Abstract
Activated cell surface and intracellular receptors lead to insulin resistance in obesity. Among these receptors, triggering receptors expressed on myeloid cells (TREM)-1, toll like receptors (TLRs), and receptors for advanced glycation end products (RAGE) play a significant role in the induction of inflammatory response in innate immunity. TREM-1 potentially amplifies TLRs and RAGE synergistically with DNA-binding high-mobility group box 1 (HMGB-1). The objective of the study was to analyze the association between TREM-1/DAP12 and HMGB-1, RAGE and TLRs in obesity-induced insulin resistance. We examined the mRNA expression by RT-PCR and protein expression by Western blotting and immunofluorescence for TREM-1, TREM-2, DAP-12, HMGB-1, RAGE, TLR-4 and TLR-2 in omentum, subcutaneous and liver biopsy tissues of obese diabetic (n=22) and non-diabetic subjects (n=24) and compared with the non-obese non-diabetic controls (n=5). There was a significantly increased expression of TREM-1, DAP-12, HMGB-1, RAGE, TLR-4 and TLR-2 and decreased expression of TREM-2 in the omentum, subcutaneous and liver biopsy of obese diabetic subjects compared to obese non-diabetics and the non-obese population. Overall, obese diabetic subjects had high expression of TREM-1 in association with HMGB1 (100% vs 58.3%, P=0.003), RAGE (77.3% vs 41.7%, P=0.045), TLR4 (100% vs 58.3%, P=0.003), and TLR2 (100% vs 50%, P=0.003) in liver biopsy samples in comparison to obese non-diabetic subjects. Obese diabetics have significantly increased TREM-1, HMGB1, RAGE, and TLRs compared to obese non-diabetics. Our findings suggest a potential pathophysiological role of TREM-1 in conjunction with HMGB1 and inflammatory cell receptors (RAGE, TLR-4 and TLR-2) in obesity-induced insulin resistance.
Keywords: Obesity, insulin resistance, inflammation, TREM-1, HMGB1, TLR
Introduction
Obesity is a major risk factor in the development of metabolic syndrome. Adipocyte-induced pro-inflammatory cytokines [interleukin (IL)-1, IL-6 and tumor necrosis factor alpha (TNF-α)], and chemokines [monocyte chemoattractant proteins (MCP-1) and macrophage inflammatory protein (MIP-1)] attract and activate pathogenic inflammatory cells, resulting in chronic low-grade inflammation [1]. The activated pro-inflammatory immune effector cells lead to impaired insulin signalling and insulin resistance (IR) secondary to altered immune homeostasis [2]. These pro-inflammatory immune cells accumulate in insulin-dependent tissues (adipose, liver and muscle) producing chronic inflammatory state, which plays a key role in the pathophysiology of obesity-induced IR [3].
Toll like receptors (TLRs) and receptors for advanced glycation end products (RAGE) are common receptors, which play a significant role in the induction of inflammatory response in innate immunity [4]. TLRs belong to a family of pattern recognition receptors (PRRs) that recognize self-molecular patterns originating from damaged cells, namely Damage Associ-ated Molecular Patterns (DAMPs) [5]. TLR2 and TLR4 play a major role in recognizing fatty acids, glucose, lipid peroxidation products, advanced glycated end (AGE) products and reactive oxygen species (ROS) and are involved in IR and Type 2 diabetes mellitus (T2DM) [6,7]. RAGE also plays a significant role in regulating chronic inflammation, which is regarded as a key factor in the pathophysiology of obesity and metabolic syndrome [8]. RAGE is a cell surface receptor that binds many ligands, including AGE products [9] and DAMPs [10], and induces oxidative stress and inflammation [11]. DNA-binding high-mobility group box 1 (HMGB1), which is primarily in the DAMPs superfamily, is released due to cell damage/necrosis, which subsequently induces pro-inflammatory cytokines IL-1 and TNF-α [12] and activates TLRs and RAGE receptors.
Triggering receptors expressed on myeloid cells (TREM)-1 (a cell surface receptor) is recognized as a potent amplifier of acute and chronic inflammation. In contrast to TREM-1, TREM-2 acts as an anti-inflammatory. The activation of TREM-1 through the trans-membrane adaptor protein, DNAX activation protein of 12 kDa (DAP12) [13], plays an important role in the pathogenesis of chronic inflammation. Chronic inflammation in obesity may lead to insulin resistance, and TREM-1 may play a potential role in inducing obesity induced insulin resistance as documented in our previous work [14]. Recently, HMGB1 has been recognized as a major ligand to activate TREM-1 and to further enhance the proinflammatory effect of TREM1 [15]. TREM-1 also interacts with TLRs [16] and RAGE [17] to amplify inflammation. Thus, TREM-1, RAGE and TLRs, although acting through different receptors, share common properties to induce the pathogenesis of obesity-induced IR. Metabolic syndrome resulting from chronic exposure to elevated levels of free fatty acids (FFA) and lipopolysaccharides (LPS) stimulates TREM1-induced activation of TLRs receptor cascade in lipid rafts. HMGB1 is a ligand for TREM-1, TLRs and RAGE.
In this study, we examined an association between TREM-1/DAP12, HMGB1, TLRs and RAGE in the chronic inflammation in obese patients, and interaction between these molecules could be the major underlying cause of insulin resistance.
Materials and methods
Patient selection
This prospective study was performed after approval from the Institutional Review Board (IRB) of Creighton University, Omaha, Nebraska, USA. Study population was selected from the patients undergoing bariatric surgery at Creighton University Medical Center (CUMC) and Immanuel Medical Center (IMC). Following the explanation of the study in layman language, the participation was voluntary, and if elected to participate, an IRB-approved informed consent together with the HIPPA forms was signed by the participants in the study. A total of 46 patients were enrolled as per inclusion criteria comprised of patients who were obese with BMI of 35 to 65 and aged between 21 and 65 years. Exclusion criteria included the patients who had prior systemic inflammatory diseases, including systemic lupus erythematosis, rheumatoid arthritis, and systemic sclerosis. Patients who were on immunosuppressant medications were also excluded from the study. Patients on non-steroidal anti-inflammatory agents (NSAIDs) were also excluded if they were not willing to discontinue them at least 1 week prior to surgery or willing to switch to other pain medications 1 week before the surgery. Of these 46 patients in the study, 22 were obese diabetic, 24 were obese and non-diabetic. Five non-obese and non-diabetic patients who had elective surgeries (ex-hernia, fundoplication) were included in the study as controls. The control population to participate is limited in this study.
Tissue and blood acquisition
Intraoperative surgical specimens of liver, omentum and subcutaneous tissue were collected in the University of Wisconsin (UW) solution maintained at 4°C and immediately transported to the research laboratory. Blood samples were collected pre-operatively into ethylenediaminetetraacetic acid (EDTA) tubes for biochemical studies.
Clinical data and biochemical analysis
The clinical and biochemical data were collected by reviewing patients’ charts. Data for demographic information like age, sex, weight, height and co-morbidities like hypertension, sleep apnea and hyperlipidemia were obtained for all 51 participants, including five controls. Apart from these demographics and co-morbidities, we have also measured biochemical parameters in the blood, including glucose levels (fasting and postprandial), HbA1c, cholesterol, VLDL, HDL, LDL, cholesterol:HDL and LDL:HDL ratio. HOMA-IR index (calculated as Glucose x Insulin/405), HOMA-β% (calculated as 360 x Insulin/Glucose-63%), FFA (ELISA kit #ab65341) and insulin (ELISA kit #ab100578).
Preparation and staining of specimen
Surgical specimens obtained from the liver, omentum and subcutaneous tissue were fixed in 4% formalin. Each specimen was then transversely sectioned at 2 mm and embedded in paraffin. Microtome was used to obtain thin sections (5 μm) and those sections were then stained with hematoxylin and eosin (H&E) following manufacturer’s standard protocol (Newcomer/supply).
Hematoxylin and eosin staining
H&E staining was done on the sectioned slides as per the standard protocol. Stained slides were used for the histological analysis of liver, omentum and subcutaneous tissues. Presence of inflammation, fibrosis, cirrhosis, hepatosteatosis and fatty liver grading was done blindly by a board-certified pathologist. The following criteria were used for inflammation (0-4-point scale); 0=no inflammation, 1=minimal, 2=mild, 3=moderate, and 4=severe; hepatosteatosis (0-3-point scale): 0=no steatosis, 1=0-33%, 2=33-66%, 3=66-100%; fibrosis (0-4-point scale): 0=no fibrosis, 1=portal fibrosis, 2=periportal fibrosis, 3=septal fibrosis, 4=cirrhosis.
RNA isolation, cDNA synthesis, and real-time PCR
Total RNA from the biopsied tissue was isolated by using TRI reagent (Trizol reagent, Sigma, St Louis, MO, USA) as per manufacturer’s instructions. The yield of RNA was measured by Nanodrop (Thermo Scientific, Rockford, IL, USA). Improm II reverse transcription kit (Promega, Madison, WI, USA) was used for the cDNA synthesis following the manufacturer’s instructions. Real-time PCR (qRT-PCR) was performed in triplicate using SYBR Green Master Mix and a Real-time PCR system (CFX96, BioRad Laboratories, and Hercules, CA, USA). The PCR cycling conditions were 5 min at 95°C for initial denaturation, 40 cycles of 30 s at 95°C, 30 s at 55-60°C (according to the primer annealing temperatures) and 30 s at 72°C followed by melting curve analysis. The primers for different genes were obtained from Integrated DNA Technologies (Coralville, IA, USA). GAPDH was used to normalize differences in RNA isolation, RNA degradation, and the efficiencies of the reverse transcription. Fold-expression of mRNA transcripts relative to controls was determined after normalizing to GAPDH. The relative expressions of target genes were stated as fold-ratio using 2-ΔΔCT method.
The oligonucleotide primer sequences for TREM-1 were: Forward 5’-AGT TAC AGC CCA AAA CAT GC-3’ Reverse 5’-CAG CCC CCA CAA GAG AAT TA-3’; TREM-2 Forward 5’-ACA GAA GCC AGG GAC ACA TC-3’, Reverse 5’-CCT CCC ATC ATC TTC CTT CA-3’; DAP12 Forward 5’-TCA GAG GTC GGA TGT CTA CAG-3’, Reverse 5’-ATC CAG GTA TCA TGT TGC TGA C-3’; HMGB1 Forward 5’-AAG CAC CCA GAT GCT TCA GT-3’, Reverse 5’-TCC GCT TTT GCC ATA TCT TC-3’; RAGE Forward 5’-GCA GGT GAG GGG TTT GAT AA-3’, Reverse 5’-TCC TCC TCT TCC TCC TGG TT-3’; TLR4 Forward 5’-GCC GAA AGG TGA TTG TTG TGG TGT-3’, Reverse 5’-TAC CAG CAC GAC TGC TCA GAA ACT-3’; TLR2 Forward 5’-ACC TGT CCA ACA ACA GGA TCA CCT-3’, Reverse 5’-TGT TCA AGA CTG CCC AGG GAA GAA-3’ and GAPDH Forward 5’-GGT GAA GGT CGG AGT CAA CGG ATT TGG TCG-3’, Reverse 5’-GGA TCT CGC TCC TGG AAG ATG GTG ATG GG-3’. The target genes expression fold changes in obese diabetics and obese non-diabetics were calculated; when compared to non-obese subjects’ fold-ratio was stated as 1. Increased fold changes of TREM-1, TREM-2, TLRs, HMGB-1, DAP-12, RAGE expressions were regarded as increased expression, likewise decrease in fold changes as down-regulation.
Protein isolation and western blotting study
The protein samples were isolated from the biopsy tissues and quantified using Bicinchoninic acid assay. The protein samples (50 µg/well) were electrophoresed by using SDS-PAGE and transferred to nitrocellulose membrane. Immunoblot analysis was carried out rabbit anti-TREM-1 (1:1000; ab93717), rabbit anti-TREM-2 (1:1000; ab175262), rabbit anti-DAP-12 (1:1000; ab124834), mouse anti-HMGB-1 (1:1000; ab77302), rabbit anti-RAGE (1:1000; ab37647), rabbit anti-TLR4 (1:1000; ab13556), goat anti-TLR2 (1:1000; ab1655), mouse anti-GAPDH (1:1000; NB300221) as primary antibody and HRP conjugated goat anti-rabbit IgG and HRP conjugated rabbit anti-mouse IgG as secondary antibodies, respectively. The protein expression was detected by incubating the membrane to chemiluminescence solution. The exposure time was adjusted to keep the integrated optical densities within a linear and non-saturated range. The band intensity was measured by densitometric analysis using Image J software. GAPDH was used to normalize the target and the relative expressions were stated as fold ratio.
Immunofluorescence (IF) study
Immunofluorescence staining was done after deparaffinization, rehydration and antigen retrieval as per the protocol. Goat anti-TREM-1 (sc-19309), rabbit (sc-48764) or mouse (sc-373828) anti-TREM-2, mouse anti-TLR2 (sc-21759), mouse anti-TLR4 (sc-52962), rabbit anti-HMGB-1 (ab191583), rabbit anti-DAP-12 (sc-20783), mouse anti-RAGE (sc-365154) antibodies were used as primary antibodies. SantaCruz biotech antibodies were used at 1:50 dilution and Abcam antibody was used at 1:200 dilution for primary antibodies. Alexa Fluor 594 (red) and Alexa Fluor 488 (green) (Invitrogen, Grand Island, NY, USA) at 1:200 dilution were used as conjugated secondary antibodies. DAPI (4,6-diamidino-2-phenylindole) was used to stain nuclei. Negative controls were run by using isotypes for each fluorochrome. Immunofluorescence microscopy was done with an Olympus inverted fluorescent microscope (Olympus BX51) and images were scanned at 20× magnification. Fluorescence intensity for TREM-1, TREM-2, TLRs, HMGB-1, DAP12, and RAGE was measured in the stained slides (20× images for each measurement) by using Image-J software (NIH) and mean fluorescence intensity (MFI) was calculated.
Statistical analysis
Demographics, co-morbidities and biochemical data were compared between obese non-diabetics and obese diabetics using student t-test for continuous variables and Fisher’s exact test or Pearson’s χ2 for categorical variables. The mRNA and protein expression of TREM-1, TREM-2, DAP12, TLR2, TLR4, HMGB1 and RAGE in biopsy tissues were compared between non-obese, obese non-diabetics and obese diabetics using One-way ANOVA for continuous variables. Among the categorical variables of the subjects, the increased expression of TREM-1, DAP-12, TLR2, TLR4, HMGB1 and RAGE and downregulation of TREM-2 in biopsy tissues were analyzed between obese non-diabetics and obese diabetics. Subjects’ categorical variables with strong correlation between TREM-1 and DAP12, TLR2, TLR4, HMGB1 and RAGE were also analyzed among obese non-diabetics and obese diabetics using Fisher’s exact test or Pearson’s χ2 test. Data are presented as mean ± SD or number (percentage) of patients. All the data were analyzed by SPSS v21 and GraphPad Prism. A value of P<0.05 (*P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001) was considered statistically significant.
Results
Demographics and co-morbid conditions of the study populations
Obese diabetic patients had significantly high incidence of hypertension (r=0.480; P=0.001) and sleep apnea (r=0.392; P=0.015) compared to obese non-diabetics (Table 1). There were no significant differences in demographics (BMI, weight and height) between obese non-diabetics and obese diabetics.
Table 1.
Demographics and co-morbid conditions of study population
| Clinical data | Non-obese (5) | Obese non-diabetics (24) | Obese diabetics (22) | Correlation (R); P value |
|---|---|---|---|---|
| Demographics | ||||
| Gender (Male/Female) | 1/4 | 2/22 | 8/14 | |
| Age (year) | 44.4 ± 15.02 | 40.08 ± 9.94 | 47.28 ± 10.36 | |
| BMI | 26.46 ± 1.71 | 46.23 ± 6.33 | 49.04 ± 10.51 | |
| Height (feet’ inches”) | 5.64 ± 0.20 | 5.41 ± 0.26 | 5.60 ± 0.31 | NS |
| Weight (pounds) | 150.70 ± 8.92 | 279.83 ± 58.47 | 312.10 ± 76.79 | |
| Co-morbid conditions | ||||
| Hypertension | 1 (20%) | 11 (45.8%) | 20 (90.9%)** | R=0.480; P=0.001 |
| Hyperlipidemia | -- | 10 (41.7%) | 11 (50%) | NS |
| Sleep apnea | -- | 5 (20.8%) | 13 (59.1%)* | R=0.392; P=0.015 |
Demographics and co-morbidities were compared between obese non-diabetics and obese diabetics using student t-test for continuous variables and Fisher’s exact test or Pearson’s χ2 for categorical variables. Not significant (NS), data for age, body mass index (BMI), height and weight are presented as mean values ± SD number (percentage) of patients. Since, the participation of non-obese subjects were limited in this study, we have used the tissue biopsies of non-obese subjects from our previous work [14] for this study. Data in co-morbid condition show number of subjects (%) in that group.
P<0.05;
P<0.01.
Biochemical profiles of the study populations
Among the biochemical profiles, the levels of FFA (P=0.0002), glucose (P<0.0001), HbA1c (P<0.0001), insulin (P<0.0001) and HOMA-IR (P<0.0001) were found to be significantly elevated in obese diabetics compared to obese non-diabetics. The LDL levels were significantly (P=0.021) higher in obese non-diabetics (Table 2).
Table 2.
Biochemical profile of obese patient population
| Biochemical Profile | Obese non-diabetics (24) | Obese diabetics (22) | P value | ||
|---|---|---|---|---|---|
|
| |||||
| Biochemical levels | N>normal values | Biochemical levels | N>normal values | ||
| Cholesterol (mg/dl), >200 | 169.70 ± 36.26 | 6 | 157.27 ± 34.47 | 4 | NS |
| Triglycerides (mg/dl), >149 | 148.33 ± 58.84 | 9 | 192.77 ± 128.69 | 16 | NS |
| FFA (µM/ml), >0.65 | 0.85 ± 0.28 | 16 | 1.34 ± 0.52*** | 19 | P=0.0002 |
| VLDL (mg/dl), >30 | 30.37 ± 11.95 | 8 | 40.36 ± 22.74 | 14 | NS |
| HDL (mg/dl), <40 | 43.37 ± 9.02 | 11 | 45.04 ± 16.85 | 14 | P=0.021 |
| LDL (mg/dl), >99 | 95.62 ± 31.23* | 9 | 73.95 ± 30.16 | 6 | NS |
| Cholesterol:HDL >4.4 | 4.03 ± 1.10 | 6 | 3.84 ± 1.55 | 8 | NS |
| LDL:HDL >3.2, | 2.28 ± 0.89 | 4 | 2.00 ± 1.11 | 4 | NS |
| HbA1c (%), >6 | 5.51 ± 0.57 | 7 | 7.3 ± 0.92**** | 22 | P<0.0001 |
| Glucose (mg/dl) >100 | 97.79 ± 15.86 | 11 | 150.95 ± 43.21**** | 22 | P<0.0001 |
| Insulin (µIU/mL) >8.4 | 13.79 ± 6.05 | 16 | 26.98 ± 13.70**** | 22 | P<0.0001 |
| HOMA-IR, >2 | 3.47 ± 1.92 | 12 | 10.23 ± 6.59**** | 22 | P<0.0001 |
| HOMA-β%, >100 | 160.00 ± 89.05 | 22 | 130.47 ± 79.41 | 13 | NS |
Biochemical profile comparison was done between obese non-diabetics and obese diabetics subjects using student t-test for continuous variables. All data are presented as mean values ± SD, (normal physiological levels), p values for significance. Not significant (NS), Free fatty acids (FFA), high density lipoprotein (HDL), low density lipoprotein (LDL), very low density lipoprotein (VLDL), glycosylated hemoglobin (HbA1c), homeostatic model assessment (HOMA)-Insulin resistance (IR).
P<0.001;
P<0.0001.
Grading of fatty liver
In fatty liver grading, the obese diabetic group had a significantly 86.4% (r=0.425; P=0.005) higher incidence of hepatosteatosis compared to the obese non-diabetic group. Fibrosis (45.5% vs 20.8%) and inflammation (68.2% vs 45.8%) was higher in obese diabetics, but not significant. Fatty liver grading showed no evidence of inflammation, hepatosteatosis or fibrosis in non-obese subjects (Table 3).
Table 3.
Grading of Fatty liver in obese subjects
| Fatty liver grading | Obese (46) | ||
|---|---|---|---|
|
| |||
| Obese non-diabetics (24) | Obese diabetics (22) | Correlation (R); P value | |
| Inflammation | 11 (45.8%) | 15 (68.2%) | NS |
| No inflammation | -- | -- | |
| Minimal | 8 (33.3%) | 5 (22.7%) | |
| Mild | 2 (8.3%) | 8 (36.3%) | |
| Moderate | 1 (4.1%) | 2 (9.0%) | |
| Severe | -- | -- | |
| Hepatosteatosis | 11 (45.8%) | 19 (86.4%)** | R=0.425; P=0.005 |
| 0% | -- | -- | |
| 0-33% | 8 (33.3%) | 10 (45.4%) | |
| 33-66% | 3 (12.5%) | 3 (13.6%) | |
| 66-100% | -- | 6 (27.2%) | |
| Fibrosis | 5 (20.8%) | 10 (45.5%) | NS |
| Portal fibrosis | 5 (20.8%) | 6 (27.2%) | |
| Periportal fibrosis | -- | 1 (4.5%) | |
| Septal fibrosis | -- | 1 (4.5%) | |
| Cirrhosis | -- | 2 (9.0%) | |
Fatty liver grading was compared between obese non-diabetics and obese diabetics using Fisher’s exact test or Pearson’s χ2 for categorical variables. Inflammation in liver biopsy was categorized with no inflammation, minimal, mild, moderate and severe inflammation; hepatosteatosis was categorized as 0%, 0-33%, 33-66% and 66-100%; and fibrosis was categorized as portal, periportal and septal fibrosis and cirrhosis for classification. Data show number of subjects in respective group (% subjects).
P<0.01.
Increased expression of TREM-1, DAP12, HMGB1, RAGE, TLR4 and TLR2 mRNA transcripts in the tissues of obese diabetic subjects
Obese diabetic and non-diabetic subjects had significant increased expression of TREM1, DAP12, HMGB1, RAGE, TLR4 and TLR2 and downregulation of TREM-2 compared to non-obese subjects. The mRNA expression levels (fold-change) of TREM-1, TREM-2, DAP12, HMGB1, RAGE, TLR4 and TLR2 in the biopsy tissues are shown in Figure 1. TREM-1, DAP12, HMGB1, RAGE, TLR4 and TLR2 mRNA expression folds were significantly (P<0.05) higher in the omentum (Figure 1A), subcutaneous (Figure 1B) and liver (Figure 1C) tissue biopsies of obese diabetic subjects compared to the obese non-diabetics and non-obese patients. The mRNA expression of TREM-1, DAP12, HMGB1, RAGE and TLR4 were also found to be significantly increased only in the omentum and no significant difference in the liver and subcutaneous tissue biopsies of obese non-diabetics compared to non-obese patients. At the same time, TREM-2 mRNA expressions were significantly (P<0.05) down regulated in the biopsy tissues of obese diabetic subjects compared to non-obese subjects. The significant (P<0.05) down regulation of TREM-2 was also observed in omentum and liver between obese diabetic and obese non-diabetics.
Figure 1.
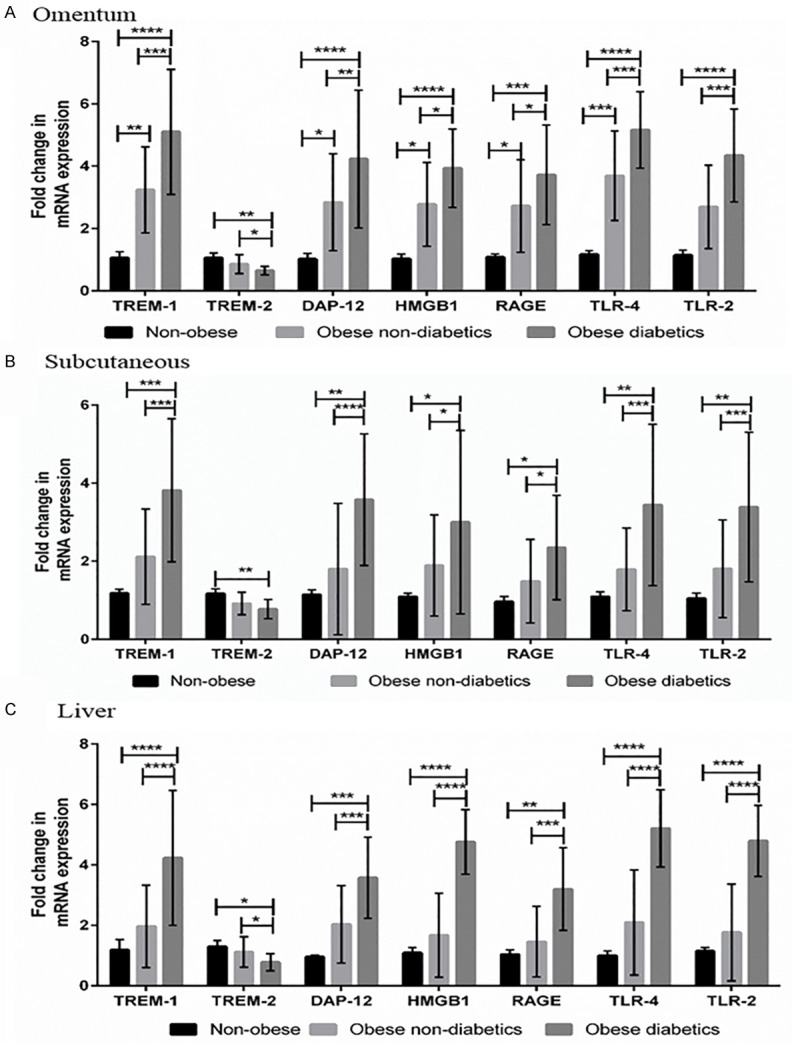
TREM-1, TREM-2, DAP12, HMGB1, RAGE, TLR-4 and TLR-2 mRNA expression studies using qPCR in the biopsy samples of study subjects. A: Gene Expression study in omentum biopsy samples. B: Gene Expression study in subcutaneous biopsy samples. C: Gene Expression study in liver biopsy samples. Results were expressed as fold change (relative to GAPDH) in obese non-diabetics and obese diabetics compared to non-obese subjects. The mRNA expression of these target genes levels in biopsy samples were compared between non-obese, obese non-diabetics and obese diabetics using One-way ANOVA for continuous variables. This study is an extended work to our previous published articles on TREM-1 and TREM-2 gene expressions in non-obese and obese populations [14]. Data were shown as mean ± SD (N=5 non-obese; 24 obese non-diabetics; 22 obese diabetics); *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001.
Increased expression of TREM-1, DAP12, HMGB1, RAGE, TLR4 and TLR2 protein levels in the tissues of obese diabetic subjects
The protein levels of TREM1, DAP12, HMGB1, RAGE, TLR4 and TLR2 was significantly higher in obese diabetic and non-diabetic subjects compared to non-obese subjects; at the same time and TREM-2 protein expression was significantly decreased. The protein expression levels (fold-change) of TREM-1, TREM-2, DAP12, HMGB1, RAGE, TLR4 and TLR2 in the biopsy tissues are shown in Figure 2. TREM-1, DAP12, HMGB1, TLR4 and TLR2 mRNA expression folds were significantly (P<0.05) higher in the omentum (Figure 2A), subcutaneous (Figure 2B) and liver (Figure 2C) tissue biopsies of obese diabetic subjects compared to the obese non-diabetics and non-obese patients. The RAGE expression levels were significantly (P<0.05) higher in omentum and liver tissues (not in subcutaneous) of obese diabetics compared to obese non-diabetics and non-obese subjects. The TREM-1, DAP12, HMGB1, TLR4 and TLR2 were also found to be significantly increased only in the omentum, and there was no significant difference in the liver and subcutaneous tissue biopsies of obese non-diabetics compared to non-obese patients. At the same time, TREM-2 protein levels were significantly (P<0.05) down regulated in the biopsy tissues of obese diabetic subjects compared to obese non-diabetics and non-obese subjects. The significant (P<0.05) down regulation of TREM-2 was also observed in omentum (not in liver and subcutaneous) biopsy tissues of obese non-diabetics compared to non-obese subjects.
Figure 2.
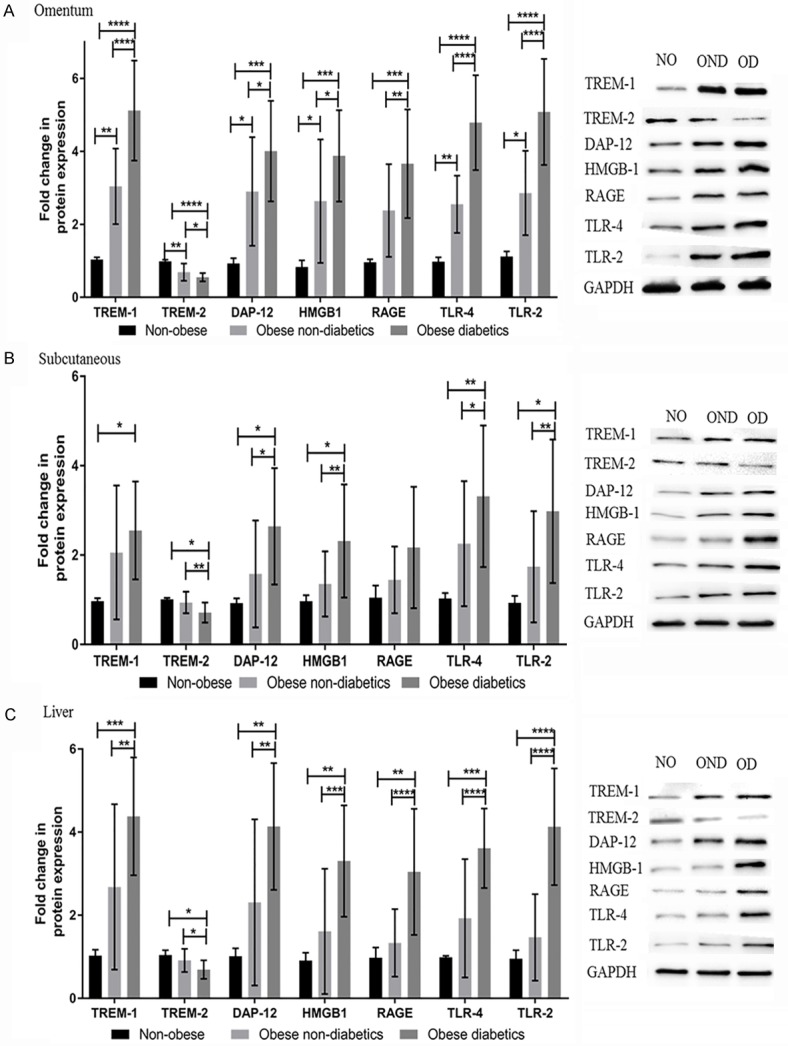
TREM-1, TREM-2, DAP12, HMGB1, RAGE, TLR-4 and TLR-2 protein expression studies using western blotting in the biopsy samples of study subjects. A: Gene Expression study in omentum biopsy samples. B: Gene Expression study in subcutaneous biopsy samples. C: Gene Expression study in liver biopsy samples. The band intensity was measured by densitometry analysis using Image J software. Results were expressed as fold change (relative to GAPDH) in obese non-diabetics and obese diabetics compared to non-obese subjects. The protein expression of these target genes levels in biopsy samples were compared between non-obese, obese non-diabetics and obese diabetics using One-way ANOVA for continuous variables. Data were shown as mean ± SD (N=5 non-obese (NO); 24 obese non-diabetics (OND); 22 obese diabetics (OD); *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001.
High immunoreactivity of TREM1, DAP12, HMGB1, RAGE, TLR4 and TLR2 in the tissue biopsies of obese diabetic subjects
The dual immunofluorescence study was performed to examine the association of TREM1 or TREM2 with DAP12, HMGB1, RAGE, TLR4 and TLR2 in the omentum, subcutaneous and liver biopsy samples. Figure 3 depicts the co-localization of TREM-1 with TREM-2, DAP12, HMGB1, RAGE, TLR-4 and TLR-2 in the omentum (Figure 3A), subcutaneous (Figure 3B), and liver (Figure 3C), biopsy samples. The increased immunoreactivity of TREM1 in obese subjects was highly associated with increased expression of DAP12, HMGB1, RAGE, TLR4 and TLR2. The decreased immunoreactivity of TREM-2 was inversely associated with the expression of DAP12, HMGB1, RAGE, TLR4 and TLR2 in omentum, subcutaneous and liver biopsy samples of obese subjects.
Figure 3.
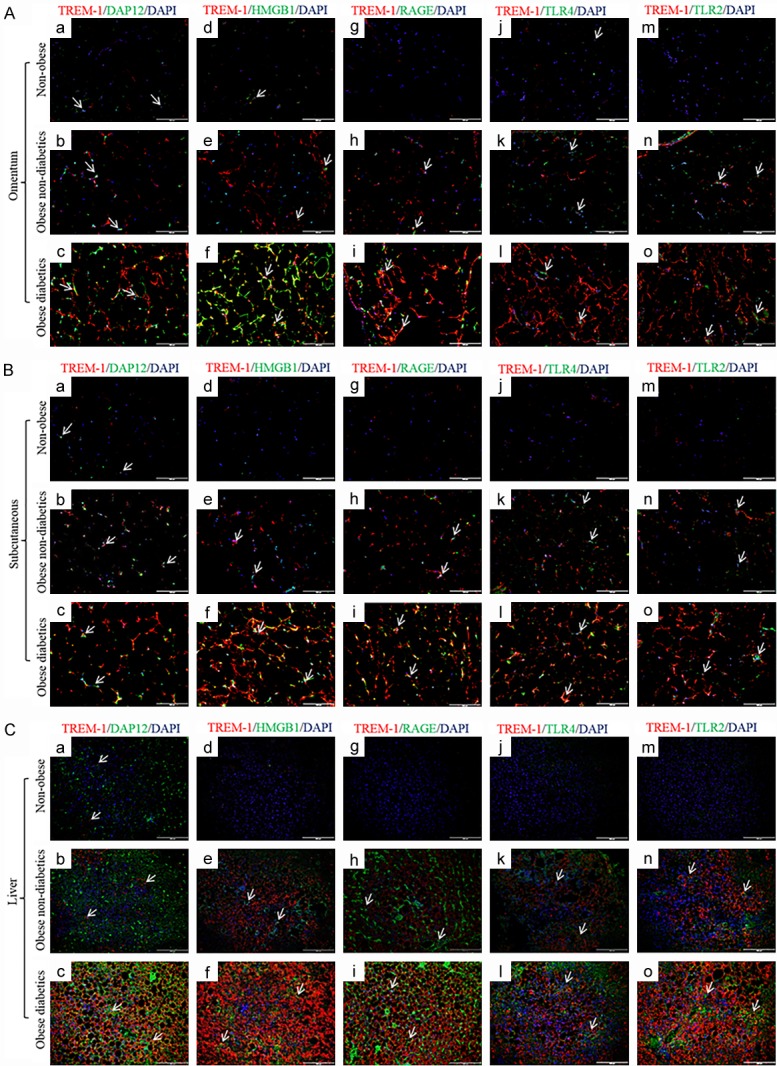
Immunofluorescence staining for TREM-1, DAP12, HMGB1, RAGE, TLR-4 and TLR-2 in the biopsy samples of study subjects. Dual fluorescence staining was done using anti-human TREM-1 (Alexa 594-Red) and DAP12, HMGB1, RAGE, TLR-4 and TLR-2 (Alexa 488-green) antibodies to co-localize TREM-1 with DAP12, HMGB1, RAGE, TLR-4 and TLR-2 respectively, counterstained with DAPI. Negative controls were run by using isotypes for each fluorochrome. A: Immunofluorescence staining in omentum biopsy samples. B: Immunofluorescence staining in subcutaneous biopsy samples. C: Immunofluorescence staining in liver biopsy samples. (N=5 non-obese; 24 obese non-diabetics; 22 obese diabetics).
The MFI levels of TREM-1, DAP12, HMGB1, and TLR4 were significantly (P<0.05) higher in omentum (Figure 4A), subcutaneous (Figure 4B) and liver (Figure 4C) tissues of obese diabetic subjects compared to obese non-diabetics and non-obese patients. The RAGE and TLR2 MFI levels were significantly (P<0.05) higher in omentum and liver tissues (not in subcutaneous) of obese diabetics compared to obese non-diabetics and non-obese subjects. The MFI levels of TREM-1, DAP12, HMGB1, RAGE, TLR4 and TLR2 were also found to be increased only in omentum tissue biopsies of obese non-diabetics compared to non-obese patients. At the same time, obese diabetics had significant (P<0.05) reduction in the TREM-2 MFI levels in the biopsy tissues when compared to non-obese subjects. The significant (P<0.05) down regulation in the TREM-2 MFI levels was also observed in omentum and liver tissues between obese diabetic and obese non-diabetics. Obese non-diabetic subjects showed significant down regulation in TREM-2 MFI levels in omentum samples compared to non-obese subjects.
Figure 4.
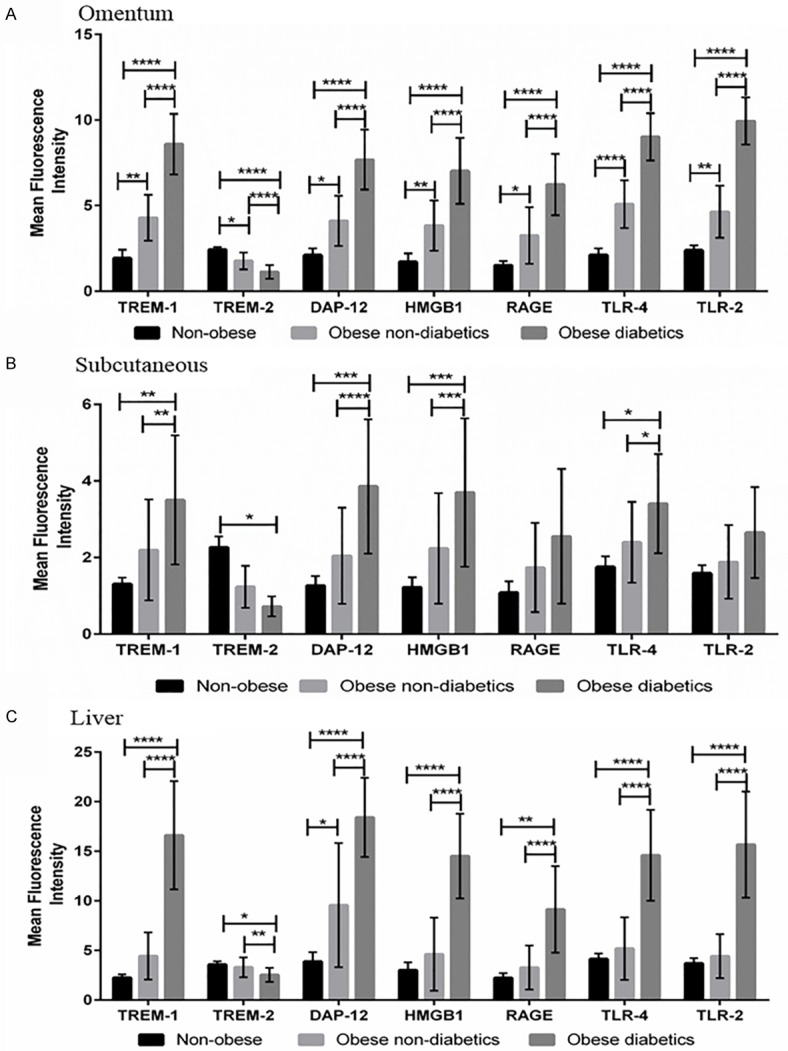
Mean fluorescence intensity (MFI) of TREM-1, TREM-2, DAP12, HMGB1, RAGE, TLR-4 and TLR-2 using immunofluorescence staining in the biopsy samples of study subjects. A: MFI of target genes in omentum biopsy samples. B: MFI of target genes in subcutaneous biopsy samples. C: MFI of target genes in liver biopsy samples. MFI was calculated using Image-J software. The MFI levels of these targets in biopsy samples were compared between non-obese, obese non-diabetics and obese diabetics using One-way ANOVA for continuous variables. Data were shown as mean ± SD (N=5 non-obese; 24 obese non-diabetics; 22 obese diabetics); *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001.
Higher number of patients with increased expression of TREM1, DAP12, HMGB1, RAGE, TLR4 and TLR2 in liver samples of obese diabetics
A higher number of patients with increased TREM-1, DAP12, HMGB1, RAGE, TLR4 and TLR2 were found in both obese non-diabetics and obese diabetics compared to non-obese subjects (Table 4). In the obese diabetic group, there was a significantly (P<0.05) higher number of patients with increased expression of TREM1, DAP12, HMGB1, RAGE, TLR4 and TLR2 in liver biopsy tissues compared to obese non-diabetics. Omentum samples of the obese diabetic group had a significantly (P<0.05) higher number of patients with increased expression of HMGB1 compared to obese non-diabetics. We found that the obese diabetic group showed a significantly (P<0.05) higher number of patients with increased expression of DAP12, HMGB1 and TLR2 compared to obese non-diabetics in subcutaneous tissues. At the same time, subcutaneous tissues of the obese diabetic group had a significantly higher number of patients with under expression of TREM2 compared to obese non-diabetics. The association graphs for TREM-1, TREM-2 DAP12, HMGB1, RAGE, TLR4 and TLR2 in liver, omentum and subcutaneous samples between obese non-diabetics and obese diabetics are also shown in Figure 5.
Table 4.
Expression of TREM-1, TREM-2, DAP-12, TLR2, TLR4, HMGB-1 and RAGE in obese subjects compared to non-obese subjects
| Target genes expression in obese biopsy samples (46) | Obese non-diabetics (24) | Obese diabetics (22) | Correlation (R); P value | |
|---|---|---|---|---|
| Omentum | ||||
| TREM-1 | 44 (95.6%) | 22 (91.7%) | 22 (100%) | NS |
| TREM-2 | 41 (89.1%) | 19 (79.2%) | 22 (100%) | NS |
| DAP-12 | 42 (93.1%) | 20 (83.3%) | 22 (100%) | NS |
| HMGB1 | 40 (86.9%) | 18 (75%) | 22 (100%)* | R=0.371; P=0.022 |
| RAGE | 34 (73.9%) | 16 (66.7%) | 19 (79.2%) | NS |
| TLR-4 | 45 (97.8%) | 23 (95.8%) | 22 (100%) | NS |
| TLR-2 | 42 (91.3%) | 20 (83.3%) | 22 (100%) | NS |
| Subcutaneous | ||||
| TREM-1 | 30 (65.2%) | 13 (54.2%) | 17 (77.3%) | NS |
| TREM-2 | 28 (60.8%) | 11 (45.8%) | 17 (77.3%)* | R=0.322; P=0.038 |
| DAP-12 | 26 (56.5%) | 9 (37.5%) | 17 (77.3%)** | R=0.401; P=0.001 |
| HMGB1 | 24 (52.1%) | 9 (37.5%) | 15 (68.2%)* | R=0.307; P=0.045 |
| RAGE | 19 (41.3%) | 8 (33.3%) | 11 (50%) | NS |
| TLR-4 | 30 (65.2%) | 13 (54.2%) | 17 (77.3%) | NS |
| TLR-2 | 24 (52.1%) | 9 (37.5%) | 15 (68.2%)* | R=0.307; P=0.045 |
| Liver | ||||
| TREM-1 | 34 (73.9%) | 12 (50%) | 22 (100%)**** | R=0.569; P<0.0001 |
| TREM-2 | 31 (67.3%) | 13 (54.2%) | 18 (81.8%) | NS |
| DAP-12 | 34 (73.9%) | 12 (50%) | 22 (100%)**** | R=0.569; P<0.0001 |
| HMGB1 | 29 (63.0%) | 7 (29.2%) | 22 (100%)**** | R=0.733; P<0.0001 |
| RAGE | 22 (47.8%) | 5 (20.8%) | 17 (77.3%)**** | R=0.564; P<0.0001 |
| TLR-4 | 29 (63.0%) | 7 (29.2%) | 22 (100%)**** | R=0.733; P<0.0001 |
| TLR-2 | 28 (60.8%) | 6 (25%) | 22 (100%)**** | R=0.768; P<0.0001 |
Expression of TREM-1, TREM-2, DAP-12, TLR2, TLR4, HMGB-1 and RAGE in obese subjects compared to non-obese subjects. Higher number of subjects with increased expression of TREM-1, DAP-12, TLR2, TLR4, HMGB-1 and RAGE and down regulation of TREM-2 were analyzed between obese non-diabetics and obese diabetics using Fisher’s exact test or Pearson’s χ2 for categorical variables. Data show number of subjects having higher values of these compared to control non-obese subjects. Values show number of subjects (% subjects of total), not significant (NS).
P<0.05;
P<0.01;
***P<0.001;
P<0.0001.
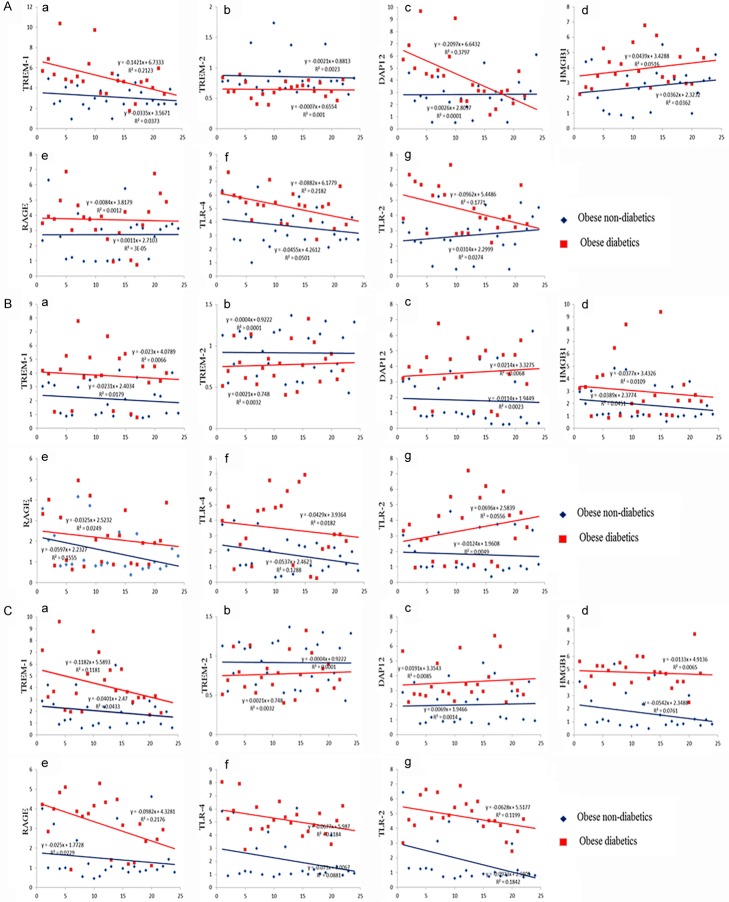
Figure 5.
Correlation of TREM-1, TREM-2, DAP12, HMGB1, RAGE, TLR-4 and TLR-2 expression using scatter blot between obese non-diabetics and obese diabetics. A: Correlation analysis in omentum biopsy samples. B: Correlation analysis in subcutaneous biopsy samples. C: Correlation analysis in liver biopsy samples. Number (n) of patients is given in Y axis (24 obese non-diabetics; 22 obese diabetics). Increased TREM-1, DAP-12, HMGB-1, RAGE, TLR4 and TLR2 in the biopsy tissues was positively associated with obese diabetics compared to obese non-diabetics. Blue linear trend line indicates the expression levels in obese non-diabetics; Red linear trend line indicates the expression levels in obese diabetics.
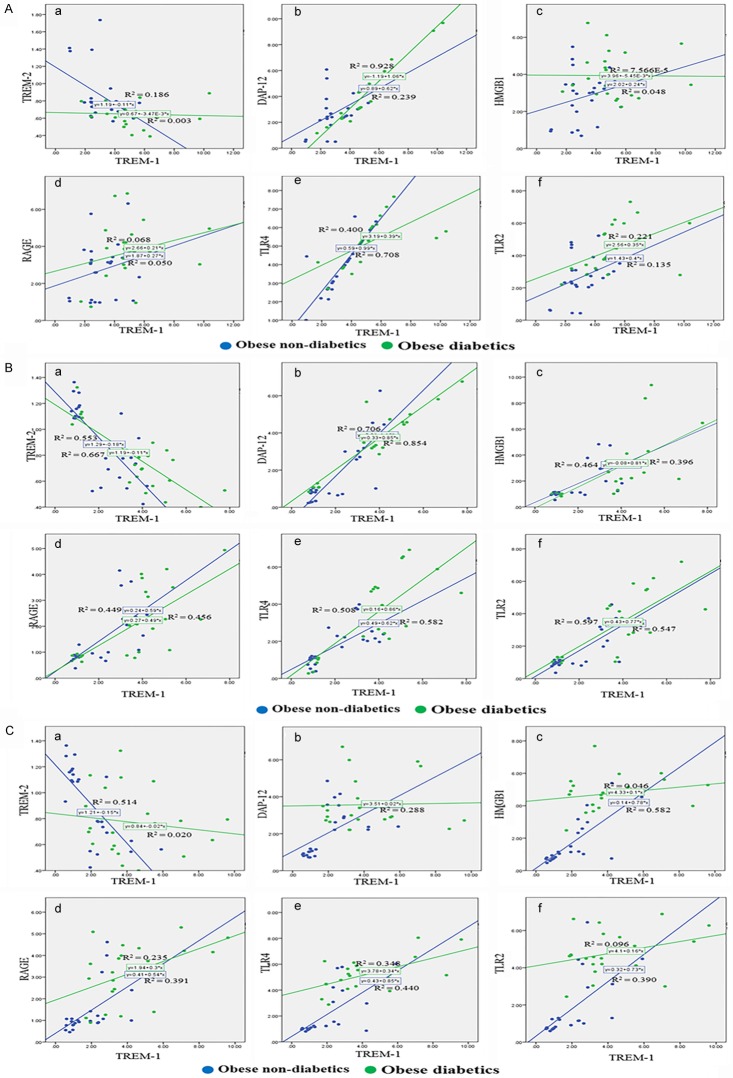
Strong correlation between TREM-1 and HMGB-1, RAGE, TLR2 and TLR4 in liver samples of obese diabetics
The association graph between TREM-1 and TREM-2, DAP12, HMGB-1, RAGE, TLR2 and TLR4 among obese non-diabetics and obese diabetics is shown in Figure 6. A significantly (P<0.05) high association was found between increased TREM-1 and HMGB1, RAGE, TLR4 and TLR2 in the liver biopsy of obese diabetic patients. However, there was no significant association between increased omentum TREM-1 and other target molecules, when compared between obese diabetics and obese non-diabetics. We found a significant (P<0.05) association of increased subcutaneous TREM-1 with DAP12 in the obese diabetic group compared to obese non-diabetics (Table 5).
Figure 6.
Graphical representation of the analysis of TREM-1 in association with TREM-2, DAP12, HMGB1, RAGE, TLR-4 and TLR-2 using scatter blot between obese non-diabetics and obese diabetics. A: Correlation analysis in omentum biopsy samples. B: Correlation analysis in subcutaneous biopsy samples. C: Correlation analysis in liver biopsy samples. Analysis showed strong association of the increased TREM-1 with DAP12, HMGB1, RAGE, TLR-4 and TLR-2 in obese diabetics. Blue linear trend line indicates the expression levels in obese non-diabetics; Green linear trend line indicates the expression levels in obese diabetics.
Table 5.
Correlation between TREM-1 with DAP-12, TLR2, TLR4, HMGB-1 and RAGE in omentum, subcutaneous and liver tissues of study subjects
| Target genes correlation | Obese non-diabetics (24) | Obese diabetics (22) | Correlation (R); P value |
|---|---|---|---|
| Increased TREM-1 association with other genes in omentum biopsy samples | |||
| DAP-12 | 20/22 (90.9%) | 22/22 (100%) | NS |
| HMGB1 | 18/22 (81.8%) | 22/22 (100%) | NS |
| RAGE | 16/22 (72.7%) | 22/22 (100%) | NS |
| TLR-4 | 22/22 (100%) | 22/22 (100%) | NS |
| TLR-2 | 20/22 (90.9%) | 22/22 (100%) | NS |
| Increased TREM-1 association with other genes in subcutaneous biopsy samples | |||
| DAP-12 | 9/13 (69.2%) | 17/17 (100%)* | R=0.449; P=0.026 |
| HMGB1 | 9/13 (69.2%) | 15/17 (88.2%) | NS |
| RAGE | 8/13 (61.5%) | 11/17 (67.4%) | NS |
| TLR-4 | 13/13 (100%) | 17/17 (100%) | NS |
| TLR-2 | 9/13 (69.2%) | 15/17 (88.2%) | NS |
| Increased TREM-1 association with other genes in liver biopsy samples | |||
| DAP-12 | 12/12 (100%) | 22/22 (100%) | NS |
| HMGB1 | 7/12 (58.3%) | 22/22 (100%)** | R=0.562; P=0.003 |
| RAGE | 5/12 (41.7%) | 17/22 (77.3%)* | R=0.356; P=0.045 |
| TLR-4 | 7/12 (58.3%) | 22/22 (100%)** | R=0.562; P=0.003 |
| TLR-2 | 6/12 (50%) | 22/22 (100%)** | R=0.627; P=0.001 |
Subject’s categorical variables with correlation between TREM-1 and DAP-12, TLR2, TLR4, HMGB-1 and RAGE were analyzed among obese non-diabetics and obese diabetics using Fisher’s exact test or Pearson’s χ2 test. Data show number of subjects having higher values of these compared to control non-obese subjects. Values show number of subjects (% subjects of total), not significant (NS).
P<0.05;
P<0.01.
Discussion
Obesity is a chronic inflammatory disease characterized by adipocyte hypertrophy, marked infiltration of macrophages and lymphocytes in adipose tissues that correlate with chronic low-grade systemic inflammation [18]. Obesity-associated chronic low-grade inflammation has been proposed to be an important reason for the development of insulin resistance and possibly type II DM [19]. TREM-1 is a recently recognized cell surface marker that activates myeloid cells which regulate innate immune system. Our previous study suggests an increase in the expression of TREM-1 in the pathogenesis of obesity-induced insulin resistance. In this study, we further analyzed the interaction of TREM-1 with other mediators of inflammation, which may be involved in this process. The activation of TREM-1/DAP12 results in the increased secretion of TNF-α, GM-CSF, chemokines (MPO, MIP-1 and MCP-1), and cytokines (IL-6, IL-8 and IL-12) from neutrophils or monocytes/macrophages [20]. Our findings suggest that TREM-1/TREM-2 cell surface receptor family plays a key role in the pathogenesis of insulin resistance in severe obese (BMI>35) patients. Obese patients with increased expression of TREM-1 also had increased DAP12 expression in tissue biopsy tissue samples (omentum, subcutaneous and liver). We also found increased expression of TREM-1 and DAP12 in the liver biopsy samples of all pre-diabetic (HOMA-IR>2; 12/12, 100%) and obese diabetic patients (22/22, 100%). The increased downstream signalling of TREM-1 may lead to increased production of inflammatory cytokines, further augmenting the inflammatory cascade. Thus, TREM-1/DAP12 pathway could be a promising therapeutic strategy, and it is important to get further insight into the signalling cascade downstream of TREM-1 [21].
DAP-12 is a common trans-membrane adaptor protein for TREM-1 and TREM-2. However, the activation of TREM-2/DAP-12 is associated with TREM-1 and TLRs inhibition [22]. Thus, TREM-2 is involved in the anti-inflammatory responses and negatively regulates the inflammatory response [23]. The role of TREM-2 in metabolic syndrome or obesity induced insulin resistance has not been yet defined in the literature. Our results showed that the TREM-2 is significantly down-regulated with minimal immune-reactivity in liver, omentum and subcutaneous fat tissues in obese diabetic population compared to non-obese population. The decreased expression of TREM-2 is reversely correlated with the increased expression of TREM-1 and DAP-12 in obese diabetic populations. Interestingly, there was no difference in TREM-2 expression between obese non-diabetics and non-obese patients. The differential expression of TREM-2 may be due to increased inflammation in obese diabetics compared to the non-obese subjects. This indicates that increased expression of TREM-1 corresponds to the TREM-2 down-regulation in obese patients with diabetics. Thus, TREM-2 can be a potential target to attenuate the development of TREM-1 and TLR-induced insulin resistance.
TREM-1 is activated through multiple ligands such as HMGB-1 and HSP-70 [15,17]. In the chronic inflammatory stage, the levels of HMGB-1 are elevated secondary to necrosis/cell damage in adipose tissue [24]. In this study, we found significantly increased expression of HMGB-1 in omentum, subcutaneous and liver biopsy tissues of obese diabetic subjects compared to obese non-diabetic and non-obese subjects. Furthermore, increased HMGB1 activates TREM-1 downstream signalling, leading to increased secretion of inflammatory cytokines (IL-6, TNF-α and IL-1β) [25]. We also found that significantly increased expression of HMGB-1 in association with TREM-1 in biopsied tissues of obese subjects, suggesting that HMGB-1 might have an important role in TREM-1 activation. Our results further support the findings from animal studies suggesting that increased secretion of HMGB-1 and TREM1 with inflammation [26]. Although the association of HMGB1 and TREM-1 has been evaluated in this study, there is a need to further investigate the significant role of necrotic and immune inflammatory cells that actively secrete HMGB1 and TREM-1 in the development of obesity-induced insulin resistance. Animal studies also reported that apart from TREM1, HMGB1 also reported to induce RAGE. In mice studies, RAGE was reported to contribute to insulin resistance when mice were fed high fat diet to induce tissue inflammation [27]. In our study, we found significantly increased expression of RAGE in liver tissues (100%) of obese-diabetic population along with an increased expression in the omentum and subcutaneous fat in obese-diabetic compared to non-obese and obese non-diabetic population. Furthermore, co-localization and increased expression of RAGE in association with TREM-1 suggest that RAGE and its ligand HMGB-1 could play a crucial role in inducing inflammatory cytokines and inflammation induced by TREM-1 activation [15].
TREM-1 activation can lead to TLRs activation and both act synergistically to increase inflammation [28]. In our study, there was increased expression of TLR2 and TLR4 in omentum, subcutaneous and liver biopsy tissues of obese-diabetic subjects. The increased expression of TLRs and HMGB-1 in association with TREM-1 indicates the synergistic effect of TLRs and HMGB1 in TREM-1 activation and in the pathogenesis of insulin resistance. Although all patients who had increased TREM-1 did not have increased HMGB-1, this suggests that TLRs-dependent TREM-1 activation might have a different ligand [26,29]. These findings suggest that TLRs also play a critical role in initiating the insulin resistance through TREM-1.
Obesity-induced chronic inflammation in visceral adipose tissues (VAT) plays a vital role in the pathogenesis of insulin resistance, diabetes, and metabolic syndrome [30]. VAT is also more metabolically active than the subcutaneous adipose tissue [31]. There was significantly increased expression of all inflammatory markers (TREM1, DAP12, HMGB1, RAGE, TLR4 and TLR2) in obese diabetics compared to obese non-diabetics in omentum and liver tissues. VAT inflammation has been implicated as the primary reason in the pathogenesis of adipose tissue-induced insulin resistance. Insulin resistance in the adipose tissue promotes lipolysis due to uninhibited hormone-sensitive lipase with release of excessive amounts of free fatty acids (FFAs) in the circulation [32] that inhibit insulin signalling in insulin-dependent tissues such as liver and muscle. Mobilization of excessive FFAs and pro-inflammatory cytokines from the visceral fat to liver through the portal vein infiltrates the liver tissues and inhibits insulin signalling and promotes hepatic insulin resistance [33]. We found significantly increased hepatosteatosis (86.4% vs 45.8%) with 27.2% of obese-diabetic patient population having 66-100% of hepatosteatosis.
In our study, we found a strong correlation between TREM-1 with HMGB1, TLR2, TLR4 and RAGE in liver biopsies of the obese diabetics compared to obese non-diabetics. These results further support our hypothesis that chronic inflammation of the liver plays a role in insulin resistance. Of the 24 obese non-diabetic patients, 12 had increased expression of TREM-1 and elevated HOMA-IR index >2 indicating these patients were pre-diabetic at this stage. In these pre-diabetic patients, TREM-1 expression also precedes the changes in the expression of other inflammatory markers like HMGB1, RAGE, TLR4 and TLR2 in the liver. The results of our study support that lipid-sensing receptor TREM-1/DAP12 association-induced HMGB-1, RAGE and TLRs in obesity play a key role in ultimately inducing insulin resistance. Thus, our study suggests that targeting the TREM-1/DAP12 pathway could be a promising therapeutic strategy in obesity-induced insulin resistance.
In conclusion, the TREM-1/DAP12 along with HMGB1 is significantly increased in obese diabetics and prediabetic patients. Along with TREM1/DAP12 and its ligand HMGB1, TLRs and RAGE were also significantly increased in obese diabetics and can potentially play a significant role in obesity-induced insulin resistance (Figure 7). However, future studies are warranted to identify the role of pathogenic inflammatory cells associated with expression of TREM-1, TREM-2, DAP-12 and other molecules in the pathogenesis of obesity-induced insulin resistance using in vitro and in vivo model. Our findings suggest that TREM-1-induced HMGB1 ligand activation and inflammatory cell receptors (RAGE, TLR-4 and TLR-2) might ultimately lead to obesity-induced insulin resistance.
Figure 7.
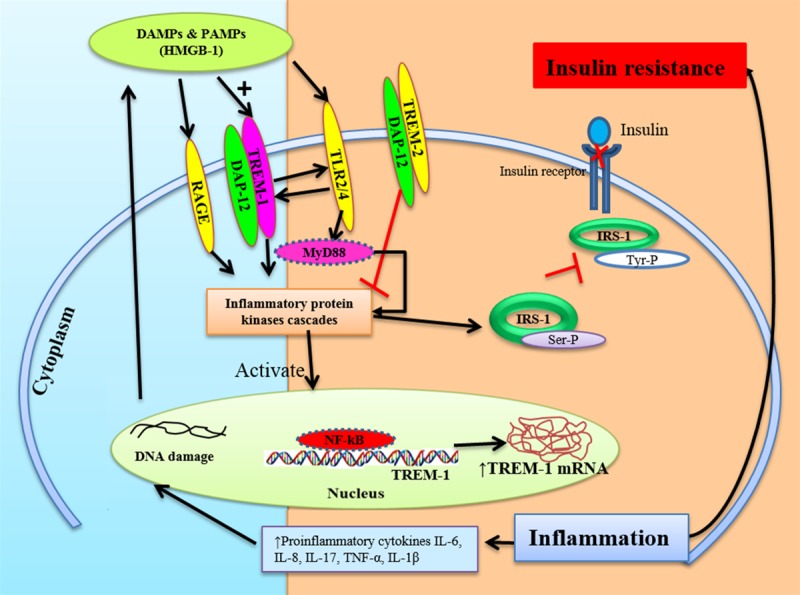
Schematic representation of the Inflammation mediated TREM-1 induced insulin resistance. Fat deposition in obesity results in chronic inflammation leading to TREM-1 activation and thereby increased secretion of DAMPs. TREM-1 activation and DAMPs will further activate TLRs and RAGE leading to activation of inflammatory protein kinases and downstream signaling. Activated TLRs will further activate the TREM-1 expression, contributing to the cascade of chronic inflammation. This results in increased secretion of pro-inflammatory cytokines like IL-6, IL-1β and TNF-alpha, and decreased TREM-2 expression, leading to increased inflammation, TREM-1 expression, and obesity induced insulin resistance. Increased expression of the inflammatory protein kinases reduces results in insulin resistance. The TREM1/DAP12 along with HMGB1, TLRs and RAGE significantly increased in obese diabetics and pre-diabetic patients. Thus TREM-1/DAP12 may potentially play a significant role in obesity-induced insulin resistance.
Acknowledgements
This study was supported by a research grant to Dr. Kalyana C Nandipati from the LB692 Nebraska Tobacco Settlement Funds to Creighton University, and by the research grants R01HL116042 and R01HL128063 from the National Institutes of Health, USA to DK Agrawal. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Disclosure of conflict of interest
None.
References
- 1.Ballak DB, van Diepen JA, Moschen AR, Jansen HJ, Hijmans A, Groenhof GJ, Leenders F, Bufler P, Boekschoten MV, Müller M, Kersten S, Li S, Kim S, Eini H, Lewis EC, Joosten LA, Tilg H, Netea MG, Tack CJ, Dinarello CA, Stienstra R. IL-37 protects against obesity-induced inflammation and insulin resistance. Nat Commun. 2014;5:4711. doi: 10.1038/ncomms5711. [DOI] [PubMed] [Google Scholar]
- 2.Revelo XS, Tsai S, Lei H, Luck H, Ghazarian M, Tsui H, Shi SY, Schroer S, Luk CT, Lin GH, Mak TW, Woo M, Winer S, Winer DA. Perforin is a novel immune regulator of obesity-related insulin resistance. Diabetes. 2015;64:90–103. doi: 10.2337/db13-1524. [DOI] [PubMed] [Google Scholar]
- 3.Li P, Oh DY, Bandyopadhyay G, Lagakos WS, Talukdar S, Osborn O, Johnson A, Chung H, Mayoral R, Maris M, Ofrecio JM, Taguchi S, Lu M, Olefsky JM. LTB4 promotes insulin resistance in obese mice by acting on macrophages, hepatocytes and myocytes. Nat Med. 2015;21:239–247. doi: 10.1038/nm.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto Y, Yamamoto H. RAGE-mediated inflammation, type 2 diabetes, and diabetic vascular complication. Front Endocrinol (Lausanne) 2013;4:105. doi: 10.3389/fendo.2013.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santoni M, Andrikou K, Sotte V, Bittoni A, Lanese A, Pellei C, Piva F, Conti A, Nabissi M, Santoni G, Cascinu S. Toll like receptors and pancreatic diseases: from a pathogenetic mechanism to a therapeutic target. Cancer Treat Rev. 2015;41:569–576. doi: 10.1016/j.ctrv.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Andrews M, Soto N, Arredondo-Olguín M. Association between ferritin and hepcidin levels and inflammatory status in patients with type 2 diabetes mellitus and obesity. Nutrition. 2015;31:51–57. doi: 10.1016/j.nut.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 7.Vitseva OI, Tanriverdi K, Tchkonia TT, Kirkland JL, McDonnell ME, Apovian CM, Freedman J, Gokce N. Inducible Toll-like receptor and NFkappaB regulatory pathway expression in human adipose tissue. Obesity (Silver Spring) 2008;16:932–937. doi: 10.1038/oby.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prakash J, Pichchadze G, Trofimov S, Livshits G. Age and genetic determinants of variation of circulating levels of the receptor for advanced glycation end products (RAGE) in the general human population. Mech Ageing Dev. 2015;145:18–25. doi: 10.1016/j.mad.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Yan SF, Ramasamy R, Schmidt AM. The RAGE axis: a fundamental mechanism signaling danger to the vulnerable vasculature. Circ Res. 2010;106:842–853. doi: 10.1161/CIRCRESAHA.109.212217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seong SY, Matzinger P. Hydrophobicity: an ancient damage-associated molecular pattern that initiates innate immune responses. Nat Rev Immunol. 2004;4:469–478. doi: 10.1038/nri1372. [DOI] [PubMed] [Google Scholar]
- 11.Goldin A, Beckman JA, Schmidt AM, Creager MA. Advanced glycation end products: sparking the development of diabetic vascular injury. Circulation. 2006;114:597–605. doi: 10.1161/CIRCULATIONAHA.106.621854. [DOI] [PubMed] [Google Scholar]
- 12.Szczepanski MJ, Luczak M, Olszewska E, Molinska-Glura M, Zagor M, Krzeski A, Skarzynski H, Misiak J, Dzaman K, Bilusiak M, Kopec T, Leszczynska M, Witmanowski H, Whiteside TL. Molecular signaling of the HMGB1/RAGE axis contributes to cholesteatoma pathogenesis. J Mol Med (Berl) 2015;93:305–314. doi: 10.1007/s00109-014-1217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Bremen T, Drömann D, Luitjens K, Dodt C, Dalhoff K, Goldmann T, Schaaf B. Triggering receptor expressed on myeloid cells-1 (Trem-1) on blood neutrophils is associated with cytokine inducibility in human E. coli sepsis. Diagn Pathol. 2013;8:24. doi: 10.1186/1746-1596-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian S, Pallati PK, Rai V, Sharma P, Agrawal DK, Nandipati KC. Increased expression of triggering receptor expressed on myeloid cells-1 in the population with obesity and insulin resistance. Obesity (Silver Spring) 2017;25:527–538. doi: 10.1002/oby.21714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Li J, Salcedo R, Mivechi NF, Trinchieri G, Horuzsko A. The proinflammatory myeloid cell receptor TREM-1 controls Kupffer cell activation and development of hepatocellular carcinoma. Cancer Res. 2012;72:3977–3986. doi: 10.1158/0008-5472.CAN-12-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouchon A, Facchetti F, Weigand MA, Colonna M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410:1103–1107. doi: 10.1038/35074114. [DOI] [PubMed] [Google Scholar]
- 17.El Mezayen R, El Gazzar M, Seeds MC, McCall CE, Dreskin SC, Nicolls MR. Endogenous signals released from necrotic cells augment inflammatory responses to bacterial endotoxin. Immunol Lett. 2007;111:36–44. doi: 10.1016/j.imlet.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 19.Luck H, Tsai S, Chung J, Clemente-Casares X, Ghazarian M, Revelo XS, Lei H, Luk CT, Shi SY, Surendra A, Copeland JK, Ahn J, Prescott D, Rasmussen BA, Chng MH, Engleman EG, Girardin SE, Lam TKT, Croitoru K, Dunn S, Philpott DJ, Guttman DS, Woo M, Winer S, Winer DA. Regulation of obesity-related insulin resistance with gut anti-inflammatory agents. Cell Metab. 2015;21:527–542. doi: 10.1016/j.cmet.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Duan M, Wang ZC, Wang XY, Shi JY, Yang LX, Ding ZB, Gao Q, Zhou J, Fan J. TREM-1, an inflammatory modulator, is expressed in hepatocellular carcinoma cells and significantly promotes tumor progression. Ann Surg Oncol. 2015;22:3121–3129. doi: 10.1245/s10434-014-4191-7. [DOI] [PubMed] [Google Scholar]
- 21.Tessarz AS, Cerwenka A. The TREM-1/DAP12 pathway. Immunol Lett. 2008;116:111–116. doi: 10.1016/j.imlet.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 22.Paradowska-Gorycka A, Jurkowska M. Structure, expression pattern and biological activity of molecular complex TREM-2/DAP12. Hum Immunol. 2013;74:730–737. doi: 10.1016/j.humimm.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M, Piccio L, Hernandez M, Colonna M. Cutting edge: TREM-2 attenuates macrophage activation. J Immunol. 2006;177:3520–3524. doi: 10.4049/jimmunol.177.6.3520. [DOI] [PubMed] [Google Scholar]
- 24.Gunasekaran MK, Viranaicken W, Girard AC, Festy F, Cesari M, Roche R, Hoareau L. Inflammation triggers high mobility group box 1 (HMGB1) secretion in adipose tissue, a potential link to obesity. Cytokine. 2013;64:103–111. doi: 10.1016/j.cyto.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Arts RJ, Joosten LA, van der Meer JW, Netea MG. TREM-1: intracellular signaling pathways and interaction with pattern recognition receptors. J Leukoc Biol. 2013;93:209–215. doi: 10.1189/jlb.0312145. [DOI] [PubMed] [Google Scholar]
- 26.Pelham CJ, Agrawal DK. Emerging roles for triggering receptor expressed on myeloid cells receptor family signaling in inflammatory diseases. Expert Rev Clin Immunol. 2014;10:243–256. doi: 10.1586/1744666X.2014.866519. [DOI] [PubMed] [Google Scholar]
- 27.Song F, Hurtado del Pozo C, Rosario R, Zou YS, Ananthakrishnan R, Xu X, Patel PR, Benoit VM, Yan SF, Li H, Friedman RA, Kim JK, Ramasamy R, Ferrante AW, Schmidt AM. RAGE regulates the metabolic and inflammatory response to high-fat feeding in mice. Diabetes. 2014;63:1948–1965. doi: 10.2337/db13-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fortin CF, Lesur O, Fulop T. Effects of TREM-1 activation in human neutrophils: activation of signaling pathways, recruitment into lipid rafts and association with TLR4. Int Immunol. 2007;19:41–50. doi: 10.1093/intimm/dxl119. [DOI] [PubMed] [Google Scholar]
- 29.Zheng H, Heiderscheidt CA, Joo M, Gao X, Knezevic N, Mehta D, Sadikot RT. MYD88-dependent and -independent activation of TREM-1 via specific TLR ligands. Eur J Immunol. 2010;40:162–171. doi: 10.1002/eji.200839156. [DOI] [PubMed] [Google Scholar]
- 30.Odegaard JI, Chawla A. The immune system as a sensor of the metabolic state. Immunity. 2013;38:644–654. doi: 10.1016/j.immuni.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyon CJ, Law RE, Hsueh WA. Minireview: adiposity, inflammation, and atherogenesis. Endocrinology. 2003;144:2195–2200. doi: 10.1210/en.2003-0285. [DOI] [PubMed] [Google Scholar]
- 32.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 33.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]