Summary
Recent studies have used fMRI signals from early visual areas to reconstruct simple geometric patterns. Here, we demonstrate a new Bayesian decoder that uses fMRI signals from early and anterior visual areas to reconstruct complex natural images. Our decoder combines three elements: a structural encoding model that characterizes responses in early visual areas; a semantic encoding model that characterizes responses in anterior visual areas; and prior information about the structure and semantic content of natural images. By combining all these elements, the decoder produces reconstructions that accurately reflect both the spatial structure and semantic category of the objects contained in the observed natural image. Our results show that prior information has a substantial effect on the quality of natural image reconstructions. We also demonstrate that much of the variance in the responses of anterior visual areas to complex natural images is explained by the semantic category of the image alone.
Keywords: DECODING, BRAIN READING, NATURAL IMAGES, RECEPTIVE FIELD, GABOR WAVELETS, VISUAL CORTEX, SEMANTIC, fMRI
Introduction
Functional magnetic resonance imaging (fMRI) provides a measurement of activity in the many separate brain areas that are activated by a single stimulus. This property of fMRI makes it an excellent tool for brain reading, in which the responses of multiple voxels are used to decode the stimulus that evoked them (Haxby et al., 2001; Carlson et al., 2002; Cox and Savoy, 2003; Haynes and Rees, 2005; Kamitani and Tong, 2005; Thirion et al., 2006; Kay et al., 2008; Miyawaki et al., 2008). The most common approach to decoding is image classification. In classification, a pattern of activity across multiple voxels is used to determine the discrete class from which the stimulus was drawn (Haxby et al., 2001; Carlson et al., 2002; Cox and Savoy, 2003; Haynes and Rees, 2005; Kamitani and Tong, 2005).
Two recent studies have moved beyond classification and demonstrated stimulus reconstruction (Thirion et al., 2006; Miyawaki et al., 2008). The goal of reconstruction is to produce a literal picture of the image that was presented. The Thirion et al. (2006) and Miyawaki et al. (2008) studies achieved reconstruction by analyzing the responses of voxels in early visual areas. To simplify the problem, both studies used geometric stimuli composed of flickering checkerboard patterns. However, a general brain reading device should be able to reconstruct natural images (Kay and Gallant, 2009). Natural images are important targets for reconstruction because they are most relevant for daily perception and subjective processes such as imagery and dreaming. Natural images are also very challenging targets for reconstruction, because they have complex statistical structure (Field, 1987; Karklin and Lewicki, 2009; Cadieu and Olshausen, 2009) and rich semantic content (i.e., they depict meaningful objects and scenes). A method for reconstructing natural images should be able to reveal both the structure and semantic content of the images simultaneously.
In this paper, we present a Bayesian framework for brain reading that produces accurate reconstructions of the spatial structure of natural images, while simultaneously revealing their semantic content. Under the Bayesian framework used here, a reconstruction is defined as the image that has the highest posterior probability of having evoked the measured response. Two sources of information are used to calculate this probability: information about the target image that is encoded in the measured response, and pre-existing, or prior information about the structure and semantic content of natural images.
Information about the target image is extracted from measured responses by applying one or more encoding models (Nevado et al., 2004; Wu et al., 2006). An encoding model is represented mathematically by a conditional distribution, p(r|s), which gives the likelihood that the measured response r was evoked by the image s (here bold r denotes the collected responses of multiple voxels; italicized r will be used to denote the response of a single voxel). Note that functionally distinct visual areas are best characterized by different encoding models, so a reconstruction based on responses from multiple visual areas will use a distinct encoding model for each area.
Prior information about natural images is also represented as a distribution, p(s), that assigns high probabilities to images that are most natural (Figure 1, inner bands of image samples) and low probabilities to more artificial, random or noisy images (Figure 1, outermost band of image samples).
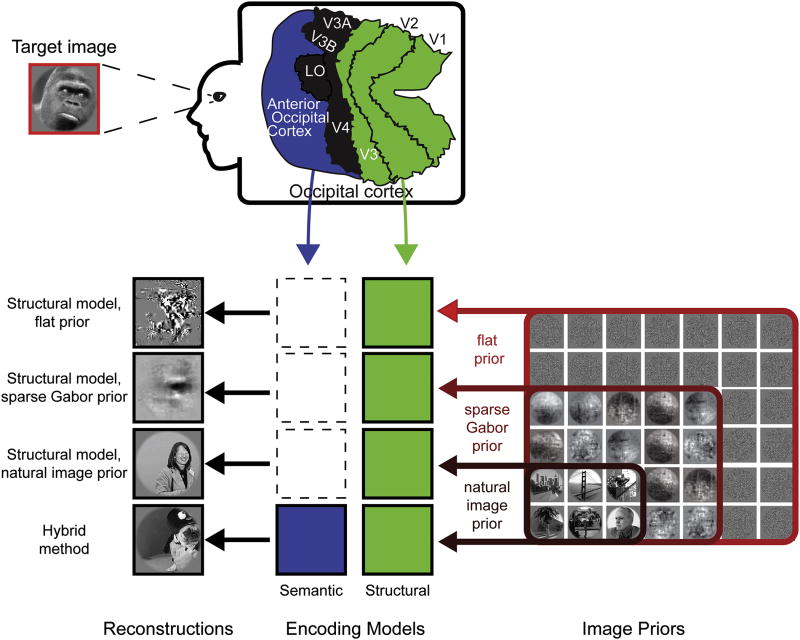
Figure 1. The Bayesian reconstruction framework.
The goal of this experiment was to reconstruct target images from BOLD fMRI responses recorded from occipital cortex. Reconstructions were obtained by using a Bayesian framework to combine voxel responses, structural and semantic encoding models and image priors. Target images were grayscale photographs selected at random from a large database of natural images. The fMRI slice coverage included early visual areas V1, V2 and V3; intermediate visual areas V3A, V3B, V4 and lateral occipital (labeled LO here); and a band of occipital cortex anterior to lateral occipital (here called AOC). Recorded voxel responses were used to fit two distinct encoding models: a structural encoding model (green) that reflects how information is encoded in early visual areas, and a semantic encoding model (blue) that reflects how information is encoded in the AOC. Three image priors were used to bias reconstructions in favor of those with the characteristics of natural images: a flat prior that does not bias reconstructions, a sparse Gabor prior that ensures reconstructions possess the lower-order statistical properties of natural images, and a natural image prior that ensures reconstructions are natural images. Several different types of reconstructions were obtained by combining the encoding models and priors in different ways: the structural model and a flat prior; the structural model and a sparse Gabor prior; the structural model and a natural image prior; and the structural model, the semantic model and a natural image prior (hybrid method). These various methods produced reconstructions with very different structural and semantic qualities, as shown in Figures 2 and 3.
The critical step in reconstruction is to calculate the probability that each possible image evoked the measured response. This is accomplished by using Bayes theorem to combine the encoding models and the image prior:
The posterior distribution, p(s|r), gives the probability that image s evoked response r. The encoding models and voxel responses from functionally distinct areas are indexed by i. To produce a reconstruction, p(s|r) is evaluated for a large number of images. The image with the highest p(s|r) (or posterior probability) is selected as the reconstruction, commonly known as the maximum a posteriori estimate (Zhang et al., 1998).
In a previous study we used the structural encoding model without invoking the Bayesian framework in order to solve image identification (Kay et al., 2008). The goal of image identification is to determine which specific image was seen on a certain trial, when that image was drawn from a known set of images. Image identification provides an important foundation for image reconstruction, but it is a much simpler problem because the set of target images is known beforehand. Furthermore, success at image identification does not guarantee success at reconstruction, because a target image may be identified on the basis of a small number of image features that are not sufficient to produce an accurate reconstruction.
In this paper we investigate two key factors that determine the quality of reconstructions of natural images from fMRI data: encoding models and image priors. We find that fMRI data and a structural encoding model are insufficient to support high-quality reconstructions of natural images. Combining these with an appropriate natural image prior produces reconstructions that, while structurally accurate, fail to reveal the semantic content of the target images. However, by applying an additional semantic encoding model that extracts the information present in anterior visual areas, we produce reconstructions that accurately reflect semantic content of the target images as well. A comparison of the two encoding models shows that they most accurately predict the responses of functionally distinct and anatomically separated voxels. The structural model best predicts responses of voxels in early visual areas (V1, V2 and so on), while the semantic model best predicts responses of voxels anterior to V4, V3A, V3B, and the posterior portion of lateral occipital. Furthermore, the accuracy of predictions of these models is comparable to the accuracy of predictions obtained for single neurons in area V1.
Results
Blood-oxygen-level-dependent (BOLD) fMRI measurements of occipital visual areas were made while three subjects viewed a series of monochromatic natural images (Kay et al., 2008). Functional data were collected from early (V1, V2, V3) and intermediate (V3A, V3B, V4, lateral occipital) visual areas, and from a band of occipital cortex directly anterior to lateral occipital that we refer to here as anterior occipital cortex (AOC). The experiment consisted of two stages: model estimation and image reconstruction. During model estimation subjects viewed 1,750 achromatic natural images while functional data were collected. These data were used to fit encoding models for each voxel. During image reconstruction functional data were collected while subjects viewed 120 novel target images. These data were used to generate reconstructions.
Reconstructions that use a structural encoding model for early visual areas and an appropriate image prior
Our Bayesian framework requires that each voxel be fit with an appropriate encoding model. In our previous study we showed that a structural encoding model based upon Gabor wavelets could be used to extract a large amount of information from individual voxels in early visual areas (Kay et al., 2008). Therefore, we began by using this model to produce reconstructions.
Under the structural encoding model, the likelihood of a voxel's response r to an image s is determined by its tuning along the dimensions of space, orientation, and spatial frequency (Kay et al., 2008). The model includes a set of weights that can be adjusted to fit the specific tuning of single voxels. These weights were fit for all of the voxels in our dataset using a coordinate-descent optimization procedure (see Experimental Procedures). This procedure produced a separate encoding model, p(r|s), for each voxel. Those voxels whose responses could be predicted accurately by the model were then selected (see Experimental Procedures for specific voxel selection criteria) for use in reconstruction. The individual models for each of the selected voxels were then combined into a single multi-voxel structural encoding model, p(r|s) (see Experimental Procedures for details on how individual models are combined into a multi-voxel model). The majority of selected voxels were located in early visual areas (V1, V2, and V3).
The Bayesian framework also requires an appropriate prior. The reconstructions reported in Thirion et al. (2006), and Miyawaki et al. (2008) used no explicit source of prior information. To obtain comparable results to theirs, we began with a flat prior that assigns the same probability to all possible images. This prior makes no strong assumptions about the stimulus, but instead assumes that noise patterns are just as likely as natural images (see Figure 1). Thus, when the flat prior is used, only the information encoded in the responses of the voxels is available to support reconstruction. (Formally, using the flat prior amounts to setting the prior, p(s), in equation (1) to a constant).
To produce reconstructions, the structural encoding model, the flat prior, and the selected voxels were used to evaluate the posterior probability (see equation 1) that an image s evoked the responses of the selected voxels. A greedy serial search algorithm was used to converge on an image with a high (relative to an initial image with all pixels set to zero) posterior probability. This image was selected as the reconstruction. Typical reconstructions are shown in the second column of Figure 2. In the example shown in row one the target image (first column, red border) is a seaside cafe and harbor. The reconstruction (second column) depicts the shore as a textured high-contrast region, and the sea and sky as smooth low-contrast regions. In row two the target image is a group of performers on a stage, but the reconstruction depicts the performers as a single textured region on a smooth background. In row three the target image is a patch of dense foliage, which the reconstruction depicts as a single textured region that covers much of the visual field.
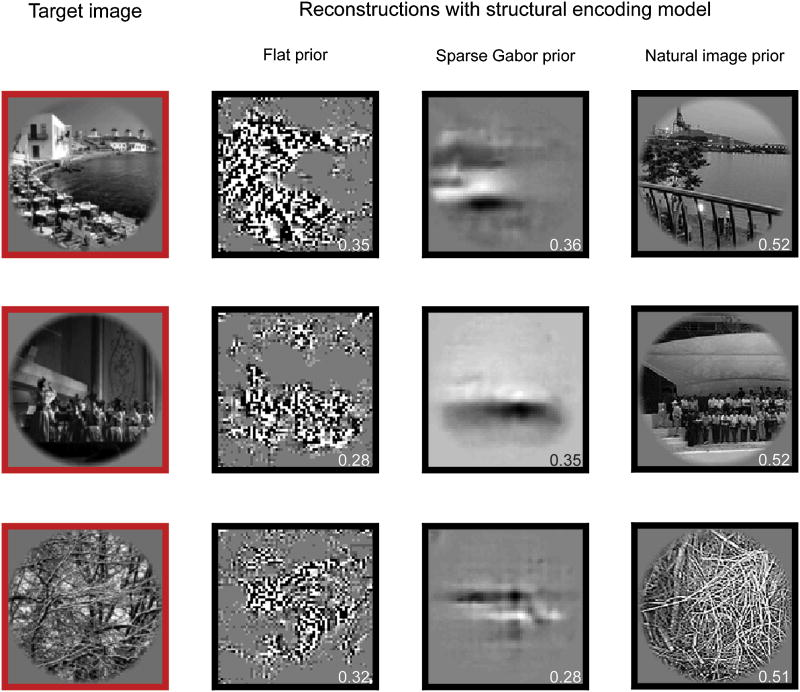
Figure 2. The effect of prior information on reconstruction with a structural encoding model.
Three target images are shown in the first column. The second through fourth columns show reconstructions obtained using the structural encoding model and three different types of prior information. Column two shows reconstructions obtained using a flat prior that does not bias reconstructions. Regions of the target images that have low texture contrast are depicted as smooth gray patches, and regions that have substantial texture contrast are depicted as textured patches. Thus, the flat prior reconstructions reveal the distribution of texture contrast in the target images, but cannot readily be interpreted. Column three shows reconstructions obtained using a sparse Gabor prior that ensures reconstructions possess the lower-order statistical properties of natural images. These reconstructions appear to be smoothed versions of those obtained with the flat prior, and they also cannot be readily interpreted. Column three shows reconstructions obtained using a natural image prior that ensures reconstructions are natural images. These reconstructions accurately reflect the structure of the target images (numbers in bottom right corner of each reconstruction indicate structural accuracy, see main text for details). The example in row one is from subject TN; rows two and three are from subject SN.
All of the example reconstructions obtained using the structural model and the flat prior have similar qualities. Regions of the target images that have low contrast or little texture are depicted as smooth gray patches in the reconstructions, and regions that have significant local contrast or texture are depicted as textured patches. Local texture and contrast are apparently the only information about natural images that can be recovered reliably from moderate-resolution BOLD fMRI measurements of activity in early visual areas. Unfortunately, reconstructions based entirely on texture and contrast do not provide enough information to reveal the identity of objects depicted in the target images.
To improve reconstructions we sought to define a more informative image prior. A distinguishing feature of natural images is that they are composed of many smooth regions, disrupted by sharp edges. These characteristics are captured by two lower-level statistical properties of natural images: they tend to have a 1/f amplitude spectrum (Field, 1987), and they are sparse in the Gabor-wavelet domain (Field, 1994). In contrast, unnatural images such as white noise patterns generally have much different power spectra and are not sparse. We therefore designed a sparse Gabor prior that biases reconstructions in favor of images that exhibit these two well-known statistical properties (see Figure 1).
To produce a new set of reconstructions, the structural encoding model, the sparse Gabor prior, and the same set of voxels selected above were used to evaluate posterior probabilities (see equation 1). The same greedy serial search algorithm mentioned above was used to converge on an image with a relatively high posterior probability. This image was selected as the reconstruction. Results are shown in the third column of Figure 2. The main effect of the sparse Gabor prior is to smooth out the textured patches apparent in the reconstruction with a flat prior. As a result, the reconstructions are more consistent with the lower-level statistical properties of natural images. However, these reconstructions do not depict any clearly identifiable objects or scenes, and thus fail to reveal the semantic content of the target images.
Because the sparse Gabor prior did not produce reconstructions that reveal the semantic content of the target images, we sought to introduce a more sophisticated image prior. Natural images have complex statistical properties that reflect the distribution of shapes, textures, objects, and their projections onto the retina, but thus far theorists have not captured these properties in a simple mathematical formalism. We therefore employed a strategy first developed in the computer vision community to approximate these complex statistical properties (Hays and Efros, 2007; Torralba et al., 2008). We constructed an implicit natural image prior by compiling a database of six million natural images selected at random from the internet (see Experimental Procedures). The implicit natural image prior can be viewed as a distribution that assigns the same probability to all images in the database and zero probability to all other images.
To produce reconstructions using the natural image prior, the posterior probability was evaluated for each of the six million images in the database (note that in this case the posterior probability is proportional to the likelihood given by the encoding model); the image with the highest probability was selected as the reconstruction. Examples are shown in the fourth column of Figure 2. In row one both the target image and the reconstruction depict a shoreline (compare row one, column one to row one, column four). In row two both the target image and the reconstruction depict a group of performers on a stage. In row three both the target image and the reconstruction depict a patch of foliage. In all three examples, the spatial structure and the semantic content of the reconstructions accurately reflect both the spatial structure and semantic content of the target images (also see Supplemental Figure 3A). Thus, these particular reconstructions are both structurally and semantically accurate.
The examples shown in Figure 2 were selected to demonstrate the best reconstruction performance obtained with the structural encoding model and the natural image prior. However, most of the reconstructions obtained this way are not semantically accurate. Several examples of semantically inaccurate reconstructions are shown in the second column of Figure 3. In row one the target image is a group of buildings, but the reconstruction depicts a dog. In row two the target image is a bunch of grapes, but the reconstruction depicts a hand against a checkerboard background. In row three the target image is a crowd of people in a corridor, but the reconstruction depicts a building. In row four, the target image is snake, but the reconstruction depicts several buildings.
Figure 3. The effect of semantic information on reconstructions.
Four target images are shown in the left hand column. The second column shows reconstructions obtained using the structural encoding model and the natural image prior. These reconstructions are structurally accurate (numbers in bottom right corner indicate structural accuracy, see main text for details). However, the objects depicted in the reconstructions are not from the same semantic categories as those shown in the target images. Thus, although these reconstructions are structurally accurate they are not semantically accurate. The third column shows reconstructions obtained using the structural encoding model, the semantic encoding model and the natural image prior (the hybrid method). These reconstructions are both structurally and semantically accurate. The examples in rows from one through three are from subject TN; row four is from subject SN.
Close inspection of the reconstructions in the second column of Figure 3 suggests that they are structurally accurate. For example, the target image depicting grapes in row two has high spatial frequency, while the reconstruction in row two contains a checkerboard pattern with high spatial frequency as well. However, the reconstruction does not depict objects that are semantically similar to grapes, so it does not appear similar to the target image. This example reveals that structural similarity alone can be a poor indicator of how similar two images will appear to a human observer. Because human judgments of similarity will inevitably take semantic content into account, reconstructions should reflect both the structural and semantic aspects of the target image. Therefore, we sought to incorporate activity from brain areas known to encode information about the semantic content of images.
A semantic encoding model
There is evidence that brain areas in anterior visual cortex encode information that is related to the semantic content of images (Kanwisher et al., 1997; Epstein and Kanwisher, 1998; Grill-Spector et al., 1998; Haxby et al., 2001; Grill-Spector and Malach, 2004; Downing et al., 2006; Kriegeskorte et al., 2008). In order to add accurate semantic content to our reconstructions, we designed a semantic encoding model that describes how voxels in these areas encode information about natural scenes. The model automatically learns—from responses evoked by a randomly chosen set of natural images—the semantic categories that are represented by the responses of a single voxel.
To fit the semantic encoding model, all 1750 natural images used to acquire the model estimation data set were first labeled by human observers with one of 23 semantic category names (see Supplemental Figure 1). These categories were chosen to be mutually exclusive yet broadly defined, so that the human observers were able to assign each natural image a single category that best described it (observers were instructed to label each image with the single category they deemed most appropriate and reasonable; see Experimental Procedures for details). Importantly, the images had not been chosen beforehand to fall into pre-defined categories; rather, the categories were designed post-hoc to provide reasonable categorical descriptions of randomly selected natural images.
After the images in the model estimation set were labeled, an Expectation Maximization optimization algorithm (EM) was used to fit the semantic model to each voxel (see Experimental Procedures and Appendix 1 for details). The EM algorithm learned the probability that each of the 23 categories would evoke a response either above, below or near the average of each voxel. The resulting semantic model reflects the probability that a voxel “likes”, “doesn't like” or “doesn't care about” each semantic category. This information is then used to calculate p(r|s)—the likelihood of the observed response, given a sampled image (see Supplemental Figure 2, and Experimental Procedures for more details). We fit the semantic model to all of the voxels in the dataset and then inspected those voxels whose responses could be predicted accurately by the model (see Experimental Procedures for specific voxel selection criteria).
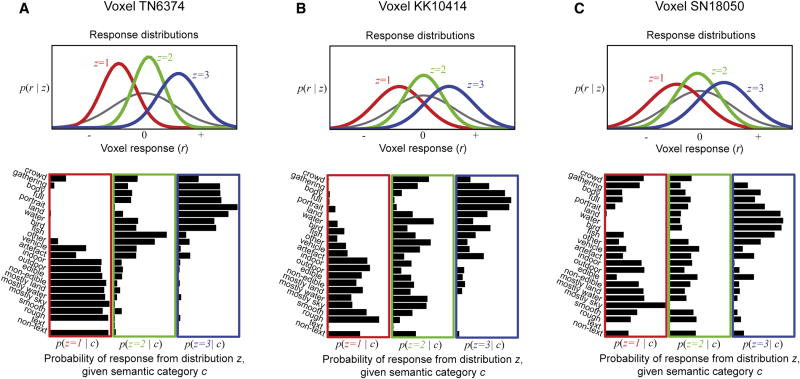
Examples of the semantic encoding model fit to three voxels (one from each of the three subjects in this study) are shown in Figure 4. Gray curves show the overall distribution of responses to all images in the model estimation set. The colored curves define responses that are above (blue curve), below (red curve), or near (green curve) the average response. The bottom boxes give the probability that an image from a specific semantic category (category names at left; names are abbreviated, see Supplemental Figure 1 for full names) will evoke a response above (blue boxes), below (red boxes) or near (green boxes) the average response. For each of these voxels most categories that pertain to non-living things—such as textures, landscapes, and buildings—are likely to evoke responses below the average. In contrast, most categories that pertain to living things—such as people, faces, and animals—are likely to evoke responses above the average. Average responses tend to be evoked by a fairly uniform distribution of categories. Thus, at a coarse level, activity in each of these voxels tends to distinguish between animate and inanimate things.
Figure 4. The semantic encoding model fit to single voxels from three subjects.
A, The top panel shows response distributions of one voxel for which the semantic encoding model produced the most accurate predictions (subject TN). The gray curve gives the distribution of z-scored responses (x-axis) evoked by all images used in the model estimation data set. This distribution was modeled in terms of three underlying Gaussian distributions (colored curves). Responses below average are shown in red (z=1), responses near average in green (z=2), and above average in blue (z=3). The black bars in the bottom panels give the probability that each semantic category, c, (abbreviated labels at left) will evoke a response below the average (red box), near the average (green box) or above the average (blue box). (Note that there are no probabilities for the text category because there were no text images in the model estimation data set.) Images depicting living things tend to evoke a large response from this voxel, while those depicting non-living things evoke a small response. Thus, this voxel discriminates between animate and inanimate semantic categories. B, The same analysis shown in A, applied to the single voxel from subject KK for which the semantic encoding model produced the most accurate predictions. Semantic tuning for this voxel is similar to the one shown in A. C, The same analysis shown in A and B, applied to the single voxel from subject SN for which the semantic encoding model produced the most accurate predictions. Semantic tuning for this voxel is similar to those shown in A and B.
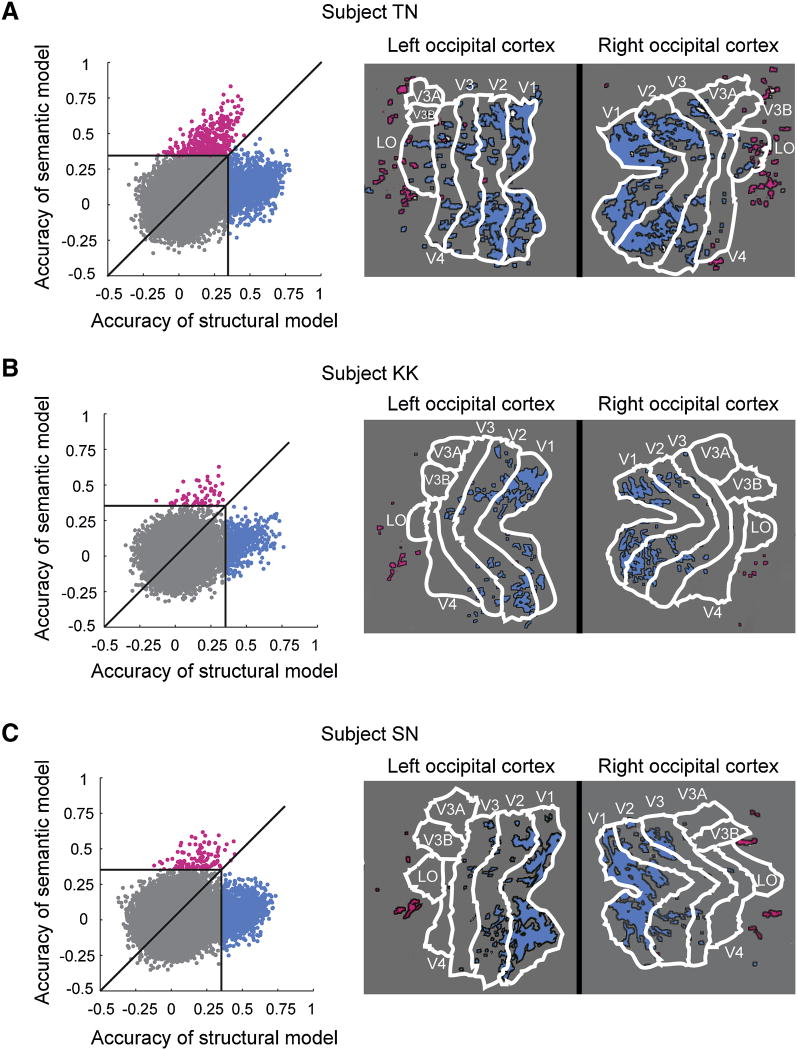
To determine how the representations of structural and semantic information are related to one another, we compared the prediction accuracy of the structural model with that of the semantic model (Figure 5, left panels). We quantified prediction accuracy as the correlation (cc) between the response observed in each voxel and the response predicted by each encoding model for all 120 images in the image reconstruction set. The points show the prediction accuracy of the structural encoding model (x-axis) and semantic encoding model (y-axis) for each voxel in our slice coverage. The distribution of points has two wings. One wing extends along the y-axis, and the other extends along the x-axis. This indicates that there are very few voxels whose responses are accurately predicted by both models. Most voxels whose responses are accurately predicted by the structural model (cc > 0.353; blue voxels; see Experimental Procedures for criteria used to set this threshold) are not accurately predicted by the semantic model. Most voxels whose responses are accurately predicted by the semantic model (cc > 0.353; magenta voxels) are not accurately predicted by the structural model. The wings have similar extents, indicating that the semantic model provides predictions that are as accurate as those provided by the structural model. Remarkably, the predictions for both the structural and semantic voxels can be as accurate as those obtained for single neurons in area V1 (David and Gallant, 2005; Carandini et al., 2005). Note that there is a large central mass of voxels (gray); these voxels either have poor signal quality or represent information not captured by either the structural or semantic models.
Figure 5. Structural versus semantic encoding models.
A, The left panel compares the accuracy of the structural encoding model (x-axis) versus the semantic encoding model (y-axis) for every voxel within the slice coverage (subject TN). Here accuracy is defined as the correlation (cc) between the response observed in each voxel and the response predicted by each encoding model for all 120 images in the image reconstruction set. The distribution of points has two wings. One wing extends along the y-axis and another extends along the x-axis, indicating that very few voxels are accurately predicted by both models. The voxels whose responses are accurately predicted by the structural model but not the semantic model are shown in blue (cc > 0.353, p < 3.9*10-5; see Experimental Procedures for criteria used to set this threshold). The voxels whose responses are accurately predicted by the semantic model but not the structural model are shown in magenta (same statistical threshold as above). Most voxels are poorly predicted by both models (gray), either because neither model is appropriate, or because of poor signal quality. The right panel shows flat maps of the left and right hemispheres of this subject. Visual areas identified using a retinotopic mapping procedure (see Experimental Procedures) are outlined in white. Voxels whose responses are accurately predicted by the structural (blue) or semantic (magenta) models are plotted on the flat maps (the few voxels for which both models are accurate are shown in white). Most structural voxels are located in early visual areas V1, V2 and V3. Most semantic voxels are located in the anterior portion of lateral occipital (labeled LO), and in the anterior occipital cortex. B, Data for subject KK, format same as in A. Most structural voxels are located in early visual areas V1, V2 and V3. Semantic voxels are located in the anterior occipital cortex. C, Data for subject SN, format same as in A and B. Structural voxels are located in early visual areas V1, V2 and V3. Semantic voxels are located in the anterior occipital cortex.
In order to determine the anatomical locations of the voxels in the two separate wings, we projected voxels whose responses are accurately predicted by the structural (blue) and semantic (magenta) models onto flat maps of the right and left occipital cortex (Figure 5, right panels). Most of the voxels whose responses are accurately predicted by the structural model are located in early visual areas V1, V2 and V3. In contrast, most of the voxels whose responses are accurately predicted by the semantic model are located in the AOC, at the anterior edge of our slice coverage.
Our results show that the semantic encoding model accurately characterizes a set of voxels in anterior visual cortex that are functionally distinct and anatomically separated from the structural voxels located in early visual cortex. The structural voxels in early visual areas encode information about local contrast and texture, while the semantic voxels in anterior portions of lateral occipital and in the AOC encode information related to the semantic content of natural images. Therefore, a reconstruction method that uses the structural and semantic encoding models to extract information from both sets of voxels should produce reconstructions that reveal both the structure and semantic content of the target images.
Reconstructions using structural and semantic models and a natural image prior
To incorporate the semantic encoding model into the reconstruction algorithm, we first selected all of the voxels for which the semantic encoding model provided accurate predictions. Most of these voxels were located in the anterior portion of lateral occipital and in the AOC (see Experimental Procedures for details on voxel selection). The individual models for each selected voxel were then combined to into a single, multi-voxel semantic encoding model, p(r|s) (see Experimental Procedures for details).
To produce reconstructions, the semantic and structural encoding models (with their corresponding selected voxels), were used to evaluate the posterior probability (see equation 1) of each the six million images in the natural image prior. For convenience, we refer to the use of the structural model, semantic model and natural image prior as the hybrid method.
Reconstructions obtained using the hybrid method are shown in the third column of Figure 3. In contrast to the reconstructions produced using the structural encoding model and natural image prior, the hybrid method produces reconstructions that are both structurally and semantically accurate. In the example shown in row one, both the target image and the reconstruction depict buildings. In row two, the target image is a bunch of grapes, and the reconstruction depicts a bunch of berries. In row three, the target image depicts a crowd of people in a corridor, and the reconstruction depicts a crowd of people on a narrow street. In row four, the target image depicts a snake crossing the visual field at an angle, while the reconstruction depicts a caterpillar crossing the visual field at a similar angle. (In Supplemental Figure 3, we also present the second and third most probable images in the natural image prior. The spatial structure and semantic content of these alternative reconstructions is consistent with the best reconstruction.)
Objective assessment of reconstruction accuracy
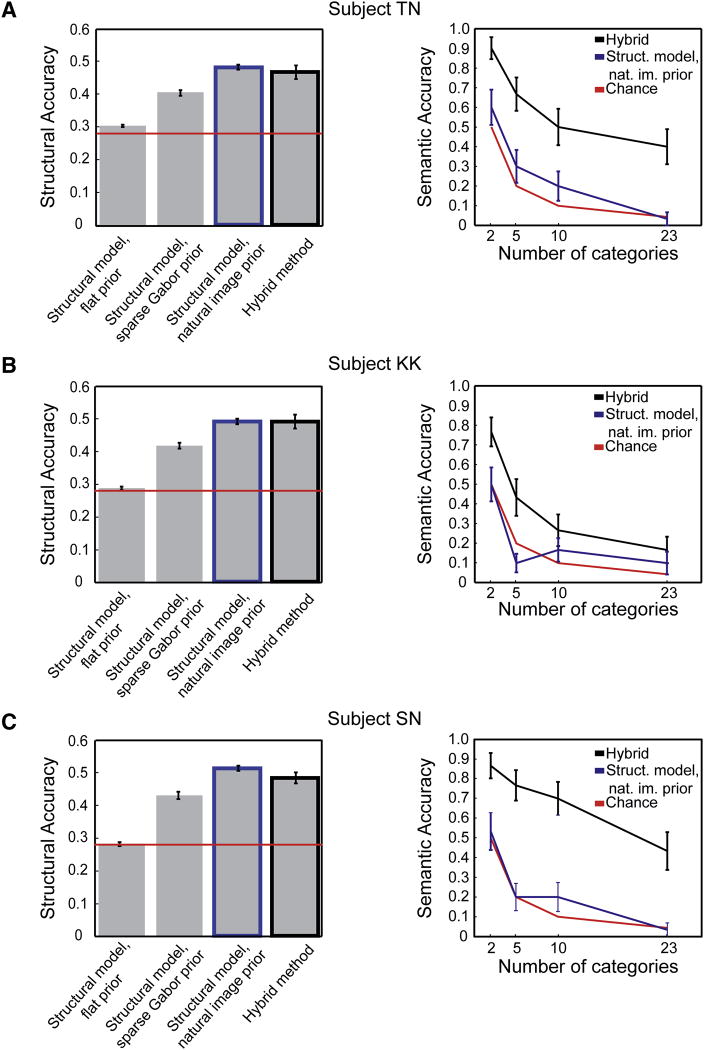
To quantify the spatial similarity of the reconstructions and the target images we used a standard image similarity metric proposed previously (Brooks and Pappas, 2006). This metric reflects the complex wavelet-domain correlation between the reconstruction and the target image. We applied this metric to the four types of reconstruction presented in Figures 2 and 3. As shown by the plots on the left side of Figure 6, the structural accuracy of all the reconstruction methods that use a non-flat prior is significantly greater than chance for all three subjects (p < 0.01, t-test; comparison is for each individual subject). Reconstruction with the structural model and the natural image prior is significantly more accurate than reconstruction with a sparse Gabor prior, (p < 0.01, t-test; comparison is for each individual subject). These results indicate that prior information is important for obtaining accurate image reconstructions. The structural accuracy of the structural model with natural image prior and the hybrid method are not significantly different (p > 0.3, t-test; comparison is for each individual subject), so structural accuracy is not affected by the addition of the semantic model.
Figure 6. Structural and semantic accuracy of reconstructions.
A, The left panel shows the structural accuracy of reconstructions using several different methods (subject TN). In each case structural reconstruction accuracy (y-axis) is quantified using a similarity metric that ranges from 0.0 to 1.0. From left to right the bars give the structural similarity between the target image and reconstruction (mean ± s.e.m, image reconstruction data set) for the structural model with a flat prior; the structural model with a sparse Gabor prior; the structural model with a natural image prior; and the hybrid method consisting of the structural model, the semantic model, and the natural image prior. The red line indicates chance performance. Reconstructions produced using the sparse Gabor or natural image prior are significantly more accurate than chance (p < 0.01, t-test; for this subject only, the reconstructions produced using a flat prior are also significant at this level). Reconstruction with the structural model and the natural image prior is significantly more accurate than reconstruction with a sparse Gabor prior (p < 0.01, t-test). These results indicate that prior information is important for obtaining structurally accurate image reconstructions. The structural accuracy of the structural model with natural image prior and the hybrid method are not significantly different (p > 0.3, t-test), so structural accuracy is not affected by the addition of the semantic model. The right panel shows semantic accuracy of reconstructions obtained using the structural model with natural image prior (blue) and the hybrid method (black). In each case semantic reconstruction accuracy (y-axis) is quantified in terms of the probability that a reconstruction will belong to the same semantic category as the target image (errorbars indicate bootstrapped estimate of s.d.). The number of semantic categories in the classification tree varies from 2 broadly defined categories to the 23 specific categories shown in Figure 4 (x-axis). The red curve indicates chance performance. The semantic accuracy of the reconstructions obtained using the structural model and natural image prior are rarely significantly greater than chance (p > 0.3, binomial test). However, the semantic accuracy of the hybrid method is significantly greater than chance regardless of the number of semantic categories (p < 10-5, binomial test). B, Data for subject KK, format same as in A. Prior information is important for obtaining structurally accurate image reconstructions (p-values of structural accuracy comparisons same as in A). The semantic accuracy of the hybrid method is significantly greater than chance (p < .002, binomial test). C, Data for subject SN, format same as in A. Prior information is important for obtaining structurally accurate image reconstructions (p-values of structural accuracy comparisons same as in A). The semantic accuracy of the hybrid method is significantly greater than chance (p < 10-5, binomial test).
To quantify the semantic similarity of the reconstructions and the target images we formulated a semantic accuracy metric. In this case we estimated the probability that a reconstruction obtained using some specific reconstruction method would belong to the same semantic category as the target image. (Because calculating semantic accuracy requires time-consuming labeling of many images, we calculated semantic accuracy for only the first 30 images in the image reconstruction set; see Experimental Procedures for details). We considered semantic categories at four different levels of specificity, from two broadly defined categories (“mostly animate” vs. “mostly inanimate”), to 23 narrowly defined categories (see Supplemental Figure 1 for complete list). Semantic accuracies for the structural model with natural image prior and the hybrid method are shown by the plots on the right side of Figure 6 (note that semantic accuracy cannot be determined for methods that did not use the natural image prior). The semantic accuracy of the hybrid method is significantly greater than chance for all three subjects, and at all levels of specificity (p < 10-5, binomial test, for subjects TN and SN; p < .002, binomial test, for subject KK). The semantic accuracy of the reconstructions obtained using the structural model and natural image prior are rarely significantly greater than chance for all three subjects (p > 0.3, binomial test). The hybrid method is quite semantically accurate. When two categories are considered accuracy is 90% (for subject TN), and when the full 23 categories are considered accuracy is still 40%. In other words, reconstructions produced using the hybrid method will correctly depict a scene whose animacy is consistent with the target image 90% of the time, and will correctly depict the specific semantic category of the target image 40% of the time.
Discussion
We have presented the first reconstructions of natural images from BOLD fMRI measurements of human brain activity. These reconstructions were produced by a Bayesian reconstruction framework that uses two different encoding models to integrate information from functionally distinct visual areas: a structural model that describes how information is represented in early visual areas, and a semantic encoding model that describes how information is represented in anterior visual areas. The framework also incorporates image priors that reflect the structural and semantic statistics of natural images. The resulting reconstructions accurately reflect the spatial structure and semantic content of the target images.
Relationship to previous reconstruction studies
Two previous fMRI decoding papers presented algorithms for reconstructing the spatial layout of simple geometrical patterns composed of high-contrast flicker patches (Thirion et al., 2006; Miyawaki et al., 2008). Both these studies used some form of structural model that reflected the retinotopic organization of early visual cortex, but neither explored the role of semantic content or prior information. Our previous study on image identification from brain activity (Kay et al., 2008) used a more sophisticated voxel-based structural encoding model that reflects the way that spatial frequency and orientation information are encoded in brain activity measured in early visual areas. However, the image identification task does not require the use of semantic information.
The study reported here is the first to present a solution to a more general problem: reconstructing arbitrary natural images from fMRI signals. It is much more difficult to reconstruct natural images than flickering geometrical patterns because natural images have a complex statistical structure and evoke signals with relatively low signal-to-noise. Our study employed a structural encoding model similar to that used in our earlier image identification study (Kay et al., 2008), but we found that this model is insufficient for reconstructing natural images, given the fMRI signals collected in our study. Successful reconstruction requires two additional components: a natural image prior and a semantic model. The natural image prior ensures that potential reconstructions will satisfy all of the lower and higher-order statistical properties of natural images. The semantic encoding model reflects the way that information about semantic categories is represented in brain responses measured in AOC. Our study is the first to integrate structural and semantic models with a natural image prior to produce reconstructions of natural images.
Under the Bayesian framework, each of the separate sources of information used for reconstruction are represented by a separate encoding model or image prior. This property of the framework makes it an efficient method for integrating information from disparate sources in order to optimize reconstruction. For example, adding the semantic model to the reconstruction process merely required adding an additional term to equation 1. However, this property also has value even beyond its use in optimizing reconstructions. Because the sources of structural and semantic information are represented by separate models, the Bayesian framework makes it possible to disentangle the contributions of functionally distinct visual areas and prior information to reconstructing the structural and semantic content of natural images (see Figure 6).
But is this really reconstruction?
Reconstruction using the natural image prior is accomplished by sampling from a large database of natural images. One obvious difference between this sampling approach and the methods used in previous studies (Thirion et al., 2006; Miyawaki et al., 2008) is that reconstructions will always correspond to an image that is already in the database. If the target image is not contained within the natural image prior then an exact reconstruction of the target image cannot be achieved. The database used in our study contains only six million images, and with a set this small it is extremely unlikely that any target image (chosen from an independent image set) can be reconstructed exactly. However, as the size of the database (i.e., the natural image prior) grows it becomes more likely that any target image will be structurally and/or semantically indistinguishable from one of the images in the database. For example, if the database contained many images of one person's personal environment, it would be possible to reconstruct a specific picture of her mother using a similar picture of her mother. In this case, the fact that the reconstruction was not an exact replica of the target image would be irrelevant.
It is important to emphasize that in practice exact reconstructions are impossible to achieve by any reconstruction algorithm on the basis of brain activity signals acquired by fMRI. This is because all reconstructions will inevitably be limited by inaccuracies in the encoding models and noise in the measured signals. Our results demonstrate that the natural image prior is a powerful (if unconventional) tool for mitigating the effects of these fundamental limitations. A natural image prior with only six million images is sufficient to produce reconstructions that are structurally and semantically similar to a target image. There are many other potential natural image priors that could be used for this process, and some of these may be able to produce reconstructions even better than those demonstrated in this study. Exploration of alternative priors for image reconstruction and other brain decoding problems will be an important direction for future research.
New insights from the semantic encoding model
Many previous fMRI studies have investigated representations in the anterior regions of visual cortex, beginning in the region we have defined as AOC and extending beyond the slice coverage used here to more anterior areas such as the fusiform face area and the para-hippocampal place area (Kanwisher et al., 1997; Epstein and Kanwisher, 1998). These anterior regions are more activated by whole images than by scrambled images (Malach et al., 1995; Grill-Spector et al., 1998), and some specialized regions appear to be most activated by specific object or scene categories (Kanwisher et al., 1997; Epstein and Kanwisher, 1998; Downing et al., 2001; Downing et al., 2006). A recent study using sophisticated multivariate techniques revealed a rough taxonomy of object representations within inferior temporal cortex (Kriegeskorte et al., 2008). Together, these studies indicate that portions of anterior visual cortex represent information related to meaningful objects and scenes—what we have referred to here as “semantic content”.
This result forms the inspiration for our semantic encoding model, which assigns a unique semantic category to each natural image in order to predict voxel responses. This aspect of the model permits us to address one very basic and important question that has not been addressed by previous studies: what proportion of the variance in the responses evoked by a natural image within a single voxel can be explained solely by the semantic category of the image? Our results show that for voxels in the region we have defined as AOC, semantic category alone can explain as much as 55% of the response variance (see Figure 5). An important direction for future research will be to apply the semantic encoding model to voxels in cortical regions that are anterior to our slice coverage. Recent work on a competing model that is conceptually similar to our semantic encoding model (Mitchell et al., 2008) suggests that the semantic encoding model will be useful for predicting brain activity in these more anterior areas .
The results in Figure 5 show that the particular structural features used to build the structural encoding model are very weakly correlated with semantic categories. However, it is important to bear in mind that all semantic categories are correlated with some set of underlying structural features. Although structural features underlying some categories of natural landscape (Greene and Oliva, 2009) have been discovered, the structural features underlying most semantic categories are still unknown (Griffin et al., 2007). Thus, it is convenient at this point to treat semantic categories as a form of representation that is qualitatively different from the structural features used for the structural encoding model.
One notable gap in our current results is that neither the structural nor semantic models can adequately explain voxel responses in intermediate visual areas such as area V4 (see Figure 5). These intermediate areas are thought to represent higher-order statistical features of natural images (Gallant et al., 1993). Because the structural model used here only captures the lower-order statistical structure of natural images (Field, 1987; Field 1994) it does not provide accurate predictions of responses in these intermediate visual areas. Development of a new encoding model that accurately predicts the responses of individual voxels in intermediate visual areas would provide an important new tool for vision research, and would likely further improve reconstruction accuracy.
Future directions
Much of the excitement surrounding the recent work on visual reconstruction is motivated by the ultimate goal of directly picturing subjective mental phenomena such as visual imagery (Thirion et al., 2006) or dreams. Although the prospect of reconstructing dreams still remains distant, the capability of reconstructing natural images is an essential step toward this ultimate goal. Future advances in brain signal measurement, the development of more sophisticated encoding models, and a better understanding of the structure of natural images will eventually make this goal a reality. Such brain reading technologies would have many important practical uses for brain-augmented communication, direct brain control of machines and computers, and for monitoring and diagnosis of disease states. However, such technology also has the potential for abuse. Therefore, we believe that researchers in this field should begin to develop ethical guidelines for the application of brain reading technology.
Experimental Procedures
Data Collection
The MRI parameters, stimuli, experimental design, and data pre-processing are identical to those presented in a previous publication from our laboratory (Kay et al., 2008). Here, we briefly describe the most pertinent details.
MRI parameters
All MRI data were collected at the Brain Imaging Center at UC-Berkeley, using a 4 T INOVA MR (Varian, Inc., Palo Alto, CA) scanner and a quadrature transmit/receive surface coil (Midwest RF, LLC, Hartland, WI). Data were acquired in 18 coronal slices that covered occipital cortex (slice thickness 2.25 mm, slice gap 0.25 mm, field-of-view 128 × 128 mm2). A gradient-echo EPI pulse sequence was used for functional data (matrix size 64 × 64, TR 1 s, TE 28 ms, flip angle 20°, spatial resolution 2 × 2 × 2.5 mm3).
Stimuli
All stimuli were grayscale natural images selected randomly from several photographic collections. The size of the images was 20° × 20° (500 px × 500 px). A central white square served as the fixation point( 0.2° × 0.2°; 4 px × 4 px). Images were presented in successive 4-s trials. In each trial, a photo was flashed at 200 ms intervals (200 ON, 200 OFF) for one second, followed by 3 s of gray background.
Experimental Design
Data for the model estimation and image reconstruction stages of the experiment were collected in the same scan sessions. Three subjects were used: TN, KK, and SN. For each subject, five scan sessions of data were collected. Scan sessions consisted of five model estimation runs and two image reconstruction runs. Runs used for model estimation were 11 minutes each, and consisted of 70 distinct images presented 2 times each. Runs used for image reconstruction runs were 12 min each, and consisted of 12 distinct images presented 13 times each. Images were randomly selected for each run and were not repeated across runs. The total number of distinct images used in the model estimation runs was 1,750. For image reconstruction runs, the total was 120.
Data preprocessing
Functional brain volumes were reconstructed and then coregistered across scan sessions. The time-series data was used to estimate a voxel-specific response timecourse; deconvolving this timecourse from the data produced, for each voxel, an estimate of the amplitude of the response (a single value) to each image used in the model estimation and image reconstruction runs. Retinotopic mapping data collected in separate scan sessions was used to assign voxels to their respective visual areas based on criteria presented in (Hansen et al., 2007).
Notation
All sections below use the same notational conventions. The response of a single voxel is denoted r. Bold notation is used to denote the collected responses of N separate voxels in an Nx1 voxel response vector: r = (r1,…,rN) T . Subscripts i applied to voxel response vectors, ri , are used to distinguish between functionally distinct brain areas. In practice, images are treated as vectors of pixel values, denoted s. These vectors are formed by columnwise concatenation of the original 2D image; unless otherwise noted, s is 1282 x 1 column vector.
Encoding models
Our reconstruction algorithm requires an encoding model for each voxel. Each encoding model describes the voxel's dependence upon a particular set of image features. Formally, this dependence is given by a distribution over the possible responses of the voxel to an image: p(r|s). We presented two different types of encoding models: a structural encoding model, and a semantic encoding model. Each model is defined by a transformation of the original image s into a set of one or more features. For the structural encoding model, these features are spatially localized orientations and spatial frequencies (which can be described using Gabor wavelets). For the semantic model, these features are semantic categories assigned to the images by human labelers. The conditional distributions p(r|s) for both models are defined by one or more Gaussian distributions. For the structural models, p(r|s) is a Gaussian distribution whose mean is a function of the image s; for the semantic model, p(r|s) is a weighted sum of Gaussian distributions, each of whose means is function of the image. Each model can be used to predict the specific response of a voxel to an image by taking the expected value of r with respect to p(r|s). Note that if a voxel has a very weak dependence on the features assumed by the model, the expected value of r with respect to p(r|s) will poorly predict the actual response of the voxel. Note also that both the structural and semantic models have a number of free parameters that must be estimated using a suitable fitting procedure.
Structural encoding model
The structural encoding model used in this work is similar to the Gabor Wavelet Pyramid model described in our previous publication (Kay et al., 2008). The model describes the spatial frequency and orientation tuning of each voxel. These attributes can be efficiently described by Gabor wavelets. A Gabor wavelet is a spatially localized filter with a specific orientation and spatial frequency. To construct the structural encoding model, all images are first filtered by a set of Gabor wavelets that cover many spatial locations, orientation, frequencies, and scales. The filtered signals are then passed through a fixed nonlinearity. This nonlinear transformation of the image defines the feature set for the structural encoding model. Formally, the features are defined as:
where f is an F x 1 vector containing the features (F =10921, the number of wavelets used for the model), and W denotes a matrix of complex Gabor-wavelets. W has as many rows as there are pixels in s, and each column contains a different Gabor wavelet; thus, it's dimension is 1282 x 10921. The features are the log of the magnitudes obtained after filtering the image by each wavelet. The log is applied because we have found that a compressive nonlinearity improves prediction accuracy.
The wavelets in W occur at six spatial frequencies: 1, 2, 4, 8, 16, and 32 cycles per field-of-view (FOV = 20°; images were presented at a resolution of 500×500 pixels, but were downsampled to 128×128 pixels for this analysis). At each spatial frequency of n cycles per FOV, wavelets are positioned on an n × n grid that tiles the full FOV. At each grid position wavelets occur at eight orientations, 0°, 22.5°, 45°, …, and 157.5°. An isotropic Gaussian mask is used for each wavelet, and its size relative to spatial frequency is such that all wavelets have a spatial frequency bandwidth of 1 octave and an orientation bandwidth of 41°. A luminance-only wavelet that covers the entire image is also included.
As mentioned above, the conditional distribution for the structural encoding model is a Gaussian distribution whose mean is defined by a weighted sum of the features f:
where h (F × 1) is the set of weighting parameters, and σ (a scalar) is the standard deviation of voxel responses.
The encoding model's predicted response to an image s is defined as the mean response with respect to p(r|s). This mean is just the feature transform (which is the same for all voxels) multiplied by the weighting parameters h (which are fit independently for all voxels):
Coordinate descent with early stopping was used to find the parameters h that minimized the sum-of-square-error between the actual and predicted responses. For each voxel, this minimization was performed on three sets of M-l training samples (M=1750, l = M*0.1), selected randomly without replacement. Each set produced a separate estimate hj (j=[1, 2, 3]). h was set equal to the arithmetic mean of (h1, h2, h3).
Semantic encoding model
The features used for the semantic encoding are quite different from those used for the structural encoding model. Instead of features that are defined by a wavelet transformation, the features for the semantic model are semantic categories assigned to each image by human labelers.
The semantic categories used for the model were drawn from a semantic basis. The semantic basis is a set of categories designed to satisfy two key properties. First, the categories are broad enough that any natural image can be assigned to at least one of them. Second, categories in the semantic basis are non-overlapping, so that a human observer can confidently assign any arbitrary image to only one of them. We developed the semantic basis using a tree of categories, shown in Supplemental Figure 1. In the first layer of the tree, all possible images are divided into mutually exclusive categories: “mostly animate” and “mostly inanimate”. In subsequent layers, each category is again divided into two or three exclusive categories. At the bottom of the tree (rightmost layer in Supplemental Figure 1) is a set of 23 categories; this is the semantic basis. The inputs to the model are natural images that have been labeled with one of these 23 categories by two human observers. The observers did not know whether the images they labeled were target images, training images, or potential reconstructions sampled from the natural image prior. Observers were instructed to label each image by working their way down the semantic tree: first they assigned the correct label from the first level, then the second level, and so on until reaching the bottom of the tree. In a few cases the labels assigned by different labelers were inconsistent, and in these cases they discussed the images between themselves in order to arrive at a consistent conclusion.
The form of the conditional distribution for the semantic encoding model is slightly more complex than for the structural model. In order to make the model easily interpretable, we designed it so that it would clearly delineate the semantic categories that a voxel likes (categories that are likely to evoke above average responses), doesn't like (categories that are likely to evoke below average responses), or doesn't care about (categories that are likely to evoke near average responses). This clear delineation is achieved by decomposing the overall voxel response distribution (the gray curves in the top panels of Figure 4) into a mixture of sub-distributions that span the above, below, and near average response ranges:
where z ∈[1, 2,3]is an indicator variable used to delineate the ranges, and c(s) denotes the semantic category assigned to the image s (this notation is used throughout to make the model's dependence on semantic categories explicit. However, the more general notation used for the structural model, p(r|s), is applicable here as well). Each of the sub-distributions, p(r|z) is a Gaussian (colored curves in top panels of Figure 4) with it's own mean and variance, μz and σz:
Each of these Gaussian sub-distributions are weighted by a multinomial mixing distribution, p(z|c(s)), that gives the probability that the voxel's response will be driven into response range z when presented with an image from category c (bar charts in bottom panels of Figure 4).
The predicted response of the semantic model to an image s is the mean of p(r|c(s)). This mean is a weighted sum of the means of each of sub-distribution:
The free parameters of the semantic encoding models are the mean and variance of p(r|z) for each value of z, and the parameters of the multinomial mixture distribution p(z|c(s)). We estimated these parameters for each voxel using an Expectation Maximization algorithm that we present in Appendix 1.
Voxel selection and multi-voxel encoding models
To perform reconstruction using the responses of many voxels, it is necessary to first select a set of voxels for use in reconstruction, and then combine the individual encoding model for each selected voxel into a single multi-voxel encoding model for the entire set.
Voxels were selected for reconstruction on the basis of the predictive accuracy of their encoding models. If the prediction accuracy of the structural encoding model was above threshold (see below), it was considered a structural voxel, and it was used for all reconstructions that involved the structural encoding model. If the prediction accuracy of the semantic encoding model for a voxel was above threshold (see below) it was considered a semantic voxel, and it was used for all reconstructions that involved the semantic encoding model. In the rare cases where both the structural and the semantic encoding models were above the selection thresholds for both models the voxel was used for both structural and semantic reconstruction.
For the structural model, the threshold was a correlation coefficient of > 0.353. This correlation coefficient corresponds to a p-value < 3.9x10-5, which is roughly the inverse of the number of voxels in our dataset. For the semantic model, the threshold was a correlation coefficient of > 0.26. This correlation coefficient was chosen because it optimized semantic accuracy on an additional set of 12 experimental trials obtained for subject TN (none of these trials were part of the model estimation or image reconstruction sets used here.)
In order to control for a possible selection bias, the correlation values for both the structural and semantic encoding models were calculated separately for each of the image reconstruction trials. To reconstruct the jth image, the correlation coefficients were calculated using the remaining 119 image reconstruction trials. Thus, a slightly different set of voxels was selected for each reconstruction trial. The average number of voxels selected by the structural model was 788 (average taken across all three subjects and all reconstruction trials). The average number of voxels selected by the semantic model was 579. The average number of voxels selected by both was 73.
Once voxels were selected for each reconstruction trial, multi-voxel versions of the structural and semantic encoding models were constructed using the univariate model for each of the selected voxels. The multi-voxel versions of the structural and semantic encoding models are given by the following distribution:
where Λ is an N x N covariance matrix. Let μˆi (s) ≔ 〈ri|s〉 be the predicted response for the ith voxel, given an image s (the predicted mean response for the structural and semantic encoding models are defined above). Let μˆ(s) = (μˆ1 (s,…,μˆN (s)) be the collection of predicted mean responses for N voxels. We define rˆ as the normalized predicted mean response vector:
where the sidebars denote vector normalization and the columns of the matrix P contain the first p principal components of the distribution over μ (p=45 for the structural model; p = 21 for the semantic model. For all subjects and both models, these values of p occur at or near the inflection point of the plot of rank-ordered eigenvalues). The prime notation denotes the same linear transformation and scaling of measured response vectors:
To estimate P for the structural encoding model, we generated predicted mean response vectors to a gallery of 12000 natural images, and applied standard principal components analysis to this sample. For the semantic encoding model, we used a smaller gallery of 3000 images labeled according to the scene categories shown in Supplemental Figure 1 (rightmost layer of the tree). The reduction of dimensionality achieved by projection onto the first p principal components of the predicted responses, and the normalization after projection, act to stabilize the inverse of the covariance matrix, Λ. The elements of Λ give the covariance of the residuals (i.e., the difference between the responses and predictions, r′ − rˆ(s)) for each pair of N voxels. We used the 1750 trials in the model estimation set to estimate Λ.
General reconstruction algorithm
All of the reconstructions presented in the paper are special cases of a general Bayesian algorithm, summarized by the following equation:
On the left hand side is the posterior distribution, p(s|r). The posterior gives the probability that an image s evoked the measured response r. The goal of reconstruction is to find the image with the highest posterior probability, given the responses (this is often referred to as maximum a posteriori decoding). The formula on the right hand side shows how the posterior probability is calculated. The first term, p(s), is the image prior. It reflects pre-existing, general knowledge about natural images, and is independent of the responses. We consider three separate priors in this study: the flat prior, the sparse Gabor prior, and natural image prior. The image prior is followed by a product of encoding models, pi, each of which is applied to the responses, ri, of voxels in a functionally distinct brain area. To produce reconstructions, we used either one (structural) or two (structural and semantic) encoding models.
The four different reconstruction methods presented in the main text differ only by the particular choice of priors and encoding models used to calculate the posterior probability. For reconstructions that use the structural model and a flat prior, the posterior is:
where p1 is the structural encoding model, and r1 are the structural voxels (selected according to the voxel selection procedure defined above: see Voxel selection and multi-voxel encoding models). Note that the image prior, p(s), does not appear here because the flat prior is simply a constant that is independent of both images and responses.
For reconstruction with the structural encoding model and sparse Gabor prior, the posterior is:
where pSG(s) is the sparse Gabor prior described in detail below.
For reconstructions with the structural model and natural image prior, the posterior is:
where pNIP(s)is the natural image prior.
Finally, for the hybrid reconstructions, the posterior incorporates two encoding models:
where p2 is the semantic encoding model, and r2 are the semantic voxels (selected according to the procedure defined above: see Voxel selection and multi-voxel encoding models).
Once a posterior distribution is defined, a reconstruction is produced by finding an image that has a high posterior probability. In general, it is not possible to determine the image that maximizes the posterior distribution analytically. Thus, a search algorithm must be applied to search the space of possible images for potential reconstructions that have high posterior probability.
Reconstructions using the structural encoding model and a flat prior
For the reconstructions presented in the second column of Figure 1, a set of voxels located primarily in the early visual areas V1, V2, and V3 (see above for explanation of how these voxel were selected), and a multi-voxel structural encoding model, were used.
The prior used for this type of reconstruction was the trivial or “flat” prior:
This prior assigns the same value to all possible images, including those with randomly selected pixel values. In this case, the posterior probability p(s|r), and the likelihood, p(r|s), are proportional.
To produce reconstructions, a greedy serial search algorithm was use to maximize the posterior distribution. At each iteration of the algorithm, a small group of pixel values in the reconstruction was updated. If the newly updated pixel values increased the posterior probability, they were retained as part of the reconstructed image. Otherwise, they were rejected. The procedure halted when the change in posterior probability remained below a small threshold (4 bits) for a number of iterations. A formal description of the procedure is given in Appendix 2 (see Supplemental Materials)
Reconstructions using the structural encoding model and a sparse Gabor prior
For the reconstructions presented in the third column of Figure 2, we used the same selected voxels and structural encoding model as above (see Reconstructions using the structural encoding model and a flat prior). Instead of a flat prior, we used a sparse Gabor prior. The sparse Gabor prior places high probability on images that have the 1/f amplitude spectrum characteristic of natural images. In other words, this prior prefers images in which nearby pixels are somewhat correlated. The distribution also assigns high probability to images that are sparse in the Gabor wavelet domain. Suppose that:
is the transformation of an image s into the Gabor domain, where G is a matrix of real-valued Gabor wavelets, and a = (a1, …, ag), is a vector whose elements, ai , denote the “activation” of the ith Gabor wavelet in G. To say that images are “sparse” in the Gabor domain means that their Gabor activations obey a distribution with a sharp peak and a steep falloff (in other words, a distribution with high kurtosis). This aspect of natural images was captured using a Laplace distribution:
where ui , and βi determine the mean and variance of the distribution.
To generate an image from the sparse Gabor prior, activations for all of the wavelets in the Gabor basis G are sampled independently from the above Laplace. The activations are then linearly transformed back into the pixel domain to obtain an image. Finally, this image is transformed again by an “unwhitening” matrix, U, and offset by μs ,the mean of all natural images:
Effectively, the application of U smooths the image so that it possesses the 1/f structure characteristic of natural images.
The Laplace distribution, along with the transformation from Gabor activations into the pixel domain, together define an explicit formula for the sparse Gabor prior:
where p(s | a) = 1 whenever s = UG−1a + μs and is set to zero otherwise. The Gabor activations are assumed to be independent of each other, so . This equation is just a formal way of stating that under the sparse Gabor prior, the probability of sampling an image s is proportional to the probability of sampling its underlying Gabor activations a. (Note that this model has a number of free parameters that must be chosen or estimated empirically. Explicit formulas for estimating these parameters are given in Appendix 3.)
Reconstructions with the structural model and sparse Gabor prior were generated using a search algorithm identical to the one used for reconstruction with a flat prior, except that in this case, reconstructions were updated at each iteration of the algorithm by incrementing the activation for a single Gabor by ± 0.1. The updated reconstruction was transformed into pixel space, and its posterior probability was evaluated using the sparse Gabor prior.
Reconstructions using the structural encoding model and a natural image prior
To produce the reconstructions shown in the fourth column of Figure 2, the second column of Figure 3, and Supplemental Figure 3A, we used the structural encoding model and corresponding structural voxels. Instead of a sparse Gabor prior, we used an implicit natural image prior, pNIP(s). Informally, the natural image prior is simply a large (6 million samples) database of natural images. Formally, it is a distribution that assigns a fixed value to all the images in the database, and a zero value to all images that are not:
where C is the total number of images in the database, and δs(i) is the delta function that returns 1 whenever s = s(i) (the ith image in the database) and a 0 otherwise.
Reconstruction was performed by simply evaluating the posterior probability for each of the images in the database (note that for images in the natural image prior, the posterior is proportional to the likelihood p(r | s)), and choosing the one that resulted in the highest posterior probability. Evaluating the posterior is computationally intensive. As a time-saving approximation, we first evaluated each image in the database using the voxel-wise correlation between the measured responses and the responses predicted by the encoding model. This metric was used in Kay et al. (2008) for image identification. For each target image, we retained the 100 images with the highest correlation. We then evaluated each of these 100 images under p (s | r). The image with the highest p (s | r) was retained as the reconstruction.
Reconstructions using the structural encoding model, the semantic encoding model, and the natural image prior (hybrid method)
To produce reconstructions shown in the third column of Figure 3 (and in Supplemental Figure 3B) the posterior probabilities were evaluated for each of the images in the natural image prior using both the structural encoding model and the semantic encoding model. Reconstruction using this hybrid method was performed for 30 of the image reconstruction trials (all of these were from the first scan session.)
To evaluate the semantic encoding model for a given image s, the image must be assigned a semantic category from the semantic basis set (Supplemental Figure 1). Because it is not feasible to label all 6 million images in the natural image prior, we labeled only those images with relatively high likelihoods under the structural encoding model. For a single reconstruction trial this set of images was defined as
where S is the database of images. βr was chosen so that Sr contained 100 images.
Reconstruction accuracy
Structural accuracy
Structural accuracy of the reconstructions was assessed using the weighted complex wavelet structural similarity metric (Brooks and Pappas, 2006). The metric uses the coefficients of a complex wavelet decomposition of two images in order to compute a single number describing the degree of structural similarity between the two images. To produce the structural accuracy metrics in Figure 6 (left panels), the similarity between each target image and its reconstruction was averaged across all 120 reconstruction trials. (For the case of the hybrid method, averages were taken over the smaller set of 30 reconstructions.) Note that this metric is not the same as the posterior probability of an image. Thus, a rank-ordering of images according to this metric may not perfectly correspond to a rank-ordering of images according to their posterior probabilities (as in Supplemental Figure 3).
Semantic accuracy
To assess the semantic accuracy of the reconstructions, the probability of a semantic category, α , given a response, r, was calculated for each of the 30 reconstruction trials used for the hybrid method. If the most probable category was also the category of the target image, the trial was considered to be semantically accurate. Semantic accuracy for each type of reconstruction (Figure 6, right panels) is the fraction of semantically accurate reconstruction trials.
The probability of a semantic category given a response, p(α | r), is calculated via a linear operation on the encoding models:
where p(r|s) can refer to either the structural encoding model, the semantic encoding model, or the product of the two (as in Equation 1). The distribution p(s | α) is the probability of an image, given a category. This probability was set to 0 if the image was not a member of the category, and a constant value otherwise. The prior on categories, p(α), was assumed to be flat. Thus, the above equation has a very intuitive explanation: the probability of a semantic category given a response is proportional to the average likelihood of all the images from that category within a subset (Sr; see above for definition) of the database.
Supplementary Material
Acknowledgments
This work was supported by an NRSA postdoctoral fellowship (T.N.), the National Institutes of Health, and University of California, Berkeley, intramural funds. We thank B. Inglis for assistance with MRI, K. Hansen for assistance with retinotopic mapping, D. Woods and X. Kang for acquisition of whole-brain anatomical data, and A. Rokem for assistance with scanner operation. We thank A. Berg for assistance with the natural image database, B. Yu and T. Griffiths for consultation on the mathematical analyses, and S. Nishimoto, and D. Stansbury for their help in various aspects of this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brooks AC, Pappas TN. Structural similarity quality metrics in a coding context: exploring the space of realistic distortions. Proc SPIE. 2006;6057:299–310. doi: 10.1109/TIP.2008.926161. [DOI] [PubMed] [Google Scholar]
- Cadieu C, Olshausen B. Learning transformational invariants from time-varying natural movies. Proc Adv Neural Inform Process Syst. 2009;21 [Google Scholar]
- Carandini M, Demb JB, Mante V, Tolhurst DJ, Dan Y, Olshausen BA, Gallant JL, Rust NC. Do we know what the early visual system does? J Neurosci. 2005;25:10577–10597. doi: 10.1523/JNEUROSCI.3726-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson TA, Schrater P, He S. Patterns of activity in the categorical representations of objects. J Cog Neurosci. 2002;15:707–717. doi: 10.1162/089892903322307429. [DOI] [PubMed] [Google Scholar]
- Cox DD, Savoy RL. Functional magnetic resonance imaging (fMRI) “brain reading”: detecting and classifying distributed patterns of fMRI activity in human visual cortex. Neuroimage. 2003;19:261–270. doi: 10.1016/s1053-8119(03)00049-1. [DOI] [PubMed] [Google Scholar]
- David SV, Gallant JL. Predicting neuronal responses during natural vision. Network. 2005;16:239–260. doi: 10.1080/09548980500464030. [DOI] [PubMed] [Google Scholar]
- Downing PE, Jiang Y, Shuman M, Kanwisher N. A cortical area selective for visual processing of the human body. Science. 2001;293:2470–2473. doi: 10.1126/science.1063414. [DOI] [PubMed] [Google Scholar]
- Downing PE, Chan AW, Peelen MV, Dodds CM, Kanwisher N. Domain Specificity in Visual Cortex. Cereb Cortex. 2006;16:1453–1461. doi: 10.1093/cercor/bhj086. [DOI] [PubMed] [Google Scholar]
- Epstein R, Kanwisher N. A cortical representation of the local visual environment. Nature. 1998;392:598–601. doi: 10.1038/33402. [DOI] [PubMed] [Google Scholar]
- Field DJ. Relations between the statistics of natural images and the response properties of cortical cells. J Opt Soc Am Optic Image Sci Vis. 1987;4:2379–2394. doi: 10.1364/josaa.4.002379. [DOI] [PubMed] [Google Scholar]
- Field DJ. What is the goal of sensory coding? Neural Comput. 1994;6:559–601. [Google Scholar]
- Gallant JL, Braun J, Van Essen DC. Selectivity for polar, hyperbolic, and Cartesian gratings in macaque visual cortex. Science. 1993;259:100–103. doi: 10.1126/science.8418487. [DOI] [PubMed] [Google Scholar]
- Griffin G, Holub A, Perona P. The caltech 256. Caltech Technical Report 2007 2007 [Google Scholar]
- Grill-Spector K, Kushnir T, Hendler T, Edelman S, Itzchak Y, Malach R. A sequence of object-processing stages revealed by fMRI in the human occipital lobe. Hum Brain Mapp. 1998;6:316–328. doi: 10.1002/(SICI)1097-0193(1998)6:4<316::AID-HBM9>3.0.CO;2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill-Spector K, Malach R. The Human Visual Cortex. Annu Rev Neurosci. 2004;27:649–677. doi: 10.1146/annurev.neuro.27.070203.144220. [DOI] [PubMed] [Google Scholar]
- Greene R, Oliva A. Recognition of natural scenes from global properties: seeing the forest without representing the trees. Cog Psych. 2009;58:137–176. doi: 10.1016/j.cogpsych.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KA, Kay KN, Gallant JL. Topographic organization in and near human visual area V4. J Neurosci. 2007;27:11896–11911. doi: 10.1523/JNEUROSCI.2991-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxby JV, Gobbini MI, Furey ML, Ishai A, Schouten JL, Pietrini P. Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science. 2001;293:2425–2430. doi: 10.1126/science.1063736. [DOI] [PubMed] [Google Scholar]
- Haynes JD, Rees G. Predicting the orientation of invisible stimuli from activity in human primary visual cortex. Nat Neurosci. 2005;8:686–691. doi: 10.1038/nn1445. [DOI] [PubMed] [Google Scholar]
- Hays J, Efros AA. Scene completion using millions of photographs. ACM Trans Graph (SIGGRAPH 2007) 2007;26:1–5. [Google Scholar]
- Kamitani Y, Tong F. Decoding the visual and subjective contents of the human brain. Nat Neurosci. 2005;8:679–685. doi: 10.1038/nn1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwisher N, McDermott J, Chun MM. The fusiform face area: a module in human extrastriate cortex specialized for face perception. J Neurosci. 1997;17:4302–4311. doi: 10.1523/JNEUROSCI.17-11-04302.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karklin Y, Lewicki MS. Emergence of complex cell properties by learning to generalize in natural scenes. Nature. 2009;457:83–86. doi: 10.1038/nature07481. [DOI] [PubMed] [Google Scholar]
- Kay KN, Gallant JL. I can see what you see. Nat Neurosci. 2009;12:245. doi: 10.1038/nn0309-245. [DOI] [PubMed] [Google Scholar]
- Kay KN, Naselaris T, Prenger RJ, Gallant JL. Identifying natural images from human brain activity. Nature. 2008;452:352–355. doi: 10.1038/nature06713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Mur M, Ruff DA, Kiani R, Bodurka J, Esteky H, Tanaka K, Bandettini PA. Matching categorical object representations in inferior temporal cortex of man and monkey. Neuron. 2008;60:1126–1141. doi: 10.1016/j.neuron.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malach R, Reppas JB, Benson RR, Kwong KK, Jiang H, Kennedy WA, Ledden PJ, Brady TJ, Rosens BR, Tootell RB. Object-related activity revealed by functional magnetic resonance imaging in human occipital cortex. Proc Nat Acad Sci USA. 1995;92:8135–8139. doi: 10.1073/pnas.92.18.8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TM, Shinkareva SV, Carlson A, Chang K, Malave VL, Mason RA, Just MA. Predicting human brain activity associated with the meanings of nouns. Science. 2008;320:1191–1195. doi: 10.1126/science.1152876. [DOI] [PubMed] [Google Scholar]
- Miyawaki Y, Uchida H, Yamashita O, Sato M, Morito Y, Tanabe HC, Sadato N, Kamitani Y. Visual image reconstruction from human brain activity using a combination of multiscale local image decoders. Neuron. 2008;60:915–929. doi: 10.1016/j.neuron.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Nevado A, Young MP, Panzeri S. Functional imaging and neural information coding. Neuroimage. 2004;21:1083–1095. doi: 10.1016/j.neuroimage.2003.10.043. [DOI] [PubMed] [Google Scholar]
- Thirion B, Duchesnay E, Hubbard E, Dubois J, Poline JB, Lebihan D, Dehaene S. Inverse retinotopy: inferring the visual content of images from brain activation patterns. Neuroimage. 2006;33:1104–1116. doi: 10.1016/j.neuroimage.2006.06.062. [DOI] [PubMed] [Google Scholar]
- Torralba A, Fergus R, Freeman WT. 80 million tiny images: a large data set for nonparametric object and scene recognition. IEEE Trans Pattern Anal Mach Intell. 2008;30:1958–1970. doi: 10.1109/TPAMI.2008.128. [DOI] [PubMed] [Google Scholar]
- Wu MCK, David SV, Gallant JL. Complete functional characterization of sensory neurons by system identification. Ann Rev Neurosci. 2006;29:477–505. doi: 10.1146/annurev.neuro.29.051605.113024. [DOI] [PubMed] [Google Scholar]
- Zhang K, Ginzburg I, McNaughton BL, Sejnowski TJ. Interpreting neuronal population activity by reconstruction: unified framework with application to hippocampal place cells. J Neurophysiol. 1998;79:1017–1044. doi: 10.1152/jn.1998.79.2.1017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.