Abstract
This highlight covers a family of enzymes of growing importance, the sedoheptulose 7-phosphate cyclases, initially of interest due to their involvement in the biosynthesis of pharmaceutically relevant secondary metabolites. More recently, these enzymes have been found throughout Prokarya and Eukarya, suggesting their broad potential biological roles in nature.
Graphical Abstract
We highlight a family of enzymes of growing importance, which are found throughout Prokarya and Eukarya and are involved in primary and secondary metabolism.
1. Scope and purpose
This highlight focuses on a subgroup of emerging importance among enzymes that catalyze the cyclization of C7-sugar phosphates to cyclic compounds as a part of primary and/or secondary metabolism. Related to the well-studied 3-dehydroquinate synthases (DHQS) from the shikimate pathway, this subgroup of cyclases use sedoheptulose 7-phosphate (SH7P) as a substrate. There are now three known sedoheptulose 7-phosphate cyclases (SH7PC), which convert their common substrate to three different cyclic products.1 They are 2-epi-5-epi-valiolone synthases (EEVS), converting SH7P to 2-epi-5-epi-valiolone; desmethyl-4-deoxygadusol synthase (DDGS), converting SH7P to desmethyl-4-deoxygadusol; and 2-epi-valiolone synthase (EVS), converting SH7P to 2-epi-valiolone. EEVS was originally thought to be present only in certain actinomycetes where it is part of the pathways to C7N-aminocyclitol natural products; however, analyses of genome sequences revealed that EEVS is broadly distributed in both prokaryotes and eukaryotes, including bacteria, fungi, stramenopiles, and animals.2 A recent study demonstrated that the role of EEVS in animals and some algae is to produce the sunscreen-like compound gadusol.3 DDGS was originally discovered based on its role in the biosynthesis of mycosporines and mycosporine-like amino acids (MAAs),4 another class of sunscreen compounds believed to play a critical role in ecology, whereas EVS was found to be involved in the biosynthesis of bioactive C7N-aminocyclitol natural products.5 In this highlight, we describe the discovery, biochemistry, and structure-function relationships of SH7PCs and discuss the distribution and the emerging roles of these less characterized sugar phosphate cyclases (SPCs) in biology and ecology.
2. Sugar phosphate cyclases and their roles in primary and secondary metabolism
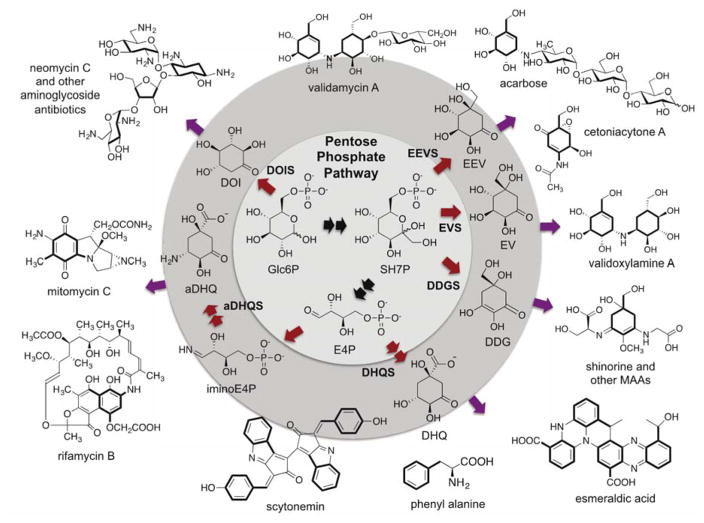
As noted above, SPCs catalyze the cyclization of six- and seven-carbon sugar phosphates to a variety of cyclitol products, which are part of primary and secondary metabolism. The one SPC superfamily member directly involved in primary metabolism is DHQS, which catalyzes one of the rate limiting steps of the shikimate pathway – the cyclization of 3-deoxy-D-arabinoheptulosonate 7-phosphate (DAHP) to 3-dehydroquinate (DHQ) – and is necessary for the synthesis of aromatic amino acids.6 Seminal work by Floss and others have shown that a close homolog of DHQS, known as aminoDHQS, is involved in secondary metabolism (e.g., in the biosynthesis of rifamycin and mitomycin), but it follows the template of DHQS in the cyclization of an amino sugar phosphate.7 AminoDHQS uses aminoDAHP to form aminoDHQ, which is further modified to 3-amino-5-hydroxybenzoic acid (AHBA).8 The latter compound is then usually used as a starter unit for polyketide synthases. In its own right, DHQS can also be linked to secondary metabolism through aromatic amino acid-derived compounds, such as esmeraldic acid and scytonemin (Figure 1).
Figure 1.
Sugar phosphate cyclases (SPCs), their products, and representative natural products resulting from their pathways. Glc6P, glucose 6-phosphate; SH7P, sedoheptulose 7-phosphate; E4P, erythrose 4-phosphate; iminoE4P, imino-erythrose 4-phosphate; EEV, 2-epi-5-epi-valiolone, EV, 2-epi-valiolone, DDG, desmethyl-4-deoxygadusol; DHQ, 3-dehydroquinate; aDHQ, 3-aminodehydroquinate; DOI, 2-deoxy-scyllo-inosose.
Other known SPCs use intermediates of the pentose phosphate pathway, which is a key primary metabolic pathway that serves as a source of NADPH, ribose 5-phosphate, and erythrose 4-phosphate. 2-Deoxy-scyllo-inosose synthase (DOIS) uses glucose 6-phosphate as substrate and generates 2-deoxy-scyllo-inosose (DOI), a key scaffold in aminoglycoside antibiotics (e.g., neomycin and kanamycin). The sedoheptulose 7-phosphate cyclases (SH7PCs) catalyze the cyclization of SH7P to give 2-epi-5-epi-valiolone (EEV), desmethyl-4-deoxygadusol (DDG), or 2-epi-valiolone (EV). These cyclitol molecules can be modified and functionalized further to generate molecules that are both diverse in terms of structure and function, such as the fungicide validamycin, the cytotoxin cetoniacytone A, and the UV-absorbant MAAs. In a sense, SPCs can be considered to be a gateway between primary and secondary metabolism, as they convert intermediates from the pentose phosphate pathway to secondary metabolites. Due to the diversity of these secondary metabolites and the organisms that synthesize them, it is also possible that aside from their functions as secondary metabolites they may have a larger role in primary metabolism than what is currently known.
3. Sedoheptulose 7-phosphate cyclases (SH7PCs)
3.1. 2-Epi-5-epi-valiolone synthases (EEVSs)
The identification of this new class of enzymes was first reported in 1999 when feeding experiments revealed that the valienamine moiety of acarbose, an α-glucosidase inhibitor used for treating type II diabetes, was derived from EEV.9 Furthermore, biochemical characterization of a DHQS-like protein (AcbC) within the acarbose pathway in Actinoplanes sp. 50/110 demonstrated that the protein could convert SH7P to EEV.10 Multiple sequence alignments and the crystal structure of DHQS, showed that AcbC and DHQS matched for 7 of 13 key residues associated with catalysis and substrate binding, metal binding, and NAD+ binding, and with notable differences postulated to be localized around the varying substituents of their SH7P and DAHP substrates. Based on this, the catalytic mechanism for EEVS was proposed to be similar to DHQS involving 5 step reactions: dehydrogenation by NAD+, elimination of the phosphate group, reduction of the C-5 ketone, ring opening and rotation along the C5 – C6 bond, and intramolecular aldol cyclization (Figure 2A).10
Figure 2.
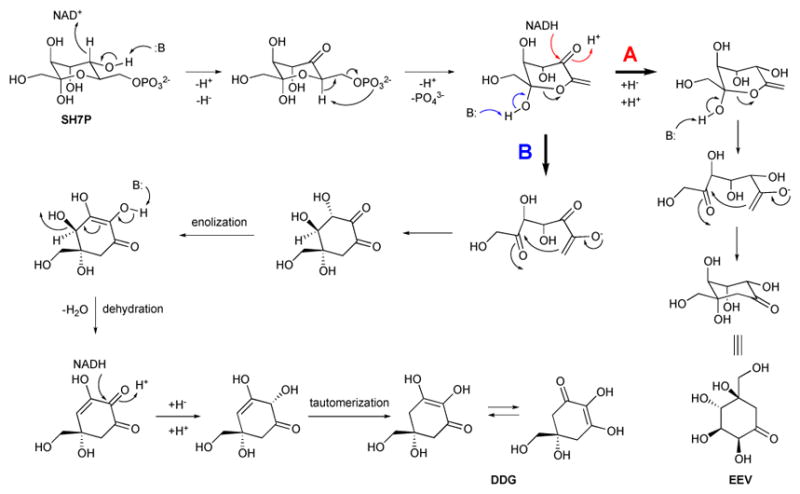
Proposed catalytic mechanisms for EEVS and DDGS. (A) Proposed mechanism for EEVS. (B) Proposed mechanism for DDGS.
After the initial identification of the AcbC protein as an enzyme that uses SH7P to make EEV, subsequent work showed 2-epi-5-epi-valiolone is incorporated into the secondary metabolites validamycin,11 cetoniacytone A,12 and pyralomicin,13 suggesting these biosynthetic pathways also contain an AcbC-ortholog. The corresponding AcbC-ortholog genes found within the biosynthetic gene clusters of validamycin (ValA),14 cetoniacytone A (CetA),12 BE-40644 (BE-Orf9),12 salbostatin (SalQ),15 acarbose in Streptomyces glaucescens (GacC),16 and pyralomicin (prlA),17 were identified and shown to be EEVSs.
While originally thought to be limited to bacteria, EEVS genes have recently been identified in algae, marine invertebrates, non-mammalian vertebrates, and a fungus, and are implicated in the de novo production of gadusol in some of these organisms.3 Based on heterologous expression and characterization of this gene from zebrafish, this too is an authentic EEVS – using SH7P to make EEV. And still more recently, a group of phylogenetically distinct, bacterial EEVS (referred to as EEVS*), have been identified and shown to have EEVS activity.2
3.2. Desmethyl-4-deoxygadusol synthases (DDGSs)
The identification of DDGSs in a number of cyanobacteria as a distinct group of SPCs was first reported in 2007.12 In 2010, Balskus and Walsh experimentally demonstrated that a DDGS, as well as an O-methyltransferase (O-MT), a ATP-grasp protein, and a NRPS-like protein, are responsible for the synthesis of MAAs, contradicting the longstanding assumption that MAAs are synthesized via the shikimate pathway.4 DDGS, e.g., Ava_3858 of Anabaena variabilis and Npun_5600 of Nostoc punctiforme, converts SH7P to DDG. A proposed mechanism for DDGS is similar to those of other SPCs, but with additional enolization, dehydration, and tautomerization steps (Figure 2B).1
In addition to these DDGSs, other putative and divergent DDGSs2 have been identified and characterized to varying degrees.18–22 Likewise, these genes are found with an O-MT or even fused with an O-MT and have been shown to be responsible for MAA production via heterologous expression and characterization.22 Many of these proteins contain N-terminal extensions or plastid targeting sequences and in some cases evidence supports horizontal gene transfer as part of the evolutionary history of these enzymes18–21, 23 (see section 6: Evolution of SH7PC by horizontal gene transfer).
3.3. 2-Epi-valiolone synthases (EVS)
EVSs appear to be the least common among the SH7PC family of enzymes and were only identified and differentiated from other SPCs in 2012.1 Only two EVSs have been heterologously expressed and characterized for their activity, showing they use SH7P as substrate to make EV, but more EVS were identified through bioinformatics studies.1–3 Prior to the characterization of EVS, its product (EV) had not been identified in nature. Despite its similarity in function to EEVS and DDGS, EVSs appear to be more similar to DHQS in their active site.1 The catalytic mechanism for EVS was proposed to be very similar to those for DHQS and EEVS, but accounted for the difference in configuration at C-5 between EEV and EV by requiring a 180° rotation prior to ring closing. In an alternative hypothesis, EEVS and EVS differ by the anomer of SH7P they bind (see section 4.2).24
EVSs continue to remain to be the most elusive and least well-understood members of the SH7PC family, in part due to their more recent identification, the greater challenge in differentiating EVS from DHQS, and the apparent limited presence of EVS in nature. However, EVS has been identified as replacing EEVS in some C7N-aminocyclitol gene clusters.5 For instance, gene knockout and comparative metabolomics with the wild-type Actinosynnema mirum identified Amir_2000 as being involved in the biosynthesis of validoxylamine A, a precursor to validamycin A. This was both the first C7N-aminocyclitol identified in A. mirum, and represented a new route for the biosynthesis of validoxylamine A.
4. Structural aspects of SH7PCs
Until recently, most inferences about structure-function relations for SH7PCs have been based on the well-studied DHQS.1, 4, 10, 12 In 2014, Kean et al.24 published the first structure of an SH7PC, that of ValA, the EEVS from S. hygroscopicus in the validamycin pathway. In 2017, the structure of a DDGS, Ava_3858 from A. variabilis involved in the production of MAAs, was published.2 These structures are highly similar to each other and reveal features shared among the SPC superfamily as well as features unique to the SH7PCs and the individual enzymes EEVS and DDGS. Unfortunately, no structural information is yet available for the third SH7PC, EVS, which surprisingly has more similarity to DHQS than to the other SH7PCs. Also no complexes of ligand-bound structures have been solved.
4.1. Overall structures and active site
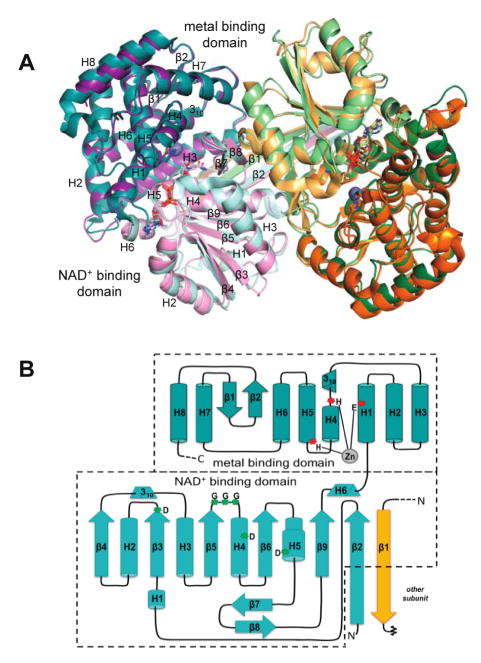
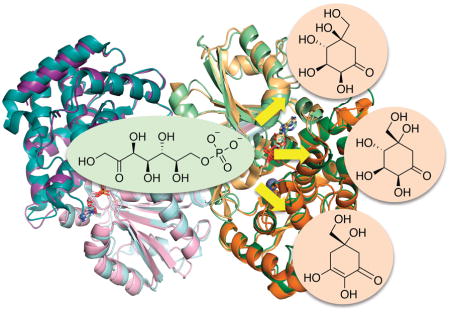
In terms of the fold and broad aspects of active site organization, EEVS and DDGS are quite similar to DHQS. Both form functionally obligatory homodimers, with both chains of the dimer contributing to each active site. Each chain contains bound NAD and Zn prosthetic groups and consists of an N-terminal NAD-binding domain with a parallel β-sheet core and a C-terminal, mostly α-helical metal-binding domain (Figure 3). The active site is in a cleft formed where N and C-terminal domains come together. Among the SPC superfamily, the identities of the metal-binding residues are conserved as 1 Glu and 2 His residues. A structural feature observed in EEVS and DDGS but not in DHQS is a domain swapping interaction in which the N-terminal residues make an extended β-strand (labeled β1/β2) that reaches across the back of the dimer and contributes a β-strand to the opposite monomer of the dimer (Figure 3), rather than forming a β-hairpin as observed in DHQS.
Figure 3.
Overall structure and topology of known SH7PCs, EEVS and DDGS. (A) Overlay of the dimers of the Ava_3858 (teal and orange; PDB code 5TPR) and ValA (purple and green; PDB code 4P53) are shown with the N-terminal NAD+-binding domains in light hues and C-terminal metal-binding domains in dark hues. The NAD+ and Zn2+ with its coordinating ligands are shown for each. Secondary structural elements in each domain of one monomer are labeled. (B) Topology diagram showing α-helices (cylinders), β-strands (arrows), 310 helices (triangular prisms) and π-helices (wider cylinder) common to EEVS and DDGS. The domains are colored light and dark teal as indicated, and helices (H) and strands (β) common to the SPCs are named within each domain. The domain swapped β-strand β1 from the other subunit of the dimer but contributing to the NAD+-binding domain of the teal subunit is colored in light orange. The three Zn2+-binding residues (red asterisks) and glycine-rich turn and conserved aspartic acids (green asterisks) important for NAD+ binding are indicated.
In the ValA active site, it was noted that many features are well conserved with DHQS. One conserved feature that had apparently not been highlighted in the DHQS literature was a conserved pair of Asp residues (Asp138 and Asp165 in ValA) which have their carboxylate oxygens positioned to electrostatically stabilize the oxidized form of the nicotinamide ring during catalysis (See Fig 2 of 24). Also, key active site differences have been identified in EEVS, DDGS, and EVS, which differentiate them from each other and from the broader SPC superfamily. Based on sequence comparisons, EEVS and DDGS are the more similar members of the SH7PC family, and the structures of ValA and Ava_3858 confirmed two previously identified pairs of residues predicted to differ in their active sites (Asp281 and His360 in ValA vs. Ala268 and Thr347 in Ava_3858), and identified a third difference (Leu267 in ValA vs. Glu254 in Ava_3858).2, 24 To assess how these differences contribute to the different catalytic activities of EEVS and DDGS, a series of mutants were generated and activity assays showed that Asp281 and Leu267 in ValA and the equivalent residues Ala268 and Glu254 in Ava_3858 are essential for activity.2 However, the specific roles these residues play in catalysis and how they lead to differential activity are not yet understood.
The comparison of the ValA structure (an EEVS) with DHQS also revealed an important surprise related to an active site Arg located at the beginning of helix 4 in the C-terminal domain.24 This Arg is also present in DDGS, but does not correspond to that found in the DHQS active site; instead it is the next residue. This alternate Arg is associated with a robust difference in a loop conformation and the sequence of EVS implies that it will conserve the Arg found in DHQS (See Fig. 7 of 24). Interestingly, these Arg residues are positioned near where we expect the substituents of the anomeric carbon (C2) of SH7P to bind, making it plausible that this arginine is involved in differential anomer recognition between EVS and EEVS/DDGS.
4.2. An anomeric selection hypothesis
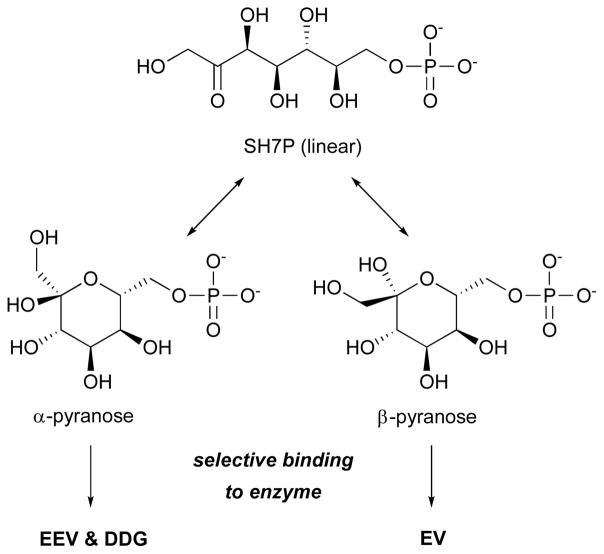
In light of these structures and conservation patterns among the SPC superfamily, we proposed a new hypothesis to explain how the SH7PCs can generate such different products from the same substrate.24 The SH7PCs had previously been viewed as binding a single substrate, the α-pyranose form of SH7P, and this meant that the mechanism proposed for EVS required a portion of the ring-open enzyme bound intermediate to undergo a 180° rotation after ring opening so that the C-C bond formation ring-closure step would yield the product with the correct stereochemistry.1 However, in reality, SH7P can readily interconvert in solution between a ring and open form and the α and β and pyranose and furanose forms. Based on the EEVS structure, and specifically the fact that EEVS and DDGS conserved a different Arg residue than did EVS, we proposed that a more structurally compelling hypothesis would be that these enzymes bind different anomers of SH7P that correspond to the stereochemistry of their final product (Figure 4). This would allow each enzyme to produce the correct product without requiring a dramatic rotation within the spatial constraints of the active site pocket.
Figure 4.
Proposed enzyme-specific selection of forms of sedoheptulose 7-phosphate (SH7P). EEV, 2-epi-5-epi-valiolone; DDG, desmethyl-4-deoxygadusol; EV, 2-epi-valiolone.
4.3. A speculative proposal for SH7P binding to DDGS/EEVS
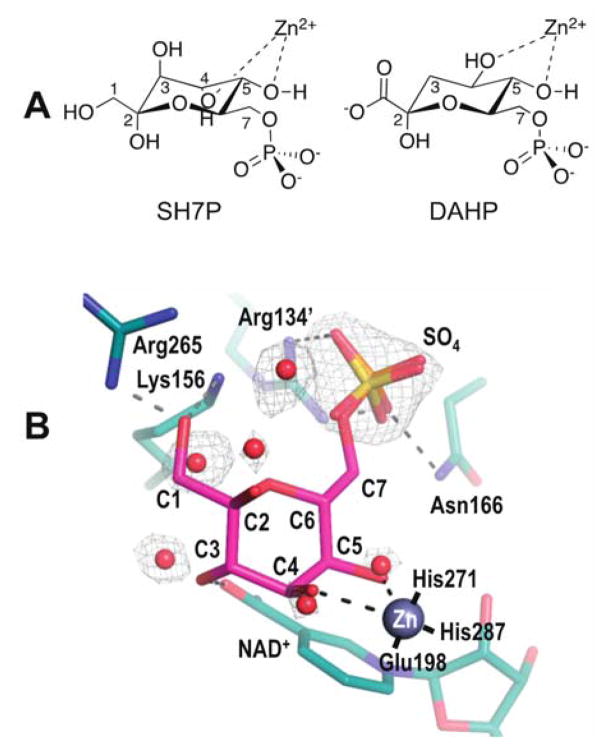
General features of substrate binding by the SH7PCs have been proposed based on what was seen for DHQS, but it has been acknowledged that the DHQS complex (PDB code 1DQS) cannot match in detail the substrate binding by the SH7PCs, in part because of the stereochemical differences in key metal-binding hydroxyls of their substrates (Figure 5A): in DAHP and its substrate analogue, CBP, both metal-coordinating hydroxyls are equatorial while in SH7P one metal-coordinating hydroxyl is equatorial while the other is axial. As part of preparing this highlight, we thought it would be valuable to better define what can be reasonably predicted about how the α-pyranose form of SH7P might bind to the DDGS and EEVS active sites. We created an idealized chair conformation of the substrate and manually docked it into the DDGS active site using the positions of ordered solvent molecules (water and a sulfate) to approximate the locations of the SH7P hydroxyl and phosphoryl substituents. Gratifyingly, this led to a unique and plausible binding mode shown in Figure 5B.
Figure 5.
Substrate recognition by SH7PCs. (A) The boat conformation of DAHP as it coordinates Zn2+ in the DHQS active site has both Zn-ligating hydroxyls equatorial, yet in SH7P one putative Zn-ligating hydroxyl is equatorial while the other Zn-ligating hydroxyl is axial. (B) Speculative binding mode of SH7P in EEVS and DDGS. The α-pyranose anomer of SH7P (magenta) manually docked into the active site of Ava_3858 (teal) with bound zinc (silver sphere). The electron density (grey, contoured at 1.3ρrms) evidence for the bound waters (red spheres) and sulfate (yellow) is shown along with proposed interactions of the docked SH7P with the active site (dashed lines). While Lys156 and Arg265 have short approach distances in this model, we expect these sidechains will move due to an anticipated conformation change upon substrate binding. The coordination of Zn with its three ligating residues are also shown (solid black lines). The carbons of SH7P the protein side chains and NAD+ are labeled, with a prime symbol meaning the side chain comes from the other subunit of the dimer.
In this docked model, both the axial C4-hydroxyl and the equatorial C5-hydroxyl of SH7P are positioned well to coordinate the Zn. Also, the phosphoryl group matches well with the bound sulfate and many of the substituent hydroxyls are positioned to form hydrogen bonds with the protein. Especially notable is that the hydroxymethyl of the anomeric carbon (C2) is positioned to hydrogen bond with both Lys156 and Arg265 (the above mentioned Arg distinguishing EEVS and DDGS from EVS), consistent with this Arg being responsible for anomer recognition. The arginine conserved in EVS would be in a different position, consistent with EVS binding the opposite anomer. Important to note is that side chain positions, especially of Lys156 and Arg265, should be taken as very rough because upon substrate binding, we expect that the DDGS structure used for modeling will undergo some domain closure similar to that observed in DHQS.25
5. Natural distribution of SH7PCs
5.1. SH7PCs in bacteria
Bacterial SH7PCs have been found in Gram-(+) and Gram-(−) bacteria isolated from soils, aquatic systems (fresh and salt), symbionts, pathogens, and extreme environments.1–3 This diversity suggests SH7P-derived natural products have many biological roles, which will be discussed later in this Highlight. The majority of identified bacterial SH7PCs are from actinobacteria (EEVS) and cyanobacteria (DDGS).2 Bacterial EEVS have been reported in a number of C7N-aminocyclitol gene clusters (e.g., acarbose and validamycin), whereas DDGS has only been found in MAA gene clusters.2 EVS appears with much fewer frequency and is limited to soil actinobacteria and myxobacteria.1 Divergent DDGS and EEVS* sequences have also been identified through phylogenetics studies, although only EEVS* was enzymatically characterized.1–3
The bulk of known EEVS sequences are from the genus Streptomyces, in which the C7N-aminocyclitol gene clusters, usually resembling the acarbose and validamycin gene clusters, are widespread.2 However, EEVS is also found in diverse Gram-(+) bacteria, including marine bacteria (e.g., Salinispora spp.), pathogenic bacteria (e.g., Rhodococcus fascians, Mycobacterium marinum, and Clostridium botulinum), other soil-dwelling bacteria (e.g., Amycolatopsis, Nonomuraea, Nocardia, Kitasatospora) and plant symbionts (e.g., Frankia alni).1–3 Of these, only C. botulinum is not an actinobacteria. Likewise, few EEVS* genes are found outside of actinobacteria and myxobacteria, most notably in Bacillus cereus. EEVS is also found in Gram-(−) bacteria, which include the genera Burkholderia, Paraburkholderia, Pseudomonas, and Rhodanobacter. The EEVS of P. kirkii is involved in the biosynthesis of kirkamide (Figure 6), an aminocyclitol with cytotoxicity towards aquatic anthropods and insects, while the gene clusters including EEVS in other Gram-(−) bacteria remain uncharacterized.2, 26
Figure 6.
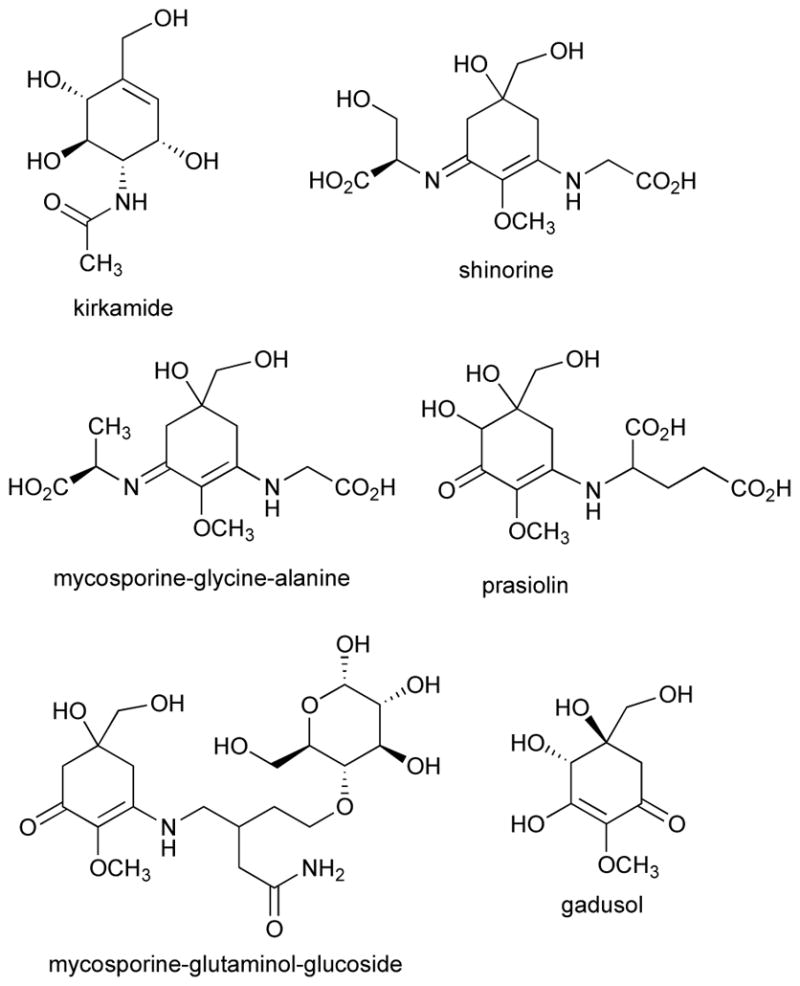
Chemical structures of kirkamide, gadusol, and representatives of mycosporines and MAAs. Some stereoconfigurations were not assigned.
Cyanobacteria have the most species with DDGS encoded in their genome and are found in MAA gene clusters. MAAs have been found in cyanobacteria from fresh and salt water, high salt conditions, and terrestrial environments.2, 22, 27 Compared to products deriving from EEVS-containing gene clusters, the products of MAA gene clusters are easier to detect due to the structural similarity that essentially only diverges by which amino acids are attached to the 4-deoxygadusol core. However, this similarity likely requires further structural elucidation to identify individual MAAs. Interestingly, to date, only one putative EEVS has been identified in cyanobacteria, Gloeocapsa sp. PCC 7428, which interestingly lacks a DDGS.2 This EEVS is located in an uncharacterized gene cluster.
In actinobacteria, DDGS and other MAA biosynthetic genes were first identified through genome mining of Actinosynnema mirum DSM 43827 and Pseudonocardia sp. P1.28 Heterologous expression of both gene clusters in S. avermitilis SUKA22 revealed that shinorine was the primary MAA made by each gene cluster, but the A. mirum cluster also produced a new MAA mycosporine-glycine-alanine (Figure 6).28 This opened the door to exploring actinobacterial MAA gene clusters as sources of novel MAAs, especially now that more DDGS have been identified in other actinobacteria.22 Although DDGS and EEVS have been identified in actinobacteria, it is rare for both to exist in the same organism. The lone exception in actinobacteria is R. fascians, a plant pathogen.
5.2. SH7PCs in fungi
Other than one putative EEVS in a yeast (Saitoella complicata), all SH7PCs in fungi are predicted to be DDGS and are present in Ascomycota and Basidiomycota.2 While mycosporines, MAA analogs with only one amino acid attached to a 4-deoxygadusol core, were first found in fungi, the corresponding gene clusters in fungi have not yet been characterized experimentally. Since mycosporines may be involved in inhibiting sporulation, it may not be surprising to see mycosporine genes broadly distributed in fungi.29 Fungi that have mycosporine genes includes many important plant fungal pathogens, such as Botrytis cinerea, Colletotrichum spp., Fusarium graminearum, F. oxysporum, Magnaporthe oryzae, and Ustilago maydis.
More recently, mycosporines and MAAs have been identified in yeast and lichen. In basidiomycetous yeast, the ability to make mycosporines appears to be taxonomically split, where five of the seven Pucciniomycotina classes produced mycosporine-glutaminol-glucoside.30 Cyanobacterial lichens and a tripartite lichen containing a green alga can make a suite of MAAs, suggesting they contain DDGS.31
5.3. SH7PCs in marine organisms
MAAs and their corresponding gene clusters have been identified in marine invertebrates, dinoflagellates, and algae.2, 18, 19, 32 These organisms have a fused divergent DDGS and O-MT, unlike traditional MAA gene clusters where the DDGS and O-MT are discrete. Phylogenetic analysis suggests that multiple horizontal gene transfer (HGT) events spread this fusion protein, with origins in cyanobacteria and dinoflagellates.18 Organisms that have this fused divergent DDGS include cyanobacteria from the genus Synechoccocus, the dinoflagellates Oxyrrhis marina and Heterocapsa triquetra, and the sea anemones Nematostella vectensis and Aiptasia pallida, and the coral Acropora digitifera. These fusion proteins also had an N-terminal addition that was proposed to target these proteins to the plastid in dinoflagellates, though all homologs seem to have this addition.20
Marine red algae are a major source of MAAs, however, few genome sequences are publically available; thus, the identification of algal SH7PCs is hindered. Nevertheless, divergent DDGS has been identified in the marine red algae Pyropia haitanensis and Chondrus crispus.2 Currently, more EEVS sequences (12) have been identified in algae than DDGS/divergent DDGS (4).2 Most organisms have EEVS or DDGS, and not both, yet two algae do have both: C. crispus also has an EEVS, while the pelagophyte Aureococcus anophagefferens, which causes harmful brown tides, has an EEVS and DDGS. The EEVS from A. anophagefferens is more closely related to the vertebrate EEVS, which is involved in gadusol biosynthesis (see below), than other algal EEVS sequences.2, 3 While there has been no direct evidence that algae can produce gadusol, the terrestrial green alga Prasiola calophylla, does produce an unusual MAA, prasiolin, which consists of glutamic acid attached to a gadusol core.33 No publically available genome exists for P. calophylla, so EEVS is not known to exist in its genome. In addition, it remains possible that prasiolin is formed from a 4-deoxygadusol core, which is then hydroxylated, rather than being derived from gadusol.
5.4. SH7PCs in vertebrates
MAAs have been found in the skin mucus and eyes of marine fishes, and since they are produced by microbes and found in marine invertebrates, are proposed to be acquired by fishes through their diet.34 A structurally similar compound named gadusol was isolated from the roes of Gadus morhua (Atlantic cod).35 Due to this similarity, it was also suggested to be acquired through the diet. More recently, however, a surprising discovery was made on the presence of EEVS-like genes in the genomes of fish, amphibians, reptiles, and birds.3 Since vertebrates lack DHQS and the shikimate pathway, it was thought they would not have any DHQS-related SPCs. Clustered with the EEVS-like gene was a methyltransferase/oxidoreductase (MT-Ox), and together they work to make gadusol, a possible sunscreen compound.3 The majority of EEVS-like sequences known so far come from fish and birds.2 To date, EEVS has been identified in 99 vertebrates.2 However, the presence of the gadusol genes does not mean they are functional in all organisms. A recent bioinformatics analysis identified mutations in the EEVS and MT-Ox genes in four crocodylians.36 These mutations in both genes were predicted to occur ~190 million years ago, and their concurrent loss is consistent with their function. The crocodilian EEVS has multiple deletions and insertions, including a chompy repeat element, two premature stop codons, and a splice-donor mutation.36 The MT-Ox also has multiple deletions. This raises the possibility that not all non-mammalian vertebrates have active gadusol genes, thus the ability to make gadusol needs be confirmed in more vertebrate species.
6. Evolution and distribution of SH7PCs by horizontal gene transfer
While HGT events are often difficult to identify and characterize, multiple independent studies looking at SH7PCs and MAA biosynthesis in cyanobacteria, dinoflagellates, and cnidarians have provided phylogenetic evidence for the transfer of what we now call divergent DDGSs across diverse genera by HGT. These enzymes have been particularly useful to study because the divergent DDGS always exist with a downstream O-MT or fused O-MT. In 2006, Waller et al.18 presented a rare and surprisingly well-defined case of HGT of a divergent DDGS and O-MT in which the donor (cyanobacteria) and recipient (dinoflagellates) are clear. Interestingly, both a fusion and subsequent fission of these two genes can be tracked through evolution of the dinoflagellates. Phylogenetic analysis also revealed a second, separate HGT event of these genes between terrestrial cyanobacteria and fungi. The physical association of terrestrial cyanobacteria with fungi and marine cyanobacteria with dinoflagellates are consistent with HGT events.18
Additional support for HGT involving a SH7PC has also been observed in some marine holobionts.19 For instance, the sea anemone Anthopleura elegantissima contain the same MAAs regardless of the absence of its dinoflagellate symbiont or diet,37, 38 suggesting that A. elegantissima is capable of synthesizing MAAs independently. Furthermore, the sea anemone Nematostella vectensis, the first basal metazoan with its genome sequenced, had a divergent DDGS:O-MT, suggesting that it could synthesize MAAs.19 Interestingly, the closest relatives of the N. vectensis divergent DDGS:O-MT were the fusion proteins from the dinoflagellates Oxyrrhis marina (~65% identical DDGS part of fusion protein) and Heterocapsa triquetra, which strongly suggests that the cnidarian divergent DDGS was derived through HGT from a symbiotic dinoflagellate to an ancestral cnidarian. This type of HGT event has also been proposed to occur between cyanobacteria, dinoflagellates, and metazoans.20, 21
Likewise, work by Osborn et al.3 suggests that the EEVS and genetically linked MT-Ox found in vertebrates has arisen by HGT. The only non-vertebrate organisms found to have orthologous adjacent EEVS and MT-Ox genes are two algae, the stramenophile A. anophagefferens and the microalgae Coccomyxa subellipsodisea. As in a phylogenetic tree these genes group closely with those of vertebrates, it is proposed that they are the source for these genes in non-mammalian vertebrates. Recently, the only known fungal EEVS (from S. complicata) was also found to clade with the algae in the vertebrate EEVS clade.2 Thus, it is possible that algae also transferred this EEVS to S. complicata. The exact time point of when vertebrates gained the genes encoding gadusol production remains unknown, but they appear to have gained the gadusol genes during the evolution of bony fishes, while certain lineages (e.g., mammals and coelacanths) lost them again. Even early diverging mammals, like the duck-billed platypus, lack the gadusol genes. If gadusol does biologically function as a UV-protectant, mammals may have lost these genes during the nocturnal bottleneck.3
7. The biology and ecology of SH7PCs and their products
While SH7PCs are widely distributed in different environments and organisms, the biological function of the final secondary metabolites is generally unknown or unclear. In soil bacteria, EEVS- and EVS-derived C7N-aminocyclitols are the most common SH7P-derived natural products. The amino-pseudooligosaccharides (e.g., acarbose, oligostatins, and trestatins), which have potent α-glucosidase inhibitory activity, could play a role in preventing the utilization of sugars by surrounding organisms, and may also function as ‘carbophores’ that tag and import extracellular glucose.39 Validoxylamine A, which strongly inhibits the fungal enzyme trehalase, can prevent the fungi from accessing its energy storage.40 Thus, some C7N-aminocyclitols seem to be used for self-defense, competition for food, or niche protection.
The most common SH7PC in marine and freshwater organisms is DDGS, and the known biological functions of MAAs are constantly expanding. MAAs have features of keystone molecules, due to their presence throughout many trophic levels, yet their function differs not only by organism but also within single organisms.32 In bacteria, production of MAAs has been associated with providing protection against UV damage, salt stress, and high ammonium due to increased MAA levels under these conditions.22, 28, 41, 42 In the cyanobacterium Microcystis aeruginosa, shinorine was found in the slime layer, where it appears to have a primary structural role in the sheath and a limited role in UV protection.43 Stronger evidence for MAAs primarily acting as sunscreen exist in other cyanobacteria, e.g., A. variabilis, where shinorine production is directly linked to UV exposure.27, 42 However, MAAs may still have multiple functions in those cyanobacteria, possibly observed only under stressful conditions (e.g., sulfate deficiency, salt shock, and UV exposure). 22, 28, 41, 42 Additionally, the location of MAAs could also signify their different roles, possibly by organism, as MAAs can be found in the cytoplasm and/or in the outer sheath.27, 43, 44
The major marine ecological source of MAAs are macro- and microalgae, which are capable of producing a wide range of MAAs.27, 45, 46 Algae can serve as the dietary source of MAAs for marine invertebrate herbivores, which is one route for MAA bioaccumulation up the food chain (Figure 7).32, 47 UV light exposure increases production of MAAs in algae, and has been reviewed previously.27, 34 This matches the tendency of elevated MAA concentrations in red algae that live closer to the ocean’s surface than those at increased depth.46 Microalgae (e.g., dinoflagellates) can be the source of MAAs during symbiosis.48
Figure 7.
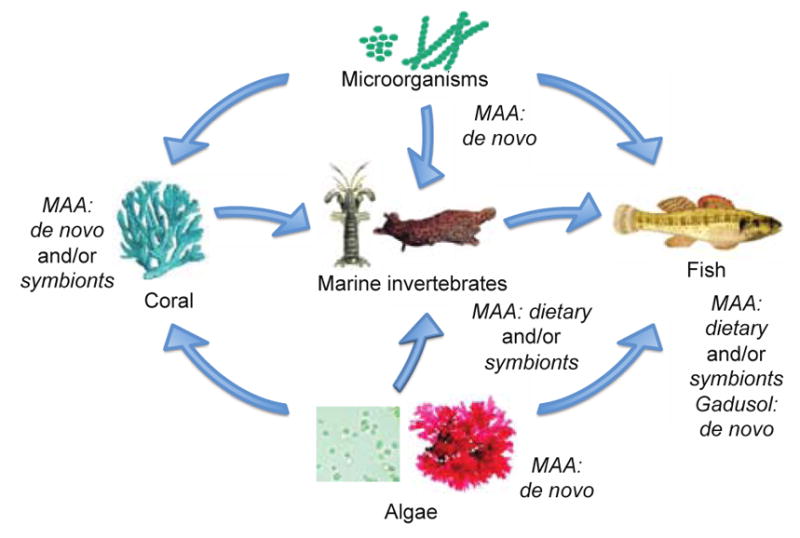
Distribution of gadusol and MAAs through different tropic levels of the marine environment. Some microorganisms, algae, and corals can produce MAA de novo, where as gadusol is mostly produced by non-mammalian vertebrates, such as fish.
The functions of MAAs in marine invertebrates are even more diverse than in bacteria and algae; in addition to UV protection, MAAs have been implicated in alarm cues and vision. MAAs can be acquired by marine invertebrates through their diet or symbionts, and can be produced de novo. 32, 47–51 In corals, MAAs accumulate with exposure to UV light, suggesting MAAs contribute to UV protection.48, 49 In the sea hare Aplysia californica, MAAs (which are acquired through diet) have been identified as a component of the sea hare’s defensive secretion, opaline, which elicited a behavior response in juvenile sea hares.32 Moreover, MAAs are found in the eyes of the mantis shrimp Neogonodactylus oerstedii, where they function as part of the shrimp’s complex color vision that includes a UV visual system.52 Beyond diet, symbiosis, and de novo synthesis, MAAs can also be inherited, as evidenced by the slipper limpets Crepipatella peruviana and C. dilatata.51 Females of these two species have different MAA levels in general and pass MAAs to their embryos through two different mechanisms, i.e., through the embryonic yolk and nurse eggs, respectively. MAAs have also been quantified in intertidal egg masses of mollusks and polychaetes, where the adult diet, phylogeny, and egg viability impact MAA composition.53 Eggs from mollusk herbivores had higher MAA concentrations than carnivores, likely because herbivores are directly consuming MAAs in their diet.53
Fish are incapable of making MAAs, yet they have been found in the skin mucus of fish and in fish eyes, where they may act as sunscreens, UV vision, and/or UV communication.34, 54, 55 Some reef fish and cleaner fish increase UV absorption in their mucus when UV-B exposure is increased. Depth and sampling location also effect MAA accumulation in the skin mucus.56 Since mollusk herbivores had more MAAs than carnivores, it is tempting to speculate that a similar phenomenon will occur with marine fishes.
Although fish cannot make MAAs, evidence implies that most can synthesize gadusol.3 Gadusol was thought to be derived from 4-deoxygadusol in MAA producing organisms and obtained only through the diet, but a recent study has shown that fish can synthesize gadusol de novo using an EEVS.3 This represents a transition from DDGS derived products to EEVS derived products in the marine environment. Like MAAs, gadusol has UV-protective and antioxidant activities and likely provides protection from UV light and/or oxidative stress, but gadusol may also play a physiological role during embryonic development. In zebra fish (Danio rerio), the EEVS and MT-Ox genes were expressed during development with maximum expression at 72 hours post fertilization. Thus, gadusol may provide some function for hatching embryos or newly hatched larvae. Other than the various live-birthing guppy species, the non-mammalian vertebrates with gadusol genes are oviparous; however, gadusol may not be linked to oviparity. The platypus, an egg laying mammal that diverged early in mammalian evolution, does not have the gadusol genes,3 and crocodylians appear to have non-functional gadusol genes.36 While MAAs have been identified in fish, it is possible that MAA-like compounds derived from gadusol exist. In fact, prasiolin, a mycosporine from the terrestrial green algae Prasiola calophylla, contains a gadusol core.33 However, homologs of the amino acid-attaching MAA genes, the ATP-grasp-like and D-Ala D-Ala ligase-like or NRPS-like genes, have not been identified in vertebrate genomes.
8. SH7PCs and Their Products in Modern Symbiotic Relationships
As described above, perhaps the most studied symbiotic relationship involving MAAs is that of corals and dinoflagellates. In this mutualistic symbiotic relationship, corals provide shelter and inorganic nutrients for dinoflagellates and dinoflagellates provide their hosts with photosynthetic and secondary metabolites, such as MAAs. Coral susceptibility to bleaching is partially dependent on the presence as well as the type of its dinoflagellate symbionts,57 the latter which may be related to the repertoire of MAAs different dinoflagellates are capable of synthesizing. Due to the threat to coral reefs posed by global warming, understanding these relationships is especially important and could aid us in developing strategies to promote coral reef health. MAAs are essential for protecting these coral from environmental factors, especially UV radiation. There is evidence that cnidarians can synthesize MAAs de novo,19 but their symbiotic dinoflagellates are also potential source of these protective compounds.49 Coral dinoflagellates also have symbiotic relationships with other marine invertebrates that involve MAAs.57 Regardless of the source, UV protection by MAAs (and therefore DDGS) is essential for maintaining this delicate but ecologically important marine habitat.
SH7PCs and their products have been implicated in other, diverse symbiotic relationships, including lichen communities, fish cleaning, leaf nodules, and kleptoplasty, where they are intimately involved in the dynamics and maintenance of these delicate relationships. The studies of these more diverse relationships are limited but we provide brief vignettes detailing the potential roles of SH7PCs and their products in symbiosis below.
Lichens are composite organisms arising from algae and/or cyanobacteria living among fungi in a symbiotic relationship. Lichens are tolerant of many different environmental conditions, such as high altitudes with high UV exposure. Cyanobacterial lichens are known to produce MAAs and four different cyanobacteria-containing lichen species belonging to the genus Peltigera, Stereocaulon, or Lobaria were analyzed for MAA production.31 Despite being collected from a similar high altitude in the Himalayas, each species produced a unique suite of MAAs. Since free-living algae, cyanobacteria, and fungi make different MAAs and mycosporines, each symbiont could be contributing to the suite of MAAs produced. The production of MAAs may protect this delicate symbiotic community and contributes to the adaptability and survival of lichens under extreme environmental conditions.
Cleaning behaviors in fish serve as a model system for understanding cooperative relationships. In these relationships, cleaner fish benefit the client fish by removing harmful gnathiid isopods but also remove epidermal mucus, perhaps at the detriment of the client fish. The dynamics surrounding these relationships are complex and one aspect of interest is what determines food preference of mucus over gnathiids by cleaner fish.55 The epidermal mucus of representative client fish contains varying amounts of MAAs, which may be sequestered to the mucus of cleaner fish. While the mucus load varies among client fish species, overall epidermal mucus has greater caloric value than gnathiid isopods and also contains more MAAs, providing the added bonus of a UV protective compound to cleaner fish which may contribute to the feeding preference of cleaner fish for mucus and the dynamics of this mutualistic relationship.
Bacteria from the genus Burkholderia and species from the Rubiaceae and Primulaceae plant families share a unique symbiotic relationship in which neither species can thrive on its own; the bacteria cannot survive independently but are critical for plant development and are vertically inherited. The secondary metabolite kirkamide, a C7N-aminocyclitol with cytotoxic effects on aquatic antropods and insects, was isolated from the leaf nodules of the plant Psychotria kirkii.26 Genomic evidence revealed that kirkamide is synthesized by P. kirkii’s bacterial symbiont, Candidatus Burkholderia kirkii, utilizing a biosynthetic gene cluster including an EEVS.23 All but one of the eight leaf nodule symbionts considered contain a functional EEVS and more than half are capable of synthesizing kirkamide, indicating kirkamide has a beneficial defensive role, but is not solely responsible for the obligate nature of the symbiosis. However, the acquisition of the kirkamide biosynthetic cluster may have enabled the shift in lifestyle in bacterial leaf nodule symbionts from commensal to mutualistic.
Kleptoplasty is the process in which non-photosynthetic host cells engulf algae and utilize their chloroplasts, referred to as kleptoplasts, for energy production. The active lifespans of these kleptoplasts vary dramatically but the reasons why are not understood. On one end of the spectrum are Ross Sea dinoflagellates, which are capable of engulfing and maintaining active kleptoplasts for a remarkably long time. A comparative study of Ross Sea dinoflagellates and free algae identified both differential profiles and localization of MAAs.58 Based on the excitation spectra of chlorophyll a, it was suggested that MAAs in kleptoplasts may serve as a non-chlorophyll sensitizer, reducing the dependence of the organism on chlorophylls for light harvesting. In fact, Ross Sea dinoflagellates cells were found to have diminished Photosystem II activity. Therefore, the reduction in photosynthetic activity and reactive oxygen species production, along with supplementary light-harvesting by MAAs, was thought to be the source of the long active lifespan of kleptoplasts involved in this symbiotic relationship.
9. Conclusions and Perspectives
The first enzyme known to cyclize the pentose phosphate pathway intermediate sedoheptulose 7-phosphate to a cyclitol product was discovered less than two decades ago, and research on these enzymes continues to thrive. Today, three highly homologous proteins have been found to catalyze the cyclization of sedoheptulose 7-phosphate, but interestingly, they produce distinct catalytic products. Recent efforts in protein crystallography and structural studies are beginning to reveal how these enzymes use the same substrate but form different products. Genome sequencing and bioinformatics studies showed that SH7PCs are widely distributed throughout prokarya and eukarya, including pathogenic bacteria, plant symbionts, nitrogen-fixing bacteria, myxobacteria, cyanobacteria, fungi, stramenopiles, and animals. Acting as a bridge between a primary metabolic pathway (the Pentose Phosphate Pathway) and secondary metabolism, SH7PCs lead to the production of various bioactive natural products. DDGS appears to be involved only in the biosynthesis of a class of natural products, 4-deoxygadusol and its related mycosporines/MAAs, and the role(s) of these compounds in symbiosis and ecology has been established. EEVSs appear to be involved in the biosynthesis of more diverse bioactive natural products. While their wide distribution in various organisms was not realized until recently, preliminary evidence has emerged to suggest their function(s) for the producing organisms, including but not limited to defense mechanism, competition for food, mutualistic symbiosis, and UV protection. While interest began with their involvement in the biosynthesis of pharmaceutically relevant natural products, their biochemistry, evolution, and expanding biological roles have continued to make SH7PCs an alluring subject to study.
Acknowledgments
This work was supported in part by grant GM112068 (to TM) from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health (NIH).
Biographies
 Andrew Osborn is a graduate student in Pharmaceutical Sciences at Oregon State University, where he also received B.S degrees in Microbiology and Bioresource Research. Andrew’s first research experience was with Dr. Taifo Mahmud as an undergraduate, where he completed a thesis project on pactamycin biosynthesis. Next, he started working with sedoheptulose 7-phosphate cyclases and joined Dr. Mahmud’s laboratory as a PhD student to continue this work. Currently, his research is focused on the evolution and function of sedoheptulose 7-phosphate cyclases and heterologous expression of their gene clusters to identify the natural products.
Andrew Osborn is a graduate student in Pharmaceutical Sciences at Oregon State University, where he also received B.S degrees in Microbiology and Bioresource Research. Andrew’s first research experience was with Dr. Taifo Mahmud as an undergraduate, where he completed a thesis project on pactamycin biosynthesis. Next, he started working with sedoheptulose 7-phosphate cyclases and joined Dr. Mahmud’s laboratory as a PhD student to continue this work. Currently, his research is focused on the evolution and function of sedoheptulose 7-phosphate cyclases and heterologous expression of their gene clusters to identify the natural products.
 Kelsey Kean is a graduate student in Biochemistry and Biophysics at Oregon State University. Kelsey received a B.S. in Biochemistry from The University of Tulsa in 2012. She got her first taste of structural biology during a summer research internship and followed this interest to graduate school where she joined the lab of Dr. Andy Karplus. Now, she uses protein crystallography to study the structure-function relationships of a variety of proteins ranging from sedoheptulose 7-phosphate cyclases involved to natural product production to peroxiredoxins involved in signaling to carbonic anhydrase with applications in carbon sequestration.
Kelsey Kean is a graduate student in Biochemistry and Biophysics at Oregon State University. Kelsey received a B.S. in Biochemistry from The University of Tulsa in 2012. She got her first taste of structural biology during a summer research internship and followed this interest to graduate school where she joined the lab of Dr. Andy Karplus. Now, she uses protein crystallography to study the structure-function relationships of a variety of proteins ranging from sedoheptulose 7-phosphate cyclases involved to natural product production to peroxiredoxins involved in signaling to carbonic anhydrase with applications in carbon sequestration.
 Andy Karplus is a Professor of Biochemistry and Biophysics at Oregon State University. He loves teaching and working with students and, guided by the view that we can better understand what we can see, his research has focused on determining the three-dimensional structures of proteins and puzzling out how they carry out their functions. Dr. Karplus has a B.S. in Biochemistry from the University of California, Davis, a Ph.D. from the University of Washington, and was an Alexander von Humboldt Postdoctoral Fellow in Freiburg, Germany. He was also a professor at Cornell University before he and his family moved to Oregon.
Andy Karplus is a Professor of Biochemistry and Biophysics at Oregon State University. He loves teaching and working with students and, guided by the view that we can better understand what we can see, his research has focused on determining the three-dimensional structures of proteins and puzzling out how they carry out their functions. Dr. Karplus has a B.S. in Biochemistry from the University of California, Davis, a Ph.D. from the University of Washington, and was an Alexander von Humboldt Postdoctoral Fellow in Freiburg, Germany. He was also a professor at Cornell University before he and his family moved to Oregon.
 Taifo Mahmud is a Professor of Natural Products and Medicinal Chemistry at Oregon State University. He received a M.S. and a Ph.D. in Pharmaceutical Sciences from Osaka University, Japan, and was a Postdoctoral Research Associate and Research Assistant Professor of Chemistry at the University of Washington. After six enjoyable years living in Seattle, he moved to Corvallis, the home of the Oregon State Beavers. Dr. Mahmud’s research interests are broadly in medicinal chemistry and natural products biosynthesis. He uses molecular genetics, enzymology, and chemistry approaches to create and develop new biologically active compounds.
Taifo Mahmud is a Professor of Natural Products and Medicinal Chemistry at Oregon State University. He received a M.S. and a Ph.D. in Pharmaceutical Sciences from Osaka University, Japan, and was a Postdoctoral Research Associate and Research Assistant Professor of Chemistry at the University of Washington. After six enjoyable years living in Seattle, he moved to Corvallis, the home of the Oregon State Beavers. Dr. Mahmud’s research interests are broadly in medicinal chemistry and natural products biosynthesis. He uses molecular genetics, enzymology, and chemistry approaches to create and develop new biologically active compounds.
References
- 1.Asamizu S, Xie P, Brumsted CJ, Flatt PM, Mahmud T. J Am Chem Soc. 2012;134:12219–12229. doi: 10.1021/ja3041866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osborn AR, Kean KM, Alseud KM, Almabruk KH, Asamizu S, Lee JA, Karplus PA, Mahmud T. ACS Chem Biol. 2017 doi: 10.1021/acschembio.7b00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osborn AR, Almabruk KH, Holzwarth G, Asamizu S, LaDu J, Kean KM, Karplus PA, Tanguay RL, Bakalinsky AT, Mahmud T. Elife. 2015;4:e05919. doi: 10.7554/eLife.05919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balskus EP, Walsh CT. Science. 2010;329:1653–1656. doi: 10.1126/science.1193637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asamizu S, Abugreen M, Mahmud T. Chem Bio Chem. 2013;14:1548–1551. doi: 10.1002/cbic.201300384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dell KA, Frost JW. J Am Chem Soc. 1993;115:11581–11589. [Google Scholar]
- 7.Kim CG, Kirschning A, Bergon P, Zhou P, Su E, Sauerbrei B, Ning S, Ahn Y, Breuer M, Leistner E, Floss HG. J Am Chem Soc. 1996;118:7486–7491. [Google Scholar]
- 8.Mahmud T. Nat Prod Rep. 2003;20:137–166. doi: 10.1039/b205561a. [DOI] [PubMed] [Google Scholar]
- 9.Mahmud T, Tornus I, Egelkrout E, Wolf E, Uy C, Floss HG, Lee S. J Am Chem Soc. 1999;121:6973–6983. [Google Scholar]
- 10.Stratmann A, Mahmud T, Lee S, Distler J, Floss HG, Piepersberg W. J Biol Chem. 1999;274:10889–10896. doi: 10.1074/jbc.274.16.10889. [DOI] [PubMed] [Google Scholar]
- 11.Dong HJ, Mahmud T, Tornus I, Lee S, Floss HG. J Am Chem Soc. 2001;123:2733–2742. doi: 10.1021/ja003643n. [DOI] [PubMed] [Google Scholar]
- 12.Wu X, Flatt PM, Schlorke O, Zeeck A, Dairi T, Mahmud T. Chem Bio Chem. 2007;8:239–248. doi: 10.1002/cbic.200600446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naganawa H, Hashizume H, Kubota Y, Sawa R, Takahashi Y, Arakawa K, Bowers SG, Mahmud T. J Antibiot. 2002;55:578–584. doi: 10.7164/antibiotics.55.578. [DOI] [PubMed] [Google Scholar]
- 14.Yu Y, Bai LQ, Minagawa K, Jian XH, Li L, Li JL, Chen SY, Cao EH, Mahmud T, Floss HG, Zhou XF, Deng ZX. Appl Environ Microbiol. 2005;71:5066–5076. doi: 10.1128/AEM.71.9.5066-5076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi WS, Wu XM, Choeng YH, Mahmud T, Jeong BC, Lee SH, Chang YK, Kim CJ, Hong SK. Appl Microbiol Biotechnol. 2008;80:637–645. doi: 10.1007/s00253-008-1591-2. [DOI] [PubMed] [Google Scholar]
- 16.Rockser Y, Wehmeier UF. J Biotechnol. 2009;140:114–123. doi: 10.1016/j.jbiotec.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Flatt PM, Wu X, Perry S, Mahmud T. J Nat Prod. 2013;76:939–946. doi: 10.1021/np400159a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waller RF, Slamovits CH, Keeling PJ. Mol Biol Evol. 2006;23:1437–1443. doi: 10.1093/molbev/msl008. [DOI] [PubMed] [Google Scholar]
- 19.Starcevic A, Akthar S, Dunlap WC, Shick JM, Hranueli D, Cullum J, Long PF. Proc Natl Acad Sci USA. 2008;105:2533–2537. doi: 10.1073/pnas.0707388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh SP, Hader DP, Sinha RP. Gene. 2012;500:155–163. doi: 10.1016/j.gene.2012.03.063. [DOI] [PubMed] [Google Scholar]
- 21.Singh SP, Klisch M, Sinha RP, Hader DP. Genomics. 2010;95:120–128. doi: 10.1016/j.ygeno.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Waditee-Sirisattha R, Kageyama H, Sopun W, Tanaka Y, Takabe T. Appl Environ Microbiol. 2014;80:1763–1769. doi: 10.1128/AEM.03729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinto-Carbo M, Sieber S, Dessein S, Wicker T, Verstraete B, Gademann K, Eberl L, Carlier A. ISME J. 2016;10:2092–2105. doi: 10.1038/ismej.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kean KM, Codding SJ, Asamizu S, Mahmud T, Karplus PA. Biochemistry. 2014;53:4250–4260. doi: 10.1021/bi5003508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichols CE, Ren J, Lamb HK, Hawkins AR, Stammers DK. J Mol Biol. 2003;327:129–144. doi: 10.1016/s0022-2836(03)00086-x. [DOI] [PubMed] [Google Scholar]
- 26.Sieber S, Carlier A, Neuburger M, Grabenweger G, Eberl L, Gademann K. Angew Chem Int Ed. 2015;54:7968–7970. doi: 10.1002/anie.201502696. [DOI] [PubMed] [Google Scholar]
- 27.Carreto JI, Carignan MO. Mar Drugs. 2011;9:387–446. doi: 10.3390/md9030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyamoto KT, Komatsu M, Ikeda H. Appl Environ Microbiol. 2014;80:5028–5036. doi: 10.1128/AEM.00727-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leite B, Nicholson RL. Exp Mycol. 1992;16:76–86. [Google Scholar]
- 30.Libkind D, Moline M, Sommaruga R, Sampaio JP, van Broock M. Yeast. 2011;28:619–627. doi: 10.1002/yea.1891. [DOI] [PubMed] [Google Scholar]
- 31.Shukla V, Kumari R, Patel DK, Upreti DK. Amino Acids. 2016;48:129–136. doi: 10.1007/s00726-015-2069-z. [DOI] [PubMed] [Google Scholar]
- 32.Kicklighter CE, Kamio M, Nguyen L, Germann MW, Derby CD. Proc Natl Acad Sci U S A. 2011;108:11494–11499. doi: 10.1073/pnas.1103906108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartmann A, Holzinger A, Ganzera M, Karsten U. Planta. 2016;243:161–169. doi: 10.1007/s00425-015-2396-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shick JM, Dunlap WC. Annu Rev Physiol. 2002;64:223–262. doi: 10.1146/annurev.physiol.64.081501.155802. [DOI] [PubMed] [Google Scholar]
- 35.Plack PA, Fraser NW, Grant PT, Middleton C, Mitchell AI, Thomson RH. Biochem J. 1981;199:741–747. doi: 10.1042/bj1990741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emerling CA. Mol Biol Evol. 2017;34:666–676. doi: 10.1093/molbev/msw265. [DOI] [PubMed] [Google Scholar]
- 37.Shick JM, Dunlap WC, Pearse JS, Pearse VB. Biol Bull. 2002;203:315–330. doi: 10.2307/1543574. [DOI] [PubMed] [Google Scholar]
- 38.Stochaj WR, Dunlap WC, Shick JM. Mar Biol. 1994;118:149–156. [Google Scholar]
- 39.Wehmeier UF, Piepersberg W. Appl Microbiol Biotechnol. 2004;63:613–625. doi: 10.1007/s00253-003-1477-2. [DOI] [PubMed] [Google Scholar]
- 40.Flatt PM, Mahmud T. Nat Prod Rep. 2007;24:358–392. doi: 10.1039/b603816f. [DOI] [PubMed] [Google Scholar]
- 41.Waditee-Sirisattha R, Kageyama H, Fukaya M, Rai V, Takabe T. FEMS Microbiol Lett. 2015;362:fnv198. doi: 10.1093/femsle/fnv198. [DOI] [PubMed] [Google Scholar]
- 42.Singh SP, Klisch M, Sinha RP, Hader DP. Photochem Photobiol. 2008;84:1500–1505. doi: 10.1111/j.1751-1097.2008.00376.x. [DOI] [PubMed] [Google Scholar]
- 43.Hu C, Voller G, Sussmuth R, Dittmann E, Kehr JC. Environ Microbiol. 2015;17:1548–1559. doi: 10.1111/1462-2920.12577. [DOI] [PubMed] [Google Scholar]
- 44.D’Agostino PM, Javalkote VS, Mazmouz R, Pickford R, Puranik PR, Neilan BA. Appl Environ Microbiol. 2016;82:5951–5959. doi: 10.1128/AEM.01633-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Llewellyn CA, Airs RL. Mar Drugs. 2010;8:1273–1291. doi: 10.3390/md8041273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan YV, Westcott ND, Hu C, Kitts DD. Food Chem. 2009;112:321–328. [Google Scholar]
- 47.Carefoot TH, Karentz D, Pennings SC, Young CL. Comp Biochem Physiol C Toxicol Pharmacol. 2000;126:91–104. doi: 10.1016/s0742-8413(00)00098-0. [DOI] [PubMed] [Google Scholar]
- 48.Rosic NN. Appl Microbiol Biotechnol. 2012;94:29–37. doi: 10.1007/s00253-012-3925-3. [DOI] [PubMed] [Google Scholar]
- 49.Shick JM, Romaine-Lioud S, Ferrier-Pages C, Gattuso JP. Limnol Oceanogr. 1999;44:1667–1682. [Google Scholar]
- 50.Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, Fujie M, Fujiwara M, Koyanagi R, Ikuta T, Fujiyama A, Miller DJ, Satoh N. Nature. 2011;476:320–323. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- 51.Paredes-Molina FJ, Cubillos VM, Montory JA, Andrade-Villagran PA. J Photochem Photobiol B. 2016;162:511–518. doi: 10.1016/j.jphotobiol.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 52.Bok MJ, Porter ML, Place AR, Cronin TW. Curr Biol. 2014;24:1636–1642. doi: 10.1016/j.cub.2014.05.071. [DOI] [PubMed] [Google Scholar]
- 53.Przeslawski R, Benkendorff K, Davis AR. J Chem Ecol. 2005;31:2417–2438. doi: 10.1007/s10886-005-7110-3. [DOI] [PubMed] [Google Scholar]
- 54.Braun C, Reef R, Siebeck UE. J Photochem Photobiol B. 2016;160:400–407. doi: 10.1016/j.jphotobiol.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 55.Eckes M, Dove S, Siebeck UE, Grutter AS. Coral Reefs. 2015;34:823–833. [Google Scholar]
- 56.Eckes MJ, Siebeck UE, Dove S, Grutter AS. Mar Ecol Prog Ser. 2008;353:203–211. [Google Scholar]
- 57.Rosic NN, Dove S. Appl Environ Microb. 2011;77:8478–8486. doi: 10.1128/AEM.05870-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stamatakis K, Vayenos D, Kotakis C, Gast RJ, Papageorgiou GC. Biochim Biophys Acta. 2017;1858:189–195. doi: 10.1016/j.bbabio.2016.12.002. [DOI] [PubMed] [Google Scholar]