRNA binding protein 1) is an RNA-binding protein involved in mRNA localization, turnover and translational regulation [30], which binds to SLIV as well as the viral 3′ NCR and has been suggested to be involved in genome circularization [31] and enhancement of IRES-mediated translation [32]. Because hnRNP D can interact with hnRNP L, SYNCRIP and IMP-1, hnRNP D may act as a scaffold to promote recruitment of other hnRNPs and IMP-1 to the HCV IRES [33-35]. Finally, Gemin5 and LSm1-7, which are RNA-binding proteins that bind snRNAs and control specific loading of Sm proteins during snRNP assembly and are involved in mRNA decapping/degradation [36, 37] interact with the HCV IRES. Specifically, both Gemin5 and LSm1-7 bind to SLIII of the HCV IRES and have opposing effects on HCV translation. While Gemin5 downregulates HCV IRES-mediated translation, LSm1-7 is required for efficient HCV RNA translation [38]. Since Gemin5 is also known to interact with LSm1-7, SYNCRIP and the La protein, it is speculated that Gemin5 may be involved in oligomeric protein interactions that control HCV IRES-mediated translation.
As mentioned above, the exact mechanism by which ITAFs modulate IRES activity is unknown. The ITAFs likely cause changes in dynamics of HCV IRES-ribosome interactions. Such changes can now be examined by Förster resonance energy transfer (FRET) in single molecule approaches that monitor changes in dynamics of the HCV IRES with ribosomes in the presence of distinct ITAFs [39].
2.3 CREs in the core-coding region
The core-coding region of the HCV genome was proposed to contain two stem-loop structures (SLV and SLVI, Fig. 1B) with high sequence conservation that have been confirmed by enzymatic structural probing in vitro [40]. Subsequent studies demonstrated that mutations disrupting SLV and SLVI reduced viral RNA translation, but were dispensable for viral RNA replication [41, 42]. However, sub-genomic replicons with a deletion of the core to NS2-coding regions are able to undergo translation, replication and packaging; indicating that these areas are devoid of RNA structures crucial to viral translation, replication or packaging [43].
3. CREs in HCV RNA replication
The majority of CREs that influence HCV RNA replication are located in the 5′ and 3′ NCRs of the positive-strand and the 3′ end of the negative-strand RNA (Fig. 1A-D) (reviewed in [44]). However, RNA sequences and structures have been described those are crucial to viral replication in the coding regions as well. These CREs have been extensively mapped and characterized both in vitro and in cell culture systems, but the precise roles and mechanistic functions of individual SL structures remain elusive in many cases.
3.1 Replication elements in the 5′ NCR
In addition to its crucial role in translation, the 5′ NCR contains CREs required for viral replication as well (Fig. 1B). Genetic analyses have mapped the first 125 nt of the 5′ NCR, containing SLI and SLII, as the minimal sequence requirement for HCV RNA replication [45]. SLI (nt 5-20) is dispensable for translation, but is essential to viral RNA replication [45]. Deletion of SLI completely abolishes replication in cell culture, but although the primary sequence of SLI plays a role, the overall structure seems to be what is critical to viral RNA replication [45-47]. Nucleotides 1 to 4 and 21 to 43 are also dispensable for translation, but are required for efficient HCV RNA replication [45]; however, this is likely due to the interaction between this region of the genome with the liver-specific microRNA, miR-122 (Fig. 1B, discussed in more detail below) [48-50]. SLII, although considered part of the HCV IRES, is also required for viral RNA replication [45]. It is unknown how these sequences direct RNA replication and is tempting to speculate that they may play a role in the switch from translation to replication; however, this has yet to be demonstrated experimentally. Notably, the nucleotide sequences of the 5′ NCR involved in initiation of positive-strand synthesis are found in the complementary negative-strand intermediate.
3.2 Replication elements in the 3′ end of the negative-strand replicative intermediate
The 3′ end of the negative-strand replicative intermediate, which is complementary to the genomic 5′ NCR, was initially predicted and subsequently structurally probed in vitro [51-53]. All three studies predicted five stem-loop structures in the 3′ terminal 220 nt of the negative-strand RNA, whereas structures upstream differed between the three studies [51-53]. More recently, computational prediction combined with genetic analyses were used to characterize the sequences and structures required for positive-strand RNA synthesis in a system that uncouples viral translation and replication [54] (Fig. 1E). The first 104 nt, containing SL-I' (nts 6 to 20) and SL-IIz' (nt 21 to 104), were shown to be absolutely required for HCV RNA replication [54]. The primary sequence of SL-I' stem was required, but not that of the loop; while for SL-IIz' preservation of the overall structure restored replication, albeit to about 10-fold lower levels than that of wild-type [54]. While a construct containing SL-I' and SL-IIz' (nts 1 to 106) was sufficient for viral RNA replication, this mutant was severely impaired (only 10-fold above background as measured with a construct containing a mutation in the NS5B RNA-dependent RNA polymerase active site), suggesting that additional sequences are required to promote efficient positive-strand synthesis [54]. The primary sequence of SL-IIy' also contributes to efficient RNA replication, whereas the remaining stem-loop structures seem to have an ancillary role [54, 55]. Similar to the 3′ terminus of the positive-sense RNA, the 3′ terminal nucleotide of the negative-strand is a uridine or cytosine residue in most HCV isolates, likely due to the preference for a purine (adenine or guanine) as the initiating nucleotide for the NS5B RNA-dependent RNA polymerase [56]. Despite recent advances in structural and sequence requirements for positive-strand RNA synthesis, the distinct mechanistic roles of the SL structures at the 3′ end of the negative-strand remain uncertain and are likely to be the focus of future studies.
3.3 Replication elements in the 3′ NCR of HCV RNA
As with most positive-strand RNA viruses, the 3′ NCR is essential for viral RNA replication, presumably due to the requirement for initiation of negative-strand synthesis at this site [57, 58]. The 3′ NCR has a tripartite structure containing a variable region, poly-U/UC region, and a 98-nt 3′ X-tail [59, 60] (Fig. 1C). The variable region is approximately 40-nt in length and is poorly conserved among HCV isolates [60-62]. Nonetheless, it is predicted to form two stem-loop structures that overlap with the 3′ terminus of the NS5B coding sequence and contain the viral polyprotein stop codon [61-63]. The variable region is not essential, but its deletion leads to severely impaired replication, suggesting that it promotes efficient viral RNA replication in cell culture [57, 64]. The poly-U/UC tract varies between 30 and 80 nt among HCV isolates and consists of a homopolyuridine stretch interspersed by single cytosine residues [59]. A minimum of 26-33 consecutive uridines is essential for HCV RNA replication in cell culture [57, 65]. It is hypothesized that the poly-U/UC region provides a platform for recruitment of host or viral proteins as introduction of cytosine residues is deleterious to viral replication, but the position of the 26-33 nt poly-U tract within the poly-U/UC stretch is flexible [57, 65]. Importantly, the NS3 helicase, NS5A and the NS5B RNA-dependent RNA polymerase have all been shown to preferentially bind to poly-U sequences in vitro [66-68]. The 98-nt 3′ X-tail is the main regulatory CRE required for negative-strand synthesis and is highly conserved across HCV isolates [59, 61]. The X-tail contains three stem-loop structures that are essential in both sequence and structure for HCV RNA replication [57, 64] (Fig. 1C). Notably, the terminal nucleotide in the HCV genome in all isolates is a uridine residue and is engaged in base-pairing interactions in the 3′ terminal SL structure (SL1) of the HCV genome (Fig. 1C). This uridine residue can be replaced with a cytosine and still direct initiation of negative-strand synthesis in HCV replicons, in accordance with the requirement of the viral polymerase to initiate RNA synthesis using a purine base (guanine or adenine) [56]. However, replication of these mutants in cell culture results in reversion or addition of uridine residues, suggesting a strong selective pressure for initiation on a terminal uridine [56, 69].
3.4 Replication elements in the NS5B coding region
In addition to the CREs located in the 3′ NCR, additional CREs have been identified in the NS5B coding region that are important to HCV RNA replication. Most notably is the stem-loop structure designated SL3.2 (or SL9266 according to H77 nomenclature, Genbank Accession #AF011753) that is part of a larger cruciform structure in the NS5B coding region [70] (Fig.1C). The loop of SL3.2 is engaged in a long range RNA-RNA kissing-loop interaction with SL2 of the 3′ X-tail creating a pseudoknot structure at the 3′ end of the HCV genome [70, 71]. Reverse genetic analyses revealed that complementarity between these two loops, rather than the primary sequence, was what was essential for viral RNA replication in cell culture [70, 71]. Interestingly, recent studies have revealed that the bulge region of SL3.2 is also engaged in a long range RNA-RNA interaction with sequences upstream around nucleotide 9110 [72, 73]. When mutations were made in region 9110, there was a significant reduction in viral titers; however, this could be restored to wild-type levels by introduction of compensatory mutations into the bulge of SL3.2 that restored base-pairing [73]. This suggests that a long range RNA-RNA interaction exists between the 3′ sub-terminal bulge of SL3.2 and the 9110 region, and that this pseudoknot promotes efficient replication [73]. Furthermore, the authors provided evidence using in vitro selective 2′ hydroxyl acylation analyzed by primer extension (SHAPE) that the kissing-loop and upstream pseudoknot interaction are mutually exclusive, suggesting that they may serve a switch function that modulates translation and replication events in the HCV life cycle [73]; however, this has yet to be verified independently.
A recent study explored the presence of additional CREs in the nonstructural protein-coding region by systematic introduction of synonymous substitutions within defined segments in the NS3-NS5B coding regions [74]. Consistent with previous computational and biochemical studies, Chu and colleagues found no significant replication or infectivity defects in viruses with synonymous mutations in the NS3, NS4A, NS4B or NS5A regions indicating that these regions do not contain critical CREs [74-78]. However, introduction of silent mutations into multiple segments of the NS5B region resulted in moderate to severe impairment of RNA replication and infectivity, even within regions upstream of the previously identified SL3.2 and pseudoknot interactions [70-74]. By combining computational prediction, mutational analyses and studies of replication and infectivity of mutant viruses, Chu and colleagues identified conserved and non-conserved stem-loop structures in the JFH-1 genome, termed SL8222 (H77-SL8157) and SL9038 (H77-9005), respectively, that are predicted to play a role in genome packaging and assembly of viral particles [74]. In addition, they experimentally verified SL8647 (H77-SL8582), a previously predicted stem-loop structure that acts as an enhancer for efficient viral RNA replication. Furthermore, the authors identified an extended stem-loop structure, termed SL9005, which plays a critical role in HCV genome replication and assembly [74]. Notably, the integrity of the stem structure was important for viral RNA replication and infectivity, and this stem-loop also contains the predicted pseudoknot residues (nt 9110 region) in the 3′ bulge at the base of the extended stem structure [73, 74] (Fig. 1C). Further studies will be required to validate these CREs in additional isolates and elucidate the mechanism of action on viral RNA replication and infectivity.
In addition to the CREs described in the 5′ and 3′ NCRs as well as the distinct stem-loop structures described in the core- and NS5B-coding regions, there is evidence that additional RNA structures exist throughout the polyprotein coding sequence [79, 80]. This includes recently described stem-loop structures in the NS5B coding region as well as the less well-defined, extensive large-scale secondary structure collectively known as genome-scale ordered RNA structure (GORS), that spans the entire coding region of the HCV genome [74, 79, 80].
4. The miR-122 binding sites in HCV RNA
MicroRNAs (miRs) are small (20-23 nt) RNAs that are processed from primary polymerase II-generated transcripts. In the cytoplasm, miRs associate with one of the four Argonaute proteins and several auxiliary factors in an RNA-induced silencing complex (RISC). The RISC binds to mRNAs that contain sequences with partial complementarity to the miR. In particular, most miRs interact with nucleotides 2-8 (seed sequence) via perfect base complementarity with seed match sequences that are usually located in 3′ NCRs of mRNAs [81]. Kinetics analyses have shown that miR-bound mRNAs are first translationally stalled and then subsequently degraded by the CNOT1/CCR4 deadenylase complex [82-84]. Many miRs are expressed in a tissue-specific manner. For example, liver cells express approximately 66,000 copies miR-122, which constitutes 70% of all miRs in the liver [85]. The normal function of miR-122 is the upregulation of fatty acid and sterol biosynthesis in animals [86, 87], most likely via downregulation of a repressor of these pathways.
4.1 miR-122 interacts with two conserved binding sites in the HCV genome and confers stability to the viral mRNA
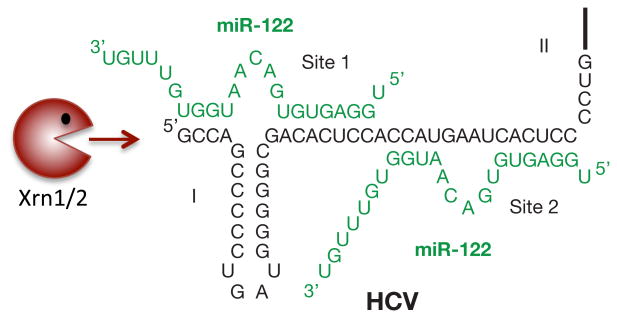
It was noted that the very 5′ end of the HCV RNA genome contains two binding sites for miR-122 that were highly conserved among all HCV genotypes (Table 1). Mutation of the miR-122 sites or sequestration of miR-122 by antisense locked nucleic acid (LNA)-containing oligomers resulted in the loss of HCV RNA [48, 49, 88]. A systematic mutational analysis performed by Machlin and coworkers showed that the miR-122 molecules bind in tandem to the viral RNA (Fig. 2; site 1 and site 2), with both miR-122 molecules exposing an internal bulge in the miR-HCV RNA duplex (Fig. 2). In addition, the six penultimate 3′ nucleotides in site 1-bound miR-122 are predicted to overhang the very 5′ terminus of the viral RNA genome (Fig. 2). As can be seen in Table 1, the two seed match sequences for miR-122 in the viral genome and the two cytidine residues, which stabilize the internal bulge in the miR-HCV RNA complex, are highly conserved among all HCV genotypes. Several studies have pointed to minor roles of the oligomeric miR-HCV RNA complex in translational control; however, the major role of this complex seems to be in the stabilization of the viral RNA [50, 89-92]. Two studies have pointed to roles of cellular 5′ to 3′ exoribonucleases XRN1 and XRN2 in miR-122-mediated protection of the viral RNA [90, 91]. XRN1 resides in the cytoplasm within processing bodies where translationally-stalled mRNAs accumulate and are degraded. Depletion of XRN1 was able to stabilize HCV RNA, but miR-122 was still required for stabilization [90]. In contrast, XRN2 resides mostly in the nucleus and participates in RNA polymerase II transcription termination. Studies by Sedano and Sarnow have shown that depletion of XRN2 greatly stabilized the viral RNA in the absence of miR-122 [91]. It was puzzling that the 5′ triphosphate-containing HCV genome was a substrate for XRN2 which prefers a 5′ monophosphate-containing RNA substrate [93]. However, XRN2 was shown to degrade 5′ triphosphate-containing RNAs, although with lesser efficiency than 5′ monophosphate-containing RNAs [91, 93]. Thus, HCV RNA likely encounters both XRN1 and XRN2 in its life cycle, perhaps at distinct steps that may or may not require miR-122.
Table 1. miR-122 sites are conserved across all HCV isolates.
| Genotype | Subtype | Accession | 5′ noncoding region |
|---|---|---|---|
| 1 | a | AF009606 (H77) | -GCCAGCCCCCUGAUGGGG-GCGACACUCCACCAUGAAUCACUCCCCUGUG |
| b | M58335 | -----------C.......-...............AG.............. | |
| c | D14853 | -..................-.......... G.................... | |
| 2 | a | D00944 | -A..C.....UAAUA....--......... G.................... |
| b | D10988 | -...C..............-..........G.................... | |
| c | D50409 | -A..C.....UAA.A....--.........G.................... | |
| k | AB031663 | U...C.....UAA...... --.........G.................... | |
| 3 | a | D17763 | -A..U...U.U.ACGA..C---..............G.............. |
| b | D49374 | -...U...U.U.UCGA..C---............................. | |
| k | D63821 | -...U...U.U.UCGA..C---............................. | |
| 4 | a | DQ295833* | --------------.....-...............AG.............. |
| 5 | a | D50466* | -A..C.....U.AU.....--...............-.............. |
| 6 | a | D88476* | -.........U.A.C....--.........G....U-.............. |
| b | D84262 | -.........U.A.C....--...............-.............. | |
| d | D84264 | -.........UAAU-....--...............-.............. | |
| f | D63822 | -.........U.AC-....--...............-.............. | |
| h | D84265 | -.........UAAU-....--...............-.............. | |
| k | D84264 | -.........UAAU-....--...............-.............. |
5′ NCRs (nts 1-49) of HCV isolates belonging to each of the confirmed genotypes/subtypes in the euHCVdb: http://euhcvdb.incp.fr/euHCVdb/jsp/nomen_tabl.jsp (Combet C, Garnier N, Charavay C, Grando D, Crisan D, Lopez J, Dehne-Garcia A, Geourjon C, Bettler E, Hulo C, Mercier PL, Bartenschlager R, Diepolder H, Moradpour D, Pawlotsky JM, Rice CM, Trepo C, Penin F and Deléage G. Nucleic Acids Res, 2007, 35:D363-66). Confirmed sequences for genotype 4a, 5a and 6a are truncated at the 5′ end; hence, provisionally assigned sequences were retrieved for analysis.
Figure 2. Interaction of miR-122 with the 5′ noncoding region of HCV.
Confirmed interactions between the 5′ NCR of Hepatitis C virus (HCV) and miR-122 molecules (green). miR-122 binding protects the HCV RNA genome from degradation by cellular exonucleases Xrn1/Xrn2.
It is also conceivable that miR-122 has additional functions during the HCV life cycle, such as shielding the triphosphate-containing terminal nucleotide of the viral RNA from being sensed by cellular sensors of RNA, like RIG-I [50, 94-96]. Moreover, miR-122 binding could modify protein-binding to the HCV genome, modify the viral RNA structure, help localize viral RNA to sites of viral replication or assembly, or alternatively, could help mediate the switch from translation to initiation of negative-strand synthesis. Despite the strong impact of miR-122 on HCV RNA accumulation, recent studies suggest that miR-122 potently enhances viral RNA replication, but is not essential for HCV RNA replication [54, 96, 97].
4.2 Sequestration of miR-122 by antisense oligonucleotides in HCV-infected patients lowers viral yield
Encouraged by the finding that miR-122 sequestration by antisense LNA-substituted oligonucleotides lowered cholesterol abundance in mice and non-human primates without obvious side effects, Santaris Pharma A/S conducted tests on the efficacy of LNAs directed against miR-122 in HCV-infected chimpanzees [86, 98, 99]. Excitingly, it was noted that the LNAs specifically lowered viral yield by several logs in the animals [99]. Subsequently, a dose ascending phase I clinical trial in healthy human volunteers showed that the antisense miR-122 LNAs had no adverse effects. In a landmark phase II clinical trial Janssen and colleagues reported that all nine patients, treated with 5 mg/kg LNA once a week for 4 weeks, lowered HCV viral yield by several logs after subcutaneous administration of LNAs directed against miR-122 [88]. Four out of the nine patients had no detectable virus at week ten after the last dosing. Most importantly, no viral revertants emerged that contained mutations in the miR-122 binding sites. This was very surprising because one could envisage that mutations in the seed match sequences that could allow binding to other cellular miRs could have evolved. Thus, miR-122 likely forms a very specific, higher-ordered oligomeric complex with the viral RNA that is essential to maintain viral RNA abundance in the infected liver.
4.3 miR-122 and hepaciviruses: one microRNA to rule them all?
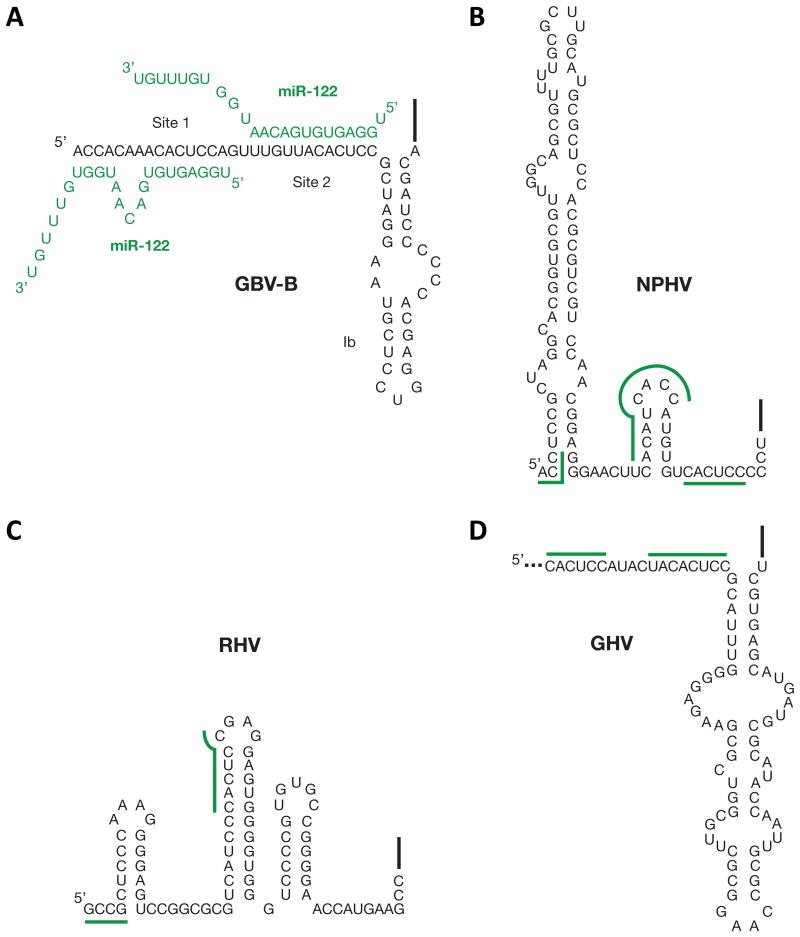
Several recent reports have identified novel HCV homologs in mammals that cluster in the hepacivirus genus [100-104]. Non-primate hepacivirus (NPHV) was first identified from domestic dogs, and subsequently was found in horses [100, 102]. Rodent hepaciviruses (RHV) were also identified in deer mice, four-striped grass mice and bank voles [101, 103]; and bat hepacivirus (BHV) in African and Central American bats [104]. A more recent study revealed a novel primate hepacivirus in an old world monkey, guereza hepacivirus (GHV), which is a close relative of GB virus B (GBV-B), another hepacivirus known to usurp miR-122 for viral RNA accumulation [105, 106]. Interestingly, all of these viruses have putative miR-122 binding sites; save for BHV, for which the complete 5′ NCR is not yet available (Fig. 3). Since miR-122 is primarily expressed in the liver, it is likely to be a key factor in regulating cell tropism. The presence of conserved miR-122 binding sites in this diverse group of viruses suggests that they may have evolved from a common hepatotropic ancestor. Furthermore, it suggests that miR-122 modulation of viral RNA accumulation is not unique to HCV and miR-122 may mediate viral RNA accumulation in diverse hepaciviruses from a variety of host species.
Figure 3. miR-122 interactions with hepacivirus 5′ non-coding regions.
A) Interactions between GB virus B (GBV-B) and miR-122 molecules (green). Predicted binding sites for miR-122 in the 5′ NCRs of B) Non-primate hepacivirus (NPHV), C) Rodent hepacivirus (RHV), and D) guereza hepacivirus (GHV). The precise 5′ terminus of GHV has not been mapped (to date) and only a partial 5′ NCR sequence is available at present. Predicted bindings sites for miR-122 are indicated.
Conclusion
CREs in the HCV genome direct important events in the HCV life cycle. Numerous studies have helped uncover the complex secondary and higher-ordered structures responsible for translation mediated by the IRES, as well as those structures in the 5′ and 3′ NCRs and the coding regions that modulate viral RNA replication. In addition, HCV recruits a cellular miR, miR-122, to the 5′ end of the viral genome that confers stability to the viral RNA from cellular exonucleases XRN1/2. Despite this progress, the precise mechanisms of the CREs in directing the switch from translation to replication, positive- and negative-strand RNA synthesis and viral packaging have yet to be elucidated. This is likely to require more sophisticated models that can reveal the distinct mechanistic functions and the complexities of individual sequences and structures in the HCV life cycle.
Highlights.
HCV contains highly structured 5′ and 3′ NCRs.
he 5′NCR contains an IRES that directs translation of the viral polyprotein.
The 5′ and 3′NCRs and the NS5B-coding region contain CREs required for replication.
The negative-strand 3′ end contains structures required for genomic RNA synthesis.
miR-122 promotes HCV RNA accumulation and stability.
Acknowledgments
S.M.S acknowledges funding provided by McGill University and the Natural Sciences and Engineering Research Council (NSERC) of Canada (RGPIN-2014-05907). P.S. acknowledges funding from the NIH (AI47365, AI069000, GM099687).
Abbreviations
- HCV
hepatitis C virus
- ORF
open reading frame
- NCR
non-coding region
- IRES
internal ribosome entry site
- SL
stem-loop
- NS
non-structural
- miR-122
microRNA-122
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36(5 Suppl 1):S21–9. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 2.Lavanchy D. Evolving epidemiology of hepatitis C virus. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2011;17(2):107–15. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 3.Casey LC, Lee WM. Hepatitis C virus therapy update 2013. Curr Opin Gastroenterol. 2013;29(3):243–9. doi: 10.1097/MOG.0b013e32835ff972. [DOI] [PubMed] [Google Scholar]
- 4.Tsukiyama-Kohara K, et al. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66(3):1476–83. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67(6):3338–44. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reynolds JE, et al. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14(23):6010–20. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol. 2007;5(1):29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 8.Perard J, et al. Structure of the full-length HCV IRES in solution. Nat Commun. 2013;4:1612. doi: 10.1038/ncomms2611. [DOI] [PubMed] [Google Scholar]
- 9.Kieft JS, et al. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292(3):513–29. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 10.Lukavsky PJ, et al. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol. 2003;10(12):1033–8. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 11.Boehringer D, et al. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13(11):1695–706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Spahn CM, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291(5510):1959–62. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 13.Collier AJ, et al. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat Struct Biol. 2002;9(5):375–80. doi: 10.1038/nsb785. [DOI] [PubMed] [Google Scholar]
- 14.Kieft JS, et al. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat Struct Biol. 2002;9(5):370–4. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 15.Wang C, et al. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1(5):526–37. [PMC free article] [PubMed] [Google Scholar]
- 16.Berry KE, et al. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19(10):1456–66. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava S, Verschoor A, Frank J. Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit-subunit interface. J Mol Biol. 1992;226(2):301–4. doi: 10.1016/0022-2836(92)90946-h. [DOI] [PubMed] [Google Scholar]
- 18.Pestova TV, et al. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12(1):67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niepmann M. Hepatitis C virus RNA translation. Curr Top Microbiol Immunol. 2013;369:143–66. doi: 10.1007/978-3-642-27340-7_6. [DOI] [PubMed] [Google Scholar]
- 20.Maraia RJ, Lamichhane TN. 3′ processing of eukaryotic precursor tRNAs. Wiley Interdiscip Rev RNA. 2011;2(3):362–75. doi: 10.1002/wrna.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci U S A. 1997;94(6):2249–54. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izumi RE, et al. A peptide from autoantigen La blocks poliovirus and hepatitis C virus cap-independent translation and reveals a single tyrosine critical for La RNA binding and translation stimulation. J Virol. 2004;78(7):3763–76. doi: 10.1128/JVI.78.7.3763-3776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pudi R, Srinivasan P, Das S. La protein binding at the GCAC site near the initiator AUG facilitates the ribosomal assembly on the hepatitis C virus RNA to influence internal ribosome entry site-mediated translation. J Biol Chem. 2004;279(29):29879–88. doi: 10.1074/jbc.M403417200. [DOI] [PubMed] [Google Scholar]
- 24.Chen HH, et al. The RNA binding protein hnRNP Q modulates the utilization of exon 7 in the survival motor neuron 2 (SMN2) gene. Mol Cell Biol. 2008;28(22):6929–38. doi: 10.1128/MCB.01332-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee KH, et al. Rhythmic interaction between Period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Mol Cell Biol. 2012;32(3):717–28. doi: 10.1128/MCB.06177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SM, et al. Translation-competent 48S complex formation on HCV IRES requires the RNA-binding protein NSAP1. Nucleic Acids Res. 2011;39(17):7791–802. doi: 10.1093/nar/gkr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahm B, et al. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J Virol. 1998;72(11):8782–8. doi: 10.1128/jvi.72.11.8782-8788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang B, et al. hnRNP L is required for the translation mediated by HCV IRES. Biochem Biophys Res Commun. 2009;378(3):584–8. doi: 10.1016/j.bbrc.2008.11.091. [DOI] [PubMed] [Google Scholar]
- 29.Paek KY, et al. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J Virol. 2008;82(24):12082–93. doi: 10.1128/JVI.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yisraeli JK. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol Cell. 2005;97(1):87–96. doi: 10.1042/BC20040151. [DOI] [PubMed] [Google Scholar]
- 31.Isken O, et al. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 2003;22(21):5655–65. doi: 10.1093/emboj/cdg562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinlich S, et al. IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3′UTR. RNA. 2009;15(8):1528–42. doi: 10.1261/rna.1578409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JH, et al. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J Mol Biol. 2000;298(3):395–405. doi: 10.1006/jmbi.2000.3687. [DOI] [PubMed] [Google Scholar]
- 34.Moraes KC, et al. Identification and characterization of proteins that selectively interact with isoforms of the mRNA binding protein AUF1 (hnRNP D) Biol Chem. 2003;384(1):25–37. doi: 10.1515/BC.2003.004. [DOI] [PubMed] [Google Scholar]
- 35.Park HG, Yoon JY, Choi M. Heterogeneous nuclear ribonucleoprotein D/AUF1 interacts with heterogeneous nuclear ribonucleoprotein L. J Biosci. 2007;32(7):1263–72. doi: 10.1007/s12038-007-0135-8. [DOI] [PubMed] [Google Scholar]
- 36.Gubitz AK, Feng W, Dreyfuss G. The SMN complex. Exp Cell Res. 2004;296(1):51–6. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 37.Khusial P, Plaag R, Zieve GW. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem Sci. 2005;30(9):522–8. doi: 10.1016/j.tibs.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Scheller N, et al. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci U S A. 2009;106(32):13517–22. doi: 10.1073/pnas.0906413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuchs G, et al. Kinetic pathway of 40S ribsomal subunit recruitment to hepatitis C virus internal ribosome entry site. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1421328111. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuplin A, Evans DJ, Simmonds P. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J Gen Virol. 2004;85(10):3037–47. doi: 10.1099/vir.0.80141-0. [DOI] [PubMed] [Google Scholar]
- 41.McMullan LK, et al. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci U S A. 2007;104(8):2879–84. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vassilaki N, et al. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J Virol. 2008;82(23):11503–15. doi: 10.1128/JVI.01640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinmann E, et al. Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles. J Virol. 2008;82(14):7034–46. doi: 10.1128/JVI.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lohmann V. Hepatitis C virus RNA replication. Curr Top Microbiol Immunol. 2013;369:167–98. doi: 10.1007/978-3-642-27340-7_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friebe P, et al. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. Journal of virology. 2001;75(24):12047–12057. doi: 10.1128/JVI.75.24.12047-12057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim YK, et al. Domains I and II in the 5′ nontranslated region of the HCV genome are required for RNA replication. Biochemical and biophysical research communications. 2002;290(1):105–112. doi: 10.1006/bbrc.2001.6167. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Jeng KS, Lai MM. Poly (C)-binding protein 2 interacts with sequences required for viral replication in the hepatitis C virus (HCV) 5′ untranslated region and directs HCV RNA replication through circularizing the viral genome. Journal of virology. 2011;85(16):7954–7964. doi: 10.1128/JVI.00339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4(1):77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jopling CL, et al. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 50.Machlin ES, Sarnow P, Sagan SM. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci U S A. 2011;108(8):3193–8. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dutkiewicz M, et al. Structural domains of the 3′-terminal sequence of the hepatitis C virus replicative strand. Biochemistry. 2008;47(46):12197–207. doi: 10.1021/bi800348g. [DOI] [PubMed] [Google Scholar]
- 52.Schuster C, et al. Secondary structure of the 3′ terminus of hepatitis C virus minus-strand RNA. J Virol. 2002;76(16):8058–68. doi: 10.1128/JVI.76.16.8058-8068.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith RM, et al. Secondary structure and hybridization accessibility of hepatitis C virus 3′-terminal sequences. J Virol. 2002;76(19):9563–74. doi: 10.1128/JVI.76.19.9563-9574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Friebe P, Bartenschlager R. Role of RNA structures in genome terminal sequences of the hepatitis C virus for replication and assembly. Journal of virology. 2009;83(22):11989–11995. doi: 10.1128/JVI.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahias K, et al. Identification of a structural element of the hepatitis C virus minus strand RNA involved in the initiation of RNA synthesis. Nucleic acids research. 2010;38(12):4079–4091. doi: 10.1093/nar/gkq109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cai Z, Liang TJ, Luo G. Effects of mutations of the initiation nucleotides on hepatitis C virus RNA replication in the cell. J Virol. 2004;78(7):3633–43. doi: 10.1128/JVI.78.7.3633-3643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friebe P, Bartenschlager R. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J Virol. 2002;76(11):5326–38. doi: 10.1128/JVI.76.11.5326-5338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Binder M, et al. Identification of determinants involved in initiation of hepatitis C virus RNA synthesis by using intergenotypic replicase chimerase. Journal of Virology. 2007;81(10):5270–5283. doi: 10.1128/JVI.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolykhalov AA, Feinstone SM, Rice CM. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70(6):3363–71. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka T, et al. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem Biophys Res Commun. 1995;215(2):744–9. doi: 10.1006/bbrc.1995.2526. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka T, et al. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70(5):3307–12. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamada N, et al. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology. 1996;223(1):255–61. doi: 10.1006/viro.1996.0476. [DOI] [PubMed] [Google Scholar]
- 63.Ito T, Lai MMC. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. Journal of Virology. 1997;71(11):8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yi M, Lemon SM. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J Virol. 2003;77(6):3557–68. doi: 10.1128/JVI.77.6.3557-3568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.You S, Rice CM. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U) J Virol. 2008;82(1):184–95. doi: 10.1128/JVI.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gwack Y, et al. Characterization of RNA binding activity and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem Biophys Res Commun. 1996;225(2):654–9. doi: 10.1006/bbrc.1996.1225. [DOI] [PubMed] [Google Scholar]
- 67.Huang L, et al. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J Biol Chem. 2005;280(43):36417–28. doi: 10.1074/jbc.M508175200. [DOI] [PubMed] [Google Scholar]
- 68.Lohmann V, et al. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71(11):8416–28. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yi M, Lemon SM. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA. 2003;9(3):331–45. doi: 10.1261/rna.2144203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.You S, et al. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J Virol. 2004;78(3):1352–66. doi: 10.1128/JVI.78.3.1352-1366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Friebe P, et al. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J Virol. 2005;79(1):380–92. doi: 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Diviney S, et al. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J Virol. 2008;82(18):9008–22. doi: 10.1128/JVI.02326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tuplin A, et al. A twist in the tail: SHAPE mapping of long-range interactions and structural rearrangements of RNA elements involved in HCV replication. Nucleic Acids Res. 2012;40(14):6908–21. doi: 10.1093/nar/gks370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chu D, et al. Systematic analysis of enhancer and critical cis-acting RNA elements in the protein-encoding region of the hepatitis C virus genome. J Virol. 2013;87(10):5678–96. doi: 10.1128/JVI.00840-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hofacker IL, et al. Automatic detection of conserved RNA structure elements in complete RNA virus genomes. Nucleic Acids Res. 1998;26(16):3825–36. doi: 10.1093/nar/26.16.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith DB, Simmonds P. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J Mol Evol. 1997;45(3):238–46. doi: 10.1007/pl00006226. [DOI] [PubMed] [Google Scholar]
- 77.Jones DM, et al. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J Virol. 2009;83(5):2163–77. doi: 10.1128/JVI.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Appel N, Herian U, Bartenschlager R. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J Virol. 2005;79(2):896–909. doi: 10.1128/JVI.79.2.896-909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simmonds P, Tuplin A, Evans DJ. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: Implications for virus evolution and host persistence. RNA. 2004;10(9):1337–51. doi: 10.1261/rna.7640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Davis M, et al. Bioinformatic and physical characterizations of genome-scale ordered RNA structure in mammalian RNA viruses. J Virol. 2008;82(23):11824–36. doi: 10.1128/JVI.01078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lewis BP, et al. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 82.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336(6078):237–40. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meijer HA, et al. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340(6128):82–5. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 84.Chen Y, et al. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell. 2014;54(5):737–50. doi: 10.1016/j.molcel.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 85.Chang J, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1(2):106–13. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 86.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 87.Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006;4(1):9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 88.Janssen HL, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 89.Henke JI, et al. microRNA-122 stimulates translation of hepatitis C virus RNA. Embo J. 2008;27(24):3300–10. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Y, et al. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc Natl Acad Sci U S A. 2013;110(5):1881–6. doi: 10.1073/pnas.1213515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sedano CD, Sarnow P. Hepatitis C virus subverts liver-specific miR-122 to protect the viral genome from exoribonuclease Xrn2. Cell Host Microbe. 2014;16(2):257–64. doi: 10.1016/j.chom.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wilson JA, et al. Human Ago2 is required for efficient microRNA 122 regulation of hepatitis C virus RNA accumulation and translation. Journal of virology. 2011;85(5):2342–50. doi: 10.1128/JVI.02046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stevens A, Maupin MK. A 5′----3′ exoribonuclease of human placental nuclei: purification and substrate specificity. Nucleic acids research. 1987;15(2):695–708. doi: 10.1093/nar/15.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saito T, et al. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454(7203):523–7. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schnell G, et al. Uridine composition of the poly-U/UC tract of HCV RNA defines non-self recognition by RIG-I. PLoS Pathog. 2012;8(8):e1002839. doi: 10.1371/journal.ppat.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wilson JA, Sagan SM. Hepatitis C virus and human miR-122: insights from the bench to the clinic. Curr Opin Virol. 2014;7:11–8. doi: 10.1016/j.coviro.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 97.Thibault PA, et al. MicroRNA-122-dependent and -independent replication of Hepatitis C Virus in Hep3B human hepatoma cells. Virology. 2013;436(1):179–90. doi: 10.1016/j.virol.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 98.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 99.Lanford RE, et al. Lack of response to exogenous interferon-alpha in the liver of chimpanzees chronically infected with hepatitis C virus. Hepatology. 2007;46(4):999–1008. doi: 10.1002/hep.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Burbelo PD, et al. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J Virol. 2012;86(11):6171–8. doi: 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Drexler JF, et al. Evidence for novel hepaciviruses in rodents. PLoS Pathog. 2013;9(6):e1003438. doi: 10.1371/journal.ppat.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kapoor A, et al. Characterization of a canine homolog of hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108(28):11608–13. doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kapoor A, et al. Identification of rodent homologs of hepatitis C virus and pegiviruses. MBio. 2013;4(2):e00216–13. doi: 10.1128/mBio.00216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Quan PL, et al. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad Sci U S A. 2013;110(20):8194–9. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lauck M, et al. A novel hepacivirus with an unusually long and intrinsically disordered NS5A protein in a wild Old World primate. J Virol. 2013;87(16):8971–81. doi: 10.1128/JVI.00888-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sagan SM, Sarnow P, Wilson JA. Modulation of GB virus B RNA abundance by microRNA-122: dependence on and escape from microRNA-122 restriction. J Virol. 2013;87(13):7338–47. doi: 10.1128/JVI.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]