Abstract
Human cytomegalovirus (hCMV) reactivation may often coincide with the development of graft-versus-host-disease (GVHD) in stem cell transplantation (SCT). Seventy seven SCT donor-recipient pairs (DRP) (HLA matched unrelated donor (MUD), n = 50; matched related donor (MRD), n = 27) underwent whole exome sequencing to identify single nucleotide polymorphisms (SNPs) generating alloreactive peptide libraries for each DRP (9-mer peptide-HLA complexes); Human CMV CROSS (Cross-Reactive Open Source Sequence) database was compiled from NCBI; HLA class I binding affinity for each DRPs HLA was calculated by NetMHCpan 2.8 and hCMV- derived 9-mers algorithmically compared to the alloreactive peptide-HLA complex libraries. Short consecutive (≥6) amino acid (AA) sequence homology matching hCMV to recipient peptides was considered for HLA-bound-peptide (IC50<500nM) cross reactivity. Of the 70,686 hCMV 9-mers contained within the hCMV CROSS database, an average of 29,658 matched the MRD DRP alloreactive peptides and 52,910 matched MUD DRP peptides (p<0.001). In silico analysis revealed multiple high affinity, immunogenic CMV-Human peptide matches (IC50<500 nM) expressed in GVHD-affected tissue-specific manner. hCMV+GVHD was found in 18 patients, 13 developing hCMV viremia before GVHD onset. Analysis of patients with GVHD identified potential cross reactive peptide expression within affected organs. We propose that hCMV peptide sequence homology with human alloreactive peptides may contribute to the pathophysiology of GVHD.
Introduction
Human cytomegalovirus and other viral infections pose a significant hurdle to successful stem cell transplantation, affecting morbidity and mortality rates among the immunocompromised populations the world over. [1,2] Human CMV seropositivity has been estimated in 50–80% of the population in the United States by age 40. [3] While the majority of infected patients are asymptomatic due to viral latency after initial viral clearance, SCT patients with newly reconstituted immune systems exhibit rates of reactivation approximately 30–65% in multiple studies. [4–7] Furthermore, hCMV reactivation often coincides with the incidence of another serious complication of SCT, i.e., GVHD. [8–11] Although many groups have historically separated GVHD incidence from hCMV infection data, even by complete exclusion in clinical trial design, there has been growing evidence that these diseases are not mutually exclusive and are likely linked in a number of ways beyond treatment. [12,13]
GVHD pathophysiology is driven by donor T cell mediated alloreactivity, directed against recipient mHA. These are recipient-derived oligopeptides presented on HLA molecules, and result from coding nucleotide sequence variation. Whole exome sequencing has been used to identify the entire library of nucleotide variation that exists between the exomes of HLA matched SCT donors and recipients. [14] From these data it is possible to derive, in silico, all the potential unmodified alloreactive peptides which bind the relevant HLA (class I for example) in a donor-recipient-pair (DRP), and in turn this metric may be used to estimate a patient’s given potential to develop alloreactivity, or GVHD. [15] Aside from helping understand GVHD pathophysiology, this library of recipient-derived mHA-HLA complexes may also be used to interrogate the relationship between pathogen antigens, as well as tumor antigens, and the disease states that may result, in particular the development of cross-reactive illness, such as GVHD triggered by CMV or other viral reactivation.
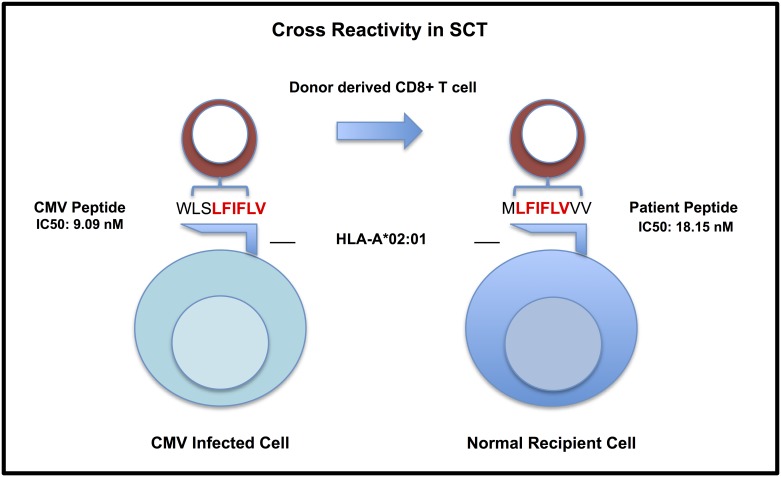
T cell cross reactivity originally was uncovered in relation to autoimmunity, especially in the context of CMV [16–19] and solid organ transplantation. [20–23] T cell cross reactivity occurs because, TCR-peptide-HLA class I bound complexes exhibit a strong recognition of 2–4 central amino acid residues in various orientations and allow for multiple amino acid substitution to the flanks in anchor positions while relying on HLA and peptide sequences simultaneously. The idea of T cells reacting to antigens with amino acid sequence homology (≥6/6 consecutive AA residues in a 9-mer for example) on different cell types with a given HLA-class I type has taken hold from multiple clinical examples. [17] Therefore, while CMV and other B-Herpesviruses have been shown to be associated with autoimmunity and contribute to oncogenic progression possibly affecting relapse rates, [24–26] CMV-specific T cell cross reactivity through adoptive lymphocyte transfer has been shown effective in the treatment of glioblastoma multiforme upon reactivation. [27,28] These clinical trials centering on confirmed CMV-specific T cell cross reactivity with infected tumor cells highlights the expectation of eliciting an immune response to CMV and ‘self’ tumor cells simultaneously. However, the development of CMV-specific T cells in that manner raises the question of eliciting a cross-reactive GVHD response against HLA bound-alloreactive recipient peptides. In theory, this may occur due to peptide polymorphisms, resulting from single nucleotide polymorphisms (SNPs). Resulting polymorphic peptides in the recipient with enough sequence homology to CMV peptides to elicit a HLA-specific immune response, may be presented as targets to donor lymphocytes (Fig 1).
Fig 1. Hypothetical CMV—Human antigenic mimicry model.
Proposed cross-reactivity model for CMV-specific T cells that may react to matched homologous peptides (≥6/6 consecutive amino acids) bound to the same HLA on normal recipient cells present in GVHD-affected tissues.
Therefore, if CMV-derived HLA class I bound oligopeptide have sequence homology with human, alloreactive peptides bound to the same HLA class I and trigger cross recognition by donor T cells, the presence of these cross-reactive peptides may constitute a risk factor for the development of GVHD following SCT. In this study, HLA bound peptide-match analysis is facilitated by a newly compiled Human CMV CROSS (Cross-Reactivity Open Source Sequence) database. There were CMV—human peptide sequence matches present, and in a majority of CMV sero-positive patients with GVHD, this was corroborated by GVHD-specific organ incidence of the human peptide source genes.
Methods
Whole exome sequencing and peptide library creation
Patients with recurrent or high-risk hematological malignancies undergoing allogeneic SCT at Virginia Commonwealth University were included in this retrospective study following Virginia Commonwealth University (VCU) Institutional Review Board approval, MCC-14012 Study. VCU IRB waived the need for informed consent to MCC-14012 on all adult participant archived samples, de-identified by clinical research staff. The demographic and clinical data were not available to the exome sequencing team, and exome sequencing data was not available to the research team, with the exception of SNP identity in a donor recipient specific manner. Exome sequencing data are maintained on secure server with the VCU Center for the Study of Biological Complexity. Previously cryopreserved DNA samples were whole exome sequenced following de-identification by the BMT research team. These recipients received an allograft from either a related or an unrelated, HLA-matched donor at 8/8 or 7/8 HLA loci S1 Table. There were no significant differences in the HLA-matched and HLA-mismatched related and unrelated donors, therefore these were considered together. The transplant DRP, were annotated for identification of all the non-synonymous single nucleotide polymorphisms (nsSNPs), across the whole exome, in the graft versus host (GVH) direction (nsSNP(GVH)), i.e., polymorphism present in the recipient but absent in the donor. [14] Each nsSNP(GVH) was analyzed using the ANNOVAR software, [29] as previously described, [15] to populate the flanking amino acids around each amino acid coded for by each of the polymorphisms. This was accomplished through sequence padding, using DB SNP130 and hg18 genome coordinates, and yielded 17-mer peptides with variant amino acids occupying the central location. This 17-mer oligopeptide library created the opportunity to derive 9 separate 9-mer recipient oligopeptides, using a sliding window method, with the polymorphism-derived amino acid positioned at position 1–9 from C- to N-terminus. The nine oligopeptides generated per nsSNP(GVH) were extended to the whole exome for each recipient, representing each DRPs’ unique peptide library.
In silico peptide-HLA binding affinity determination
Patient peptide libraries were initially analyzed by the NetMHCpan software, version 2.8. [30,31] The analytic output yielded source gene information, polymorphic peptide sequence (9-mer), and a calculated IC50 value from the NetMHCpan algorithms for each of the six HLA class I molecules, HLA-A, B and C in each DRP. IC50 values (nM) indicated the amount of peptide required to displace 50% of intended or standard peptides specific to a given HLA. Binding affinity is inversely related to IC50 values such that a smaller IC50 value indicated a stronger affinity. The variant alloreactive peptides with a cutoff value of IC50 ≤500 nM to the relevant HLA were included, and designated, presented peptides; and those that had an IC50 ≤50 nM, were termed strongly presented peptides. IC50 values in this range (IC50<500 nM) were considered relevant in previous studies of HLA-binding [32] and predictive mouse models of CTL response to vaccinia virus epitopes. [33] Further processing and interrogation of HLA binding in sliding 9-mer windows allowed for affinity sorting and HLA-specific separation along with gene information.
Determining sequence homology between human and CMV derived HLA bound peptides
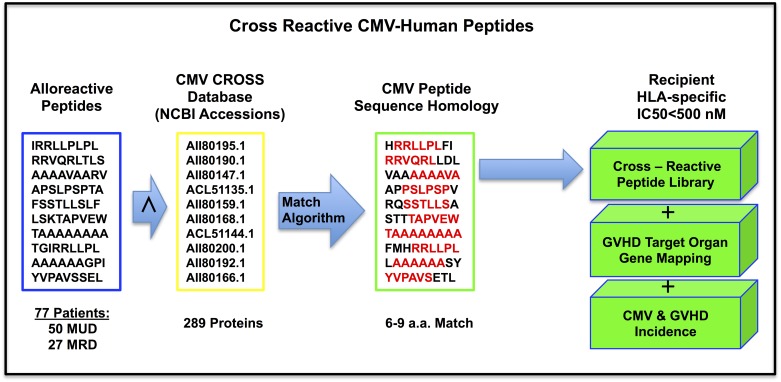
The bioinformatic pipeline utilized to interrogate SCT DRPs for HLA bound polymorphic peptides derived from nsSNP(GVH) in each DRP was extended and refined to interrogate each SCT DRP for relevant HLA class I bound CMV peptides. The binding of CMV-derived peptides to specific HLA Class I molecules was the first step towards determining their sequence homology with human HLA bound peptides. This bioinformatic pipeline is depicted in Fig 2. The initial step was compilation of the Human CMV-CROSS (Cross-Reactivity of Open Source Sequences). This is a database of 289 hCMV proteins or variants representative of the entire known multi-strain inclusive (Merlin, Towne, Toledo and AD169) hCMV proteome sourced from NCBI S1 File (Protein variants had a single or multiple amino acid difference reported for the same protein/CMV gene; Deduplication was performed to remove bias). [34] The next step was the utilization of previously created patient polymorphic peptide libraries (derived from nsSNP(GVH) in each DRP) for initial feasibility of BLAST protein sequence alignment analysis. [35] This was followed by a subsequent confirmatory CMV-Human sequence homology analysis. Sequence homology relied on a match of 6 or more continuous amino acids in a string of 9 total amino acids (6/6-9/9) by sliding window analysis to identify sequence overlap between HLA bound 9-mer alloreactive human and hCMV peptides. Next, HLA Class I binding prediction (NetMHCpan) was performed for the hCMV peptides, screening the generated hCMV 9-mer peptides for HLA binding affinity across the test patient population (n = 9).
Fig 2. Bioinformatic pipeline of the alloreactive peptide library comparison with the CMV proteome.
A determination of the potential for Pro-GVHD DRP cross-reactivity upon CMV reactivation correlated with GVHD-tissue specific distribution of peptides & immunogenicity.
A specific streamlined bioinformatics application (Table 1) was developed to directly compare the alloreactive donor-recipient peptide libraries for all the patients (n = 77) to the HLA bound hCMV peptides by sequence homology; eliminating the need for the Protein BLAST sequence alignment (≥6/6 match by 9-mer sliding window analysis using all 9-mers extracted from each database, DRP peptide library or Human CMV CROSS proteome, pre- binding affinity screening). Binding affinity was predicted (NetMHCpan) for all the 70,686 hCMV 9-mer peptides (resulting from the 289 hCMV proteins) to the known 2,915 human HLA-class I types along with collection of source gene information (hCMV and Human geneids), which were used to sort the peptides for strength of binding affinity. Immunogenicity screening was then performed to validate CMV-human matched peptides for the presence of hCMV gene products previously reported as CD8+ T cell targets known to elicit specific immune responses.
Table 1. HCMV cross-reactivity potential of 77 SCT DRP from whole exome sequencing data using sliding window match analysis.
| 1 | 289 HCMV protein sequences representative of the HCMV proteome were downloaded from NCBI by batchentrez utility |
| 2 | Downloaded CMV protein sequences were sliced into 9AA peptides and deduplicated, resulting in a library of 70686 9AA peptides |
| 3 | Each of the 70686 9AA peptides were further divided into 2 8AA, 3 7AA and 4 6AA peptides to form a lookup table for each component peptide (6,7,8) with the one or more 9AA parent peptide(s) and the associated CMV geneid |
| 4 | As a focus on Class I HLA, a list of the 2915 plausible HLA-A/B/C types interrogated by netmhcpan was compiled |
| 5 | IC50 values for all 70686 9AA CMV peptides were precalculated against all 2915 HLA types and saved in the CROSS database |
| 6 |
9-mer peptides isolated from ANNOVAR per patient were searched for match with the 9AA CMV peptide pool … if no match found then each 9-mer was split into 2 8-mers and each searched in the 8AA CMV peptide pool … if still no match found then each 9-mer was split into 3 7-mers and each searched in the 7AA CMV peptide pool … if still no match found then each 9-mer was split into 4 6-mers and each searched in the 6AA CMV peptide pool |
| 7 | If any of the above searches found a match, the search was terminated and results for that patient 9-mer peptide reported |
| 8 | Reported 9-mers were linked with geneid (Human & CMV), HLA type, IC50 (Human & CMV), partial peptide (6/7/8), CMV_peptide (9) |
| 9 | Each patient-specific peptide library generated by comparing alloreactive peptides with the CROSS database were compiled, statistically analyzed (Student's T-Test) and further screened for high affinity binding (1/IC50 or an IC50 range of 0–500 nM) |
| 10 | Binding strength (IC50<500 nM) of homologous CMV and human peptides to DRP-specific HLA was compared for pattern analysis |
| 11 | Immunogenicity of common CMV gene products as CD8+ T cell targets was screened for validation on CMV infected patients (n = 30) |
| 12 | CMV peptide matches to Human alloreactive peptides underwent multiple rounds of deduplication to ensure only unique peptide inclusion by both software programming (Excel or independent coding) and finally by human observation upon final analysis. |
| 13 | Gene Tissue Expression (GTEx) data was gathered on CMV infected patients (n = 30) to find GVHD specific organ incidence |
| 14 | Case studies were prepared, 6 patients with CMV before GVHD: 2 acute GVHD, 2 chronic GVHD and 2 aGVHD + cGVHD. |
Tissue distribution of cross-reactive peptides
Homologous CMV-Human peptide libraries were compiled for each DRP and the source genes organized for putative GVHD target tissue-specific distribution analysis using the GTEx portal for expression data. [37] Each patient with GVHD involving specific tissues (e.g., skin) had tabulation of those disease-incident tissue-specific alloreactive peptides presented by their HLA (e.g., 10 skin peptides from 8 genes expressed in the skin) and the remainder GVHD tissue peptides/genes were tabulated as other or nonspecific to the patient’s disease then tallied. Following criteria were used to assign CMV-Human peptide cross-reactivity potential: 1. Amino acid sequence homology of 6/6 to 9/9 (9-mers) between CMV and Human HLA bound peptides 2. IC50 values less than 500 nM for binding to the unique HLA in each DRP (Presented or strongly presented peptides) 3. Human Gene-GVHD Tissue Expression Connection (Patient specific, i.e. skin, GI, etc.)
Patients
CMV reactivation defined as ≥200 copies of hCMV DNA/μL of plasma was determined by quantitative polymerase chain reaction. Absolute lymphocyte counts were measured at least twice weekly following SCT during the first 100 days and at least once weekly out to 6 months following SCT. Human CMV titers, lymphocyte counts and serum immunosuppressant drug levels were collected to one year post SCT per MCC-14012 Study. Acute and chronic GVHD was diagnosed according to Glucksberg and NIH 2005 Consensus clinical criteria, respectively. Lymphoid recovery patterning was assessed to examine differences among patients with CMV alone, GVHD alone, both or neither conditions post SCT. Three patterns were observed as previously described [38]: Pattern A or early (ALC ≥ 1 Billion cells/L by Day 45 post SCT), Pattern B or middle (ALC ≥ 0.5 up to <1 Billion cells/L by Day 45 post SCT), and Pattern C (ALC <0.5 Billion cells/L by Day 45 post SCT). Student’s T test and Pearson correlation were utilized for statistical analysis of the alloreactive/CMV peptide data presented. Cox regression hazard ratio and Kaplan-Meier Survival were utilized for evaluating patient outcomes.
Results
Patient characteristics
Seventy-seven DRPs underwent exome sequencing following SCT, and were assessed by in silico analysis for alloreactivity potential derived from non-synonymous SNPs in the GVHD direction as previously described. [15] The cohort comprised of 27 MRD and 50 MUD SCT recipients underwent data collection for retrospective analysis including GVHD occurrence and hCMV reactivation information S1 Table. Of the 30 patients that experienced hCMV viremia within a median of 29 days from transplant, 26 were hCMV-seropositive or reactivated; one patient had drug-refractory hCMV infection, and succumbed to it. CMV reactivation was experienced in conjunction with GVHD in 18 of the 30 patients, 13 of whom experienced hCMV reactivation prior to GVHD onset. Multiple recurrences of hCMV viremia were experienced in 11 of the 30 patients (2 or more separate reactivation events, ≥200 copies of hCMV DNA/μL plasma per event by quantitative polymerase chain reaction).
CMV—Human HLA bound peptide homology
Based on the hypothesis that significant amino acid sequence homology between human- and viral-derived peptides presented on the same Class I HLA molecules may lead to donor CD8+ T cell cross reactivity, a bioinformatic pipeline was utilized to assess sequence homology between the patient’s putative alloreactive peptide library, and hCMV-derived oligopeptides bound to each recipient’s HLA. Whole exome sequencing of HLA matched SCT donors and recipients revealed 2,463 ± 603 nsSNPs (mean±SD) with a GVH direction per MRD and 4,287 ± 1154 nsSNPs per MUD recipient (Student’s T-test, p<0.001). Following ANNOVAR 9-mer peptide determination and completion of the sliding window analysis of the resulting 17 amino acid variant oligopeptide resulting from each nsSNP, 43,705 ± 10,938 nonameric potentially alloreactive human peptides were identified per MRD recipient, and 77,025 ± 21,170 per MUD recipient. These were organized into a library by HLA-specific binding affinity (IC50: 0–50,000 nM). Next, each alloreactive Human 9-mer peptide library was further evaluated for their degree of match with the 70686 nonamer peptides derived from the hCMV proteome, utilizing the HLA-specific algorithmic CROSS database. This comparison determined sequence homology for strings of ≥6/6 consecutive amino acids between the two sets of Human and hCMV peptides, and was termed ‘sliding window-match analysis’. This initial screen yielded an average of 29,659 ± 9039 total peptide matches per MRD patient and 52,910 ± 16122 total peptide matches per MUD patient to the hCMV proteome (Student’s T-test, p<0.001) following analysis (S1 Fig). The program output reported for each peptide match, the geneid’s, the HLA, IC50 values (Range: 0–50,000 nM) and complete peptide sequence, as well as the shared partial peptide for degree of sequence homology.
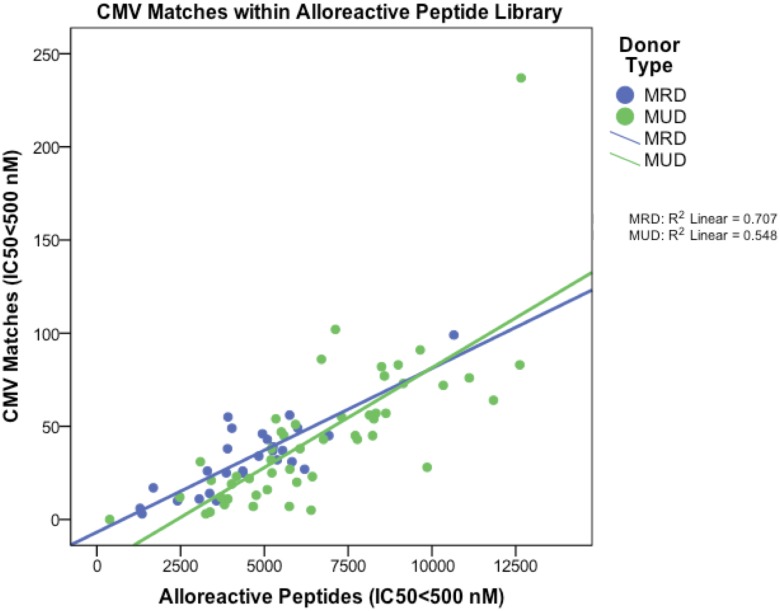
Upon confirmation that all 77 patient alloreactive peptide-HLA libraries studied had matches with the hCMV peptide-HLA arrays, the degree of sequence homology in tightly HLA bound peptides (IC50: 0.01–500 nM) was determined. Following removal of duplicate peptide sequences, the total CMV-human match (homology) library, yielded an average of 33 peptide matches per MRD patient and 44 peptide matches per MUD patient (Student’s T-test, p = 0.09), constituting approximately 1% of the total matches reported (Table 2). Considerable variability was observed in the in silico HLA presentation of human-CMV sequence homologous peptides among the different HLA in the DRP studied (S2 Fig). Analysis of the number of Human—CMV matches with IC50<500 nM yielded a lack of association between how common different HLA molecules are within a given population and the overall average or peak number of matches per HLA. The most common HLA-A (02:01) to our patient population was outside the top ten molecules to have the highest average or peak number of Human—CMV matches. The homologous sequence information per patient’s peptide library following high affinity match analysis (IC50<500) was compiled into S2 File; this includes an immunogenicity sorting performed for hCMV reactivating patients. Considering the degree of sequence homology present between hCMV and human peptides bound to HLA class I molecules in specific DRP, each MRD DRP on average had 31, 2, 0 and 0 relevant nonameric CMV-Human peptide matches with 6/6, 7/7, 8/8 and 9/9 sequence homology respectively (Table 2). Also each MUD DRP on average had 40, 3, 1, and 0 peptide matches with 6/6–9/9 sequence homology respectively. As expected from the donor type SNP differences, relevant alloreactive peptide differences, and total peptide matches with hCMV, MUD DRPs exhibited a trend for a higher number of hCMV matched peptides than MRD DRPs by peptide sequence homology differences (6/6-9/9 matching; student’s T-test, p = 0.07–0.38 respectively). To determine the correlation of the number of high affinity CMV-Human matches (IC50<500 nM) with sequence homology >6/6 amino acids in 76/77 patients, these were plotted against the pool of total alloreactive peptides per patient (Fig 3). Notably, MUD DRPs post screening (IC50<500 nM) exhibited a mean of 6,545 ± 2689 alloreactive peptides per patient which was significantly greater than the 4,522 ± 1915 mean alloreactive peptides per MRD DRPs (Student’s T-test, p<0.001) prior to interrogating for matches to the hCMV proteome, with a trend for a higher prevalence of CMV-Human matches in the MUD recipients. These results indicate that there exists a pool of homologous-CMV-derived peptides, which may be presented by the recipient HLA.
Table 2. CMV- Human match analysis per DRP (IC50 <500 nM) by donor type according to continuous amino acid sequence homology (one standard deviation from the mean value).
| Mean Relevant Matches (IC50<500) | Number of Patients (n) | Sequence Homology 6/6 (±SD) | Sequence Homology 7/7 (±SD) | Sequence Homology 8/8 (±SD) | Sequence Homology 9/9 (±SD) | Total 6/6 to 9/9 (±SD) |
|---|---|---|---|---|---|---|
| MRD | 27 | 31 ± 19 | 2 ± 2 | 0 ± 1 | 0 ± 0 | 33 ± 20 |
| MUD | 50 | 40 ± 33 | 3 ± 6 | 1 ± 2 | 0 ± 1 | 44 ± 39 |
| Overall | 77 | 37 ± 29 | 3 ± 5 | 1 ± 2 | 0 ± 1 | 40 ± 34 |
| Student's T-test (p < .05) | 0.12 | 0.09 | 0.07 | 0.38 | 0.09 | |
Fig 3. CMV+GVHD cross reactivity potential.
Patient-specific peak CMV peptide matches intersecting peak alloreactive peptides (IC50<500 nM) as a cross-reactivity potential stratified by donor type contained within each DRP alloreactive peptide library.
Potential immunogenicity of the hCMV peptides presented on recipient HLA
To compare the sequence homology analysis results and determine the presence of previously validated immunogenic hCMV peptides, [36] the specific hCMV peptide sequences and the proteins of origin were reviewed (Table 3). Twelve of the 13 patients with hCMV reactivation before GVHD onset exhibited one or more immunogenic hCMV peptide matches, derived from CMV protein previously shown to elicit a hCMV-specific T-cell response targeting the listed source genes. The remaining patient expressed CMV—Human peptide-matches derived from proteins specific to the patient’s affected GVHD tissue, which had not previously been reported in other studies. The analysis revealed multiple CMV-Human peptide matches that were, immunogenic (IC50<500 nM) and expressed in a GVHD-affected tissue-specific manner. Further, the binding affinity (reflected by the IC50 values) of hCMV peptides when plotted against the IC50 values of the alloreactive Human peptides S3 Fig, demonstrated a wide range of values, indicating the potential for varying degrees of cross reactivity. This analysis illustrates the magnitude of overlap in peptide sequence between the two sources of HLA presented peptides.
Table 3. GVHD tissue-specific immunogenic CMV peptide matches, CMV reactivation before GVHD patients.
Patient-specific process of cross comparison along with the source genes, IC50 values to indicate inverse binding affinity and GVHD organ specific tissue involvement. *Note: Twelve patients with CMV reactivation/infection before GVHD onset exhibited previously identified immunogenic CMV peptide matches with gene expression specific to the tissues affected by GVHD (above); The filter of immunogenicity provides a connection to T cell reactivity shown in vitro to the listed CMV genes in a separate patient population [36]; Patient 79 with muscle/fascia GVHD showed no muscle-specific previously known immunogenic CMV peptide matches but still had three relevant CMV peptide matches expressed in the skeletal muscle (not shown); Tissues in parentheses were also affected by GVHD but without immunogenic matches/expression by patient; Patients 67 and 71 experienced de novo CMV infection; All 13 patients exhibited multiple CMV peptide matches with unknown immunogenicity.
| Patient | HLA | Human Gene | IC50 Value | DRP Peptide | CMV Peptide | IC50 Value | CMV Gene | GVHD Organ with Target Gene Expression |
|---|---|---|---|---|---|---|---|---|
| 10* | HLA-B40:01 | TSPYL1 | 12 | SETVALPPL | QENSETVAL | 23 | UL86 | Skin |
| HLA-B40:01 | HDAC7 | 27 | EEVEAVTAL | AEREAVTAL | 20 | UL49 | ||
| HLA-B08:01 | APEH | 487 | NRRSALYSV | MWKNRRSAL | 11 | UL37 | ||
| HLA-B08:01 | WKNRRSALL | 274 | UL37 | |||||
| 27* | HLA-C03:04 | IL22RA1 | 16 | IVHPTPTPL | SSTPTPTPL | 42 | UL82 | Skin |
| HLA-A02:01 | TECR | 28 | ALFSLVVFT | YLFSLVVLV | 3 | US3 | ||
| HLA-A02:01 | LVYLFSLVV | 442 | US3 | |||||
| HLA-C03:04 | WNK1 | 41 | ITAAATAPV | TTAAATATV | 199 | UL105 | ||
| 45* | HLA-A68:02 | PPP1R15A | 149 | SSAAAAAAL | TAAAAAAAA | 58 | IRS1,TRS1 | Skin & Vagina |
| HLA-A68:02 | ATAAAAAAA | 59 | IRS1,TRS1 | |||||
| HLA-A68:02 | DAAAAAAPT | 286 | IRS1 | |||||
| HLA-A68:02 | SVSSSAAAA | 376 | UL105 | |||||
| HLA-A68:02 | SASAAAAAA | 469 | UL105 | |||||
| HLA-A68:02 | DATAAAAAA | 470 | IRS1,TRS1 | |||||
| HLA-A68:02 | MUC21 | 106 | ESSTTSSGA | TTYTTSSGA | 24 | UL86 | Vagina | |
| HLA-A68:02 | YTTSSGAKI | 140 | UL86 | |||||
| HLA-A68:02 | HOXD13 | 352 | AAAAAAAGA | TAAAAAAAA | 58 | IRS1,TRS1 | ||
| HLA-A68:02 | ATAAAAAAA | 59 | IRS1,TRS1 | |||||
| HLA-A68:02 | DAAAAAAPT | 286 | IRS1 | |||||
| HLA-A68:02 | SASAAAAAA | 469 | UL105 | |||||
| HLA-A68:02 | DATAAAAAA | 470 | IRS1,TRS1 | |||||
| 47* | HLA-B07:05 | MUC17 | 52 | TPVSHTLVA | TPVSHTQPL | 8 | UL150 | GI |
| HLA-A30:02 | CARS2 | 343 | SPASLSSLY | YFDSLSSLY | 173 | UL29,UL28 | ||
| 48* | HLA-A31:01 | PLXND1 | 200 | LFVFCTKSR | NLFVFCTER | 58 | UL34 | GI (Skin & Liver) |
| HLA-A31:01 | PLXND1 | 307 | VALFVFCTK | |||||
| 67* | HLA-A02:01 | FGFR4 | 20 | RLLLALLGV | VLLLALLLL | 209 | US29 | GI, Lung (Skin) |
| HLA-A02:01 | FGFR4 | 165 | LLLALLGVL | |||||
| 68* | HLA-A31:01 | ALCAM | 188 | TYTLTAVRR | ASLLTAVRR | 68 | IRS1,TRS1 | GI, Liver (Skin) |
| 70* | HLA-C07:02 | LCN2 | 122 | SYPGLPSYL | MRPGLPSYL | 50 | UL75 | GI |
| HLA-B35:01 | SERPINB4 | 97 | EAAAATAVL | CAAAAATAA | 393 | UL105 | ||
| HLA-A03:01 | FYCO1 | 488 | RASLKRLVK | STLKRLVKK | 83 | UL69 | ||
| 71* | HLA-C03:03 | PPP1R15A | 19 | SSAAAAAAL | AAAAAAPTV | 48 | IRS1,TRS1 | Skin |
| HLA-C03:03 | AAAAAAAAA | 343 | IRS1,TRS1 | |||||
| HLA-C03:03 | TAAAAAAAA | 389 | IRS1,TRS1 | |||||
| HLA-C03:03 | SASAAAAAA | 421 | UL105 | |||||
| HLA-A30:02 | CARS2 | 343 | SPASLSSLY | YFDSLSSLY | 173 | UL29,UL28 | ||
| 76* | HLA-A03:01 | ZZEF1 | 119 | SVLSELLKK | SVLSELLNK | 91 | UL54 | GI |
| HLA-B27:05 | MUC4 | 414 | TRHATSLPV | YQLRHATSL | 473 | US29 | ||
| 84* | HLA-A02:01 | FGFR4 | 20 | RLLLALLGV | VLLLALLLL | 209 | US29 | GI (Skin) |
| HLA-A02:01 | FGFR4 | 165 | LLLALLGVL | |||||
| HLA-C02:02 | MUC17 | 270 | TSMPVSTTV | ISYPVSTTV | 215 | UL75 | ||
| HLA-B27:05 | MUC4 | 414 | TRHATSLPV | YQLRHATSL | 473 | US29 | ||
| 90* | HLA-A02:01 | PELP1 | 78 | LLALLLAPT | LLALLLLEL | 38 | US29 | Skin & GI |
| HLA-A02:01 | VLLLALLLL | 209 | US29 | |||||
| HLA-A02:01 | AHSA1 | 321 | KTLFLAVQV | ELFLAVQFV | 145 | UL86 | ||
| HLA-C03:03 | TCF7L1 | 107 | AAASSSGQM | AAASSSSAV | 27 | UL48 | GI | |
| HLA-C03:03 | IAAASSSSA | 220 | UL48 | |||||
| HLA-A03:02 | ZZEF1 | 325 | SVLSELLKK | SVLSELLNK | 185 | UL54 | ||
| HLA-C03:03 | PLEKHA6 | 385 | AASSSLRRL | FGGAASSSL | 71 | UL122 |
Tissue expression of cross-reactive human peptides
In order to study the GVHD tissue-specific expression of the hCMV peptides matched to human alloreactive peptides (Table 3), gene expression data organized in a tissue specific manner was obtained from the GTEx Portal of the Broad Institute of MIT and Harvard (Version 6). [37] Gene expression data for all CMV-Human peptide matches in the 30 hCMV reactivating/de novo infected patients was compiled (median minimum threshold of expression: ≥10 reads/kilobase of transcript/million mapped reads), specifically focusing on GVHD target tissues including: skin, GI, liver, lung, and others (vagina, muscle, adipose and salivary gland). A focused analysis of GVHD tissue gene expression in conjunction with actual GVHD occurrence was performed on 18 patients with GVHD and hCMV reactivation, and showed that 18/18 patients had expression of hCMV-matched alloreactive peptides (IC50<500 nM) with ≥10 RPKM tissue-specific gene expression (Table 4). As depicted, combining both match data and gene expression data, a cross reactivity profile was created for the hCMV infected subset of patients to include: match number, discrete numbers of peptides (Human and CMV), gene count, immunogenic hCMV genes, and tissue specific GVHD peptides or genes. These data points taken together comprise a predictive case for potential alloreactive trigger following hCMV infection if human and CMV peptide libraries possess enough peptide sequence homology that may be derived from whole exome sequencing of transplant donors and recipients using this analytic approach.
Table 4. Human-CMV short sequence homology in GVHD tissue specific peptide and gene distribution from GTEx analysis (n = 18).
GVHD incidence denotes the specific organs affected in each patient; Peptides, lists the number of unique peptide-HLA complexes matched between human and CMV peptide library; the column, Genes lists the source genes for the aforementioned peptides; GVHD tissue specific peptides lists the number of peptides which bind HLA with an IC50 <500nM, and are expressed in tissues affected by GVHD; GVHD tissue gene expression denotes the number of genes expressed at an RPKM >10 corresponding to the GVHD tissue specific peptides. Note: *- All patients with an asterisk following their numeric representation experienced CMV reactivation prior to GVHD (except Patients 67 and 71, de novo CMV infected) and patients without an asterisk experienced GVHD prior to CMV reactivation. **- human peptides may have overlapping areas of homology yielding a higher number of matches. Immunogenic CMV genes in this context refer to the genes associated with HCMV ORF-specific CD8+ T memory cell responses measured in frequency among CMV seropositive patients previously [36].
| Patients | GVHD Incidence | Peptides** | GVHD Tissue Specific Peptides (IC50: <500 nM) | Genes | GVHD Tissue Genes (RPKM: >10) | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 18 | Skin | GI | Liver | Lung | Vaginal | Muscle | Human | Matches | CMV | Skin | GI | Liver | Lung | Vaginal | Muscle | Human | Immunogenic CMV | CMV | Skin | GI | Liver | Lung | Vaginal | Muscle |
| 10* | X | 58 | 72 | 58 | 10 | 47 | 10 | 35 | 8 | |||||||||||||||
| 27* | X | 43 | 64 | 55 | 7 | 39 | 9 | 35 | 7 | |||||||||||||||
| 45* | X | X | 17 | 45 | 26 | 7 | 9 | 15 | 5 | 15 | 6 | 8 | ||||||||||||
| 47* | X | 21 | 23 | 22 | 6 | 20 | 5 | 19 | 6 | |||||||||||||||
| 48* | X | X | X | 24 | 25 | 21 | 5 | 11 | 5 | 21 | 5 | 18 | 4 | 9 | 4 | |||||||||
| 67* | X | X | X | 28 | 43 | 36 | 3 | 8 | 9 | 24 | 4 | 24 | 3 | 7 | 7 | |||||||||
| 68* | X | X | X | 10 | 10 | 10 | 2 | 7 | 2 | 10 | 2 | 10 | 2 | 7 | 2 | |||||||||
| 70* | X | 25 | 28 | 22 | 9 | 21 | 5 | 21 | 8 | |||||||||||||||
| 71* | X | 29 | 55 | 39 | 2 | 28 | 8 | 30 | 2 | |||||||||||||||
| 76* | X | 10 | 11 | 11 | 5 | 10 | 4 | 10 | 5 | |||||||||||||||
| 79* | X | 39 | 49 | 41 | 3 | 28 | 7 | 26 | 2 | |||||||||||||||
| 84* | X | X | X | 21 | 25 | 21 | 1 | 7 | 2 | 18 | 4 | 14 | 1 | 6 | 1 | |||||||||
| 90* | X | X | 39 | 56 | 43 | 8 | 17 | 36 | 7 | 27 | 7 | 16 | ||||||||||||
| 34 | X | 38 | 57 | 47 | 8 | 32 | 12 | 31 | 7 | |||||||||||||||
| 38 | X | 32 | 47 | 41 | 6 | 30 | 6 | 29 | 6 | |||||||||||||||
| 41 | X | 12 | 12 | 11 | 5 | 11 | 2 | 11 | 4 | |||||||||||||||
| 73 | X | X | 5 | 5 | 5 | 1 | 2 | 5 | 0 | 4 | 1 | 2 | ||||||||||||
| 81 | X | 10 | 17 | 10 | 4 | 6 | 0 | 6 | 2 | |||||||||||||||
Clinical correlations of CMV reactivation with GVHD
In this cohort hCMV reactivation following SCT, was associated with poorer survival (n = 46, Log Rank: 4.5, p = 0.03, Hazard ratio: 2.31, p = 0.04) when adjusting for hCMV reactivation by donor or recipient seropositivity S4 Fig, consistent with the findings of a recent CIBMTR analysis. [2] Further we observed a trend of poor survival among CMV seropositive DRPs that experienced hCMV reactivation along with GVHD S5 Fig (n = 46, Log Rank: 3.04, p = 0.08, Hazard ratio: 2.02, p = 0.09). To determine whether there was any association between onset of hCMV reactivation and GVHD, we evaluated lymphocyte growth kinetics and large variations in calcineurin inhibitor levels as possible triggers of GVHD occurrence in the above referenced patients (Table 4). Lymphoid recovery patterns were examined among these patients with early lymphocytosis seen in patients with CMV viremia S6 Fig. GVHD was also examined for association with lymphoid recovery rate and magnitude along with CMV reactivation. CMV reactivation + GVHD patients did have a trend for more frequent occurrence of earlier and larger magnitude lymphoid recovery patterns compared with GVHD alone, CMV alone or the patients with neither. There was however no significant difference in survival between these patient groups S7 Fig. Importantly in the patients with an observed association between CMV reactivation, lymphocytosis and GVHD onset, the T cells were found to be donor-derived DNA by chimerism analysis in most of our patients.
In the 30 hCMV-reactivating patients examined S2 File, reactivation events were often coincident with GVHD onset and associated with preceding lymphocytosis, suggesting an alloreactive cellular immune response S8 Fig (Panels A, B and C). Acute GVHD onset following hCMV reactivation was seen in 7 of the 13 patients experiencing hCMV reactivation before GVHD; 3 of these patients developed grade IV gastrointestinal GVHD (2 steroid refractory) which was fatal in all cases. As seen in figure S8 Fig (Panel A), with acute GVHD in patients 47 and 68, there was evidence of hCMV reactivation events (top graph) that preceded rapid onset lymphocytosis (middle graph) and often occurred with stable CNI or immunosuppression levels (bottom graph), indicating a potential hCMV-GVHD relationship. Patients 47 and 68 with acute GI GVHD had an average of 16.5 ± 9.2 matches (±SD) derived from 15.5 ± 7.8 human peptides. Patient 68 also exhibited skin and liver GVHD. When considering the percentage of genes with GVHD-tissue specific expression on average, in these two patients 87.5 ± 17.7% peptides were derived from GI tract specific alloreactive-hCMV matched genes (primary GVHD organ).
Patients depicted in S8 Fig (Panel B, patients 10 and 27) exhibited the more stable or gradual hCMV reactivation effects seen in chronic GVHD, where lower or less frequent elevations in hCMV titers may still elicit lymphocyte growth, albeit at a slower rate, probably accounting for the observed difference in outcomes between patients with symptoms of only acute GVHD or chronic GVHD. Patients 10 and 27 with Chronic Skin GVHD had a mean of 68 ± 5.7 matches (±SD) derived from 50.5 ± 10.6 human peptides. These two patients exhibited a lower percentage of GVHD-tissue specific gene expression on average when considering all potential GVHD genes identified, in this instance skin gene expression amounted to 51.9 ± 2.7%.
The final S8 Fig group (Panel C, patients 71 and 84) exhibited symptoms of both acute and chronic GVHD at different times post transplant with differing disease patterns according to the GVHD tissues involved (i.e., acute skin or GI vs chronic skin or GI GVHD may be defined/graded differently), but patient 84’s lower grade cyclical reactivation events altered the lymphocyte growth pattern towards a recurring waveform pattern, with peaks and troughs of lymphocytes over time. Patients 71 and 84 exhibiting both acute and chronic GVHD had a mean of 40 ± 21.2 matches (±SD) derived from 25 ± 5.7 human peptides. Patient 84 also exhibited GI and liver GVHD. Patients 71 and 84 also exhibited lower GVHD-tissue specific gene expression on average when considering all potential GVHD genes identified, Skin,or GI (primary organs): 44.3 ± 27.3%. There were 3 patients who developed recipient-derived T cell chimerism following CMV reactivation and lymphocytosis (patients 2, 7 and 28), suggesting expansion of recipient derived T cell clones in response to CMV reactivation. These three patients were all CMV sero positive with CMV + donors. These variable dynamics of hCMV reactivation and GVHD onset demonstrate the complexity involved in analyzing the relationship between ongoing immunosuppression in the setting of multiple sets of potentially cross reactive antigens by affected organ system being presented to a reconstituting donor-derived immune system.
Discussion
CMV reactivation is a frequent complication of allografting, requiring frequent monitoring and associated with an increased risk of treatment related mortality, primarily in its own right, but also because it is frequently associated with GVHD. [12] Therapy and effective prophylaxis involve the use of toxic drugs and monitoring for reactivation is not straightforward. The ability to identify patients at risk of developing alloreactive complications from hCMV reactivation will therefore be a useful adjunct to the supportive care of transplant recipients, as well as an important step forward in understanding virus-induced-alloreactivity. In this paper a computational algorithm that identifies hCMV peptides homologous to human alloreactive peptides is described. This determination required three steps, whole exome sequencing of transplant donors and recipients, followed by in silico determination of the patient specific class I HLA binding of the oligopeptides resulting from the nsSNP in the exome and finally a comparison of these alloreactive peptide sequences with those of hCMV peptides predicted to bind the same HLA molecules. This algorithm identifies a number of hCMV peptides which bind the same HLA as human peptides with a similar range of binding affinities and also demonstrate a degree of sequence homology with the human peptides. We hypothesize that these peptides may potentially be cross-presented to donor T cells.
To understand how this may impact GVHD pathophysiology, consider a T cell clone (TCCMV), which recognizes an hCMV peptide-HLA complex, is activated by hCMV viremia. The T cell receptor of this clone may also recognize a human alloreactive peptide with sequence homology to the hCMV peptide and bound to the same HLA molecule. Even if it does so weakly, tissue damage may be initiated and GVHD ensue. This process can work in reverse as well, a T cell clone with high affinity for alloreactive peptides (TCmHA), which only binds the hCMV peptide-HLA complex weakly may be ‘set-off’ by a hCMV reactivation event, again leading to down-stream GVHD. This general principle may hold true for other virus derived peptides.
A mathematical model utilizing matrices has been developed to understand aggregate T cell responses to many mHA-HLA complexes the donor T cells may encounter in the recipient milieu. [39] The simplifying notion underlying this model is that each donor T cell interacts with a single recipient HLA-bound antigen, therefore an identity matrix may be used to calculate the resulting T cell response. This model requires that the antigens occupy the matrix operator (MAPO) and T cell vector gets transformed by the operator (Table 5).
Table 5. Matrix depicting the T cell clonal interaction with mHA-HLA.
| mHA1 HLA | mHA2 HLA | mHAn HLA | |
|---|---|---|---|
| TC1 | 1 | 0 | 0 |
| TC2 | 0 | 1 | 0 |
| TCn | 0 | 0 | 1 |
In the above simplified matrix T cell clone (Table 5), TC1 interacts with mHA1, and so on. The 1 in the cells means that the T cells recognize that antigen and responds, and 0 means absence of recognition, this can also be understood as probability of TCR binding to the peptide-HLA complex. In reality, the TCR-Ag-HLA interactions are not likely to be quite so simple. An important clue to this is the observation that antigen-HLA binding affinities, reported in this paper as IC50, do not take on discrete values of 0 or 1 (binding or no binding); instead there is a continuum of IC50 values. So in the case of cross-reactive antigens, a hCMV derived antigen binds the same HLA molecule and may interact with the T cell receptor, albeit with a different binding affinity. This implies that each TCR might interact with multiple antigens with different affinity, thus the 0 in the matrix above may in reality be replaced with a series of numbers between 0 and 1. These cross-reactive antigens may augment the T cell response to the primary target antigens, as shown in the matrix below where cross reactivity is depicted (Table 6).
Table 6. Matrix depicting T cell clonal cross-reactivity between CMVp-HLA and mHA-HLA.
*-indicates response of the alloreactive T cell clone to a viral pathogen peptide,bound to the same HLA as the mHA and vice versa. For example, TC1 recognizes, mHA1 HLA + CMVp2HLA, TC1CMV recognizes, mHA1 HLA + CMVp1 HLA, and so on.
| mHA1-HLA | (CMVp1-HLA) | mHA2-HLA | (CMVp2-HLA) | |
|---|---|---|---|---|
| TC1 | 1 | 0.2* | 0 | 0 |
| TC2 | 0 | 0 | 1 | 0.3 |
| TC1CMV | 0.3* | 1 | 0 | 0 |
| TC2CMV | 0 | 0 | 0.1* | 1 |
The notion of T cell receptor (TCR) cross reactivity presented here also merits further discussion. An argument may be made that mHA that do not share an entire sequence are unlikely to elicit a response from relevant TCR. However this argument assumes a rigid interaction between the TCR and the mHA-HLA complex. In reality these interactions are likely to be elastic in nature, as can be inferred from the continuum of values that the CMV peptide/mHA-HLA IC50s demonstrate. In a rigid frame work one will likely observe discrete binding affinity value sets. Such ‘elasticity’ in TCR recognition has previously been demonstrated in the context of HLA-B35:01 bound HIV-1 derived Nef epitope VY8. In this study, patient derived CD8+ T cell clones recognized index peptides despite substitution in the AA residues along the peptides, demonstrating unique cross reactivity ‘footprints’ for individual T cell clones. [40,41] The counter argument of multiple T cell clones recognizing a single antigen has also been studied in the context of tumor antigen HLA-A:0201/NY-ESO-1 specific CD8+ T cell clones, where multiple T cell clones with relatively restricted TRB V and J segment usage were identified. These T cell clones were equally efficient and dependent on recognition of the central peptide residues for activity. [42] Similar findings have been reported for melanoma tumor infiltrating CD8+ T cells which recognize variants of HLA-A2 bound MART-1 antigen variants. [43] Other models of T cell cross reactivity have also been reported in the literature for TCR recognition, such as, widely degenerate recognition of unrelated peptides in peptide-MHC complexes exhibiting docking geometry diversity and CDR Loop displacement. [16,44–46] Alternatively, TCR recognition of only closely related peptides to original pathogen-derived peptides with the same docking geometry of binding and similar CDR3 displacement patterns has been proposed more recently. [47] Another consideration is the differential effect of binding affinity and stability of CMV peptide/mHA—HLA interactions influenced by enhanced antigen availability by either greater proteolytic activity [48] or related to higher tissue expression levels. [49–51] Taken together these lines of evidence suggest that TCR cross reactivity, and stimulation of alloreactivity is certainly possible in the context of the same HLA molecules presenting both CMV and alloreactive human peptides with sequence homology. It is however important to recognize that this mechanism most likely only contributes to the initiation of GVHD in some patients, with more conventional alloreactive T cell responses occurring in the face of tissue injury and systemic inflammation being responsible for GVHD in most cases. [52] While the in silico analysis for hCMV + alloreactivity potential reveals a large body of antigens which may influence clinical outcomes. However, there are caveats to be considered in developing this peptide analysis pipeline. First, the process of creating the CMV CROSS database included some variant forms of the hCMV proteins known to have differing amino acid lengths that may allow for duplicates, which were accounted for during multiple processes of deduplication. However the slight differences to the variant forms of the proteins reported in the NCBI database may generate a few more peptide possibilities than may be realistic but would allow for strain differences of the human peptide matches during exome sequencing.
An interesting observation among alloreactive or hCMV peptides was where human genes and hCMV immunogenic genes were shared among multiple patients with many of each gene or peptide involved being common in our cohort, often with the same HLA specificity and predicted binding affinity S2 File. Epitope spreading is another phenomenon, that may be exemplified by donor lymphocytes and antigen presenting cells encountering either hCMV or recipient alloreactive peptides released upon organ damage that may be processed and presented in an HLA restricted manner and elicit an immune response. CMV has evolved in parallel with the human genome over hundreds of millions of years, potentially exchanging genetic information from virus and human randomly with each latent infection, selecting primarily for immune evasion. [53] This phenomenon shields hCMV from immune response but also exposes endogenous antigens from recipient cells simultaneously with hCMV antigens to donor immune surveillance during organ damage from lytic reactivation in athymic adults. [54] This form of epitope spreading, developing T cell antigenic experience, affects overall immune responses and may account for the robust CD8+ T cell response to hCMV infection in otherwise healthy individuals (10% of the entire T cell compartment) that inflates with age. [36] In addition, the primarily memory T cell response to hCMV accounts for the sharp responses to various hCMV antigens upon reactivation, which may be as great as 50 fold the strength of a naïve T cell response during primary infection. [19] Recently, reduced TCR repertoire diversity in naïve T cells has been reported in patients who have CMV reactivation and GVHD following MUD SCT. The authors evaluated repertoire deficiencies by comparing patient samples with reference and found that patients who had CMV reactivation, with or without GVHD had greater ‘holes’ in the repertoire, suggesting that an oligoclonal population of cross reactive T cells clones may lead to immune dysregulation. [55] Strong CD8+ and CD4+ response to soluble recombinant hCMV antigen has been demonstrated in the past. [56] This may be related to epitope spreading in post-transplant viral immunity, as has already been discovered in Multiple Sclerosis and in other autoimmune disorders. [24,57,58] Nevertheless, GVHD is a condition that is primarily dependent on the presence of an adequate alloreactive stimulus, the abrogation of which would mitigate GVHD regardless of CMV reactivation. This may be observed in recipients of T cell depleted transplants, including some of the patients reported here. Further, when considering the opportunity for hCMV to catalytically trigger GVHD apart from the potential method we propose here, we must note that hCMV is the largest of the herpesvirus family with a 235 kb DNA genome capable of encoding more than 200 potential protein products [59] and second the near ubiquity of cells it is capable of infecting in man, including parenchymal and connective tissue cells of virtually any organ along with various hematopoietic cell types. [60]
In conclusion, this paper reports sequence homology in HLA bound peptide antigens of hCMV and human origin. Given the distribution of human peptides in various tissues, and their involvement with GVHD in the patients examined, we posit that hCMV derived peptides may influence the development of GVHD in patients who develop hCMV reactivation following SCT These findings support the use of more aggressive antiviral strategies for preventing hCMV reactivation in patients undergoing allografting and argue against the use of simply monitoring as the major therapeutic strategy. We endeavor in the future to understand the supporting CD4+ T cell hCMV protective dynamics involved by interrogating HLA class II peptides, (1) looking at potential influences of human CMV mimicry and unraveling the potential GVHD/CMV cross-reactivity/Auto-immune relationship further. [13,18,24,61–65]
Supporting information
CMV CROSS database tools excel spreadsheet.
- Sheet 1. CMV proteome—NCBI accessions: hCMV/HHV5
- Sheet 2. CMV immunogenic protein—CD8+ list from Ref. #36.
(XLSX)
Match analysis excel spreadsheet.
- Sheet 1. Raw CMV match algorithm results—pre-IC50 screening
- Sheet 2. Match analysis summary—total peptide # by patient
- Sheet 3. Match analysis peptides—screened, sorted pt. peptides
- Sheet 4. All CMV case studies, tables, and figures
- Sheet 5. Peptide-HLA binding patterns sorted by CMV+GVH
- Sheet 6. Exome dataset—retrospective, de-identified info.
(XLSX)
Total number of Human—CMV sequence homologous peptides without binding affinity filtering by IC50 values for TCR relevance. Range of IC50 values: 0.01–50000 nM. Averaged number of total matches for MRD DRP (Left, Blue) versus MUD DRP (Right, Green) were displayed above the graph to indicate the observed difference of CMV peptide matches to alloreactive peptides prior to screening by donor type.
(TIFF)
Average number of HLA bound human—CMV sequence homologous peptides ordered by number of peptides presented by HLA locus. Descending order of average matches per HLA (>5 matches per HLA as threshold). Error bars indicated where HLA molecules were shared by more than one DRP and matches averaged.
(TIF)
Binding affinity differences indicated by inverse IC50 values from matched human and CMV peptides bound to the same HLA. Each data point represents the intersection of a matched peptide bound to an HLA class I molecule (CMV→Human). These peptides may be cross reactive, with varying degrees of T cell cross reactivity potential for alloreactivity trigger to ensue (towards the origin on the human peptide axis being the greatest).
(TIF)
GVHD-independent effects of CMV post SCT in CMV seropositive DRP (n = 46).
(TIF)
Cumulative effects of CMV viremia + GVHD post SCT in CMV-seropositive DRP (n = 46).
(TIF)
(Top) Lymphoid Recovery Patterns by ALC value achieved by Day 45 post SCT. Pattern A showed early lymphoid recovery (Patient 2), Pattern B showed a medium time to lymphoid recovery (Patient 48) and Pattern C showed late or very low lymphoid recovery (Patient 16). Trendlines indicated the fit to the logistic pattern previously observed. (Bottom) Lymphoid Recovery Patterns by CMV+GVHD Status (n = 77): CMV Reactivation triggers Lymphocytosis that may influence GVHD onset. Fisher’s Exact Chi Square compared CMV + GVHD groups with lymphoid recovery patterns as shown.
(TIF)
CMV reactivation with or without GVHD onset affects the survival negatively compared to the protective GVHD effect.
(TIFF)
(A) CMV-reactivation course with acute GVHD onset and progression: Patients 47 and 68. Patient 47, grade IV GVHD of the GI tract; patient 68,steroid refractory grade IV GI tract GVHD, skin and liver. Both patients showed signs of CMV reactivation and bursts of lymphocytosis prior to GVHD onset during stable immunosuppression (TAC, Tacrolimus; SIR, Sirolimus; Cyclosporin) as measured by serum levels or following taper. (B) CMV-reactivation with chronic GVHD onset: Patients 10 and 27. Patient 10 had CMV reactivation prior to a gradual lymphocyte proliferation during stable immunosuppression levels (TAC, Tacrolimus) and eventually developed relapsed malignancy. Patient 27 exhibited a CMV reactivation event prior to lymphocytosis at stable tacrolimus levels, and had mainly skin and oral GVHD. (C) CMV reactivation/de novo infection in patients with aGVHD + cGVHD: Patients 71 (new, continuous infection for 200+ days) and 84 (reactivation). Patient 71 showed poor lymphoid recovery, continuous CMV viremia and eventual relapsed malignancy, also developed aGVHD of the skin grade I and also developed mild cGVHD of skin. Patient 84 exhibited lymphocytosis following CMV reactivation events and eventually developed acute recurrent gut and skin grade IV GVHD. This patient also developed moderate cGVHD of gut, skin and liver. Both patients received tacrolimus.
(TIF)
(TIF)
Acknowledgments
This study conducted at Virginia Commonwealth University’s Massey Cancer Center was supported, in part, by research funding from the NIH-NCI Cancer Center Support Grant (P30-CA016059) to Amir Toor and by research funding from Virginia’s Commonwealth Health Research Board Grant #236-11-13 to Michael Neale. Sequencing and Bioinformatics Analysis was performed in the Genomics Core of the Nucleic Acids Research Facilities at VCU. We appreciate the critical review of our work provided by Jeremy Meier, VCU student MD-PhD program, and all others who supported this work in some way.
Data Availability
Human CMV accessions from the NCBI protein database (with full accession numbers and sequences) used to determine amino acid sequence homology with patient alloreactive peptide libraries (Previously published with VCU MCC-14012 Whole Exome Sequencing Study) for cross-reactivity potential determination were listed in S1 File. All raw patient data necessary to analyze the resulting peptide matches between CMV & patient peptide libraries were de-identified and compiled into S2 File with sorting based on the screening parameters set for the HLA-specific peptide binding affinity, IC50<500 nM, using netMHCpan version 2.8. These binding data were acquired for use in the bioinformatics pipeline from IEDB as previously described. GVHD tissue-specific peptide expression data was obtained from GTEx through third-party public download for CMV-reactivation cross-sectional analyses. The predictive binding affinity classification along with the data from the Gene-Tissue Expression Database (GTEx) were used to sort patient data by GVHD-tissue for statistical correlation and precluded patient alloreactive peptide library publication in the data repositories, dbSNP/ClinVar, per their own specifications. All other previously published alloreactivity data from the earlier VCU Whole Exome Sequencing Study MCC-14012 may be available upon request by contacting: amir.toor@vcuhealth.org.
Funding Statement
This study conducted at Virginia Commonwealth University’s Massey Cancer Center was supported, in part, by research funding from the NIH-NCI Cancer Center Support Grant (P30-CA016059, Director, Gordon Ginder) to Amir Toor in the form of a Massey Pilot Project (https://projectreporter.nih.gov/reporter_SearchResults.cfm?icde=31442271) and by research funding from Virginia’s Commonwealth Health Research Board Grant #236-11-13 to Michael Neale (http://www.chrb.org/linked_documents/2014%20CHRB%20Grant%20Awards%20and%20Recipients.pdf). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ljungman P, Hakki M, Boeckh M. Cytomegalovirus in hematopoietic stem cell transplant recipients. Hematol Oncol Clin North Am 2011. February;25(1):151–169. doi: 10.1016/j.hoc.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teira P, Battiwalla M, Ramanathan M, Barrett AJ, Ahn KW, Chen M, et al. Early cytomegalovirus reactivation remains associated with increased transplant-related mortality in the current era: a CIBMTR analysis. Blood 2016. May 19;127(20):2427–2438. doi: 10.1182/blood-2015-11-679639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.U.S. National Library of Medicine. Cytomegalovirus Infections. 2016; https://www.nlm.nih.gov/medlineplus/cytomegalovirusinfections.html. Accessed July 15, 2016.
- 4.Gotoh M, Yoshizawa S, Katagiri S, Suguro T, Asano M, Kitahara T, et al. Human herpesvirus 6 reactivation on the 30th day after allogeneic hematopoietic stem cell transplantation can predict grade 2–4 acute graft-versus-host disease. Transpl Infect Dis 2014. June;16(3):440–449. doi: 10.1111/tid.12229 [DOI] [PubMed] [Google Scholar]
- 5.Jaskula E, Bochenska J, Kocwin E, Tarnowska A, Lange A. CMV Serostatus of Donor-Recipient Pairs Influences the Risk of CMV Infection/Reactivation in HSCT Patients. Bone Marrow Res 2012;2012:375075 doi: 10.1155/2012/375075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaskula E, Dlubek D, Sedzimirska M, Duda D, Tarnowska A, Lange A. Reactivations of cytomegalovirus, human herpes virus 6, and Epstein-Barr virus differ with respect to risk factors and clinical outcome after hematopoietic stem cell transplantation. Transplant Proc 2010. October;42(8):3273–3276. doi: 10.1016/j.transproceed.2010.07.027 [DOI] [PubMed] [Google Scholar]
- 7.Wang LR, Dong LJ, Zhang MJ, Lu DP. Correlations of human herpesvirus 6B and CMV infection with acute GVHD in recipients of allogeneic haematopoietic stem cell transplantation. Bone Marrow Transplant 2008. November;42(10):673–677. doi: 10.1038/bmt.2008.238 [DOI] [PubMed] [Google Scholar]
- 8.Miller W, Flynn P, McCullough J, Balfour HH Jr, Goldman A, Haake R, et al. Cytomegalovirus infection after bone marrow transplantation: an association with acute graft-v-host disease. Blood 1986. April;67(4):1162–1167. [PubMed] [Google Scholar]
- 9.Varani S, Landini MP. Cytomegalovirus-induced immunopathology and its clinical consequences. Herpesviridae 2011. April 7;2(1):6-4280-2-6. doi: 10.1186/2042-4280-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ljungman P. Immune reconstitution and viral infections after stem cell transplantation. Bone Marrow Transplant 1998. April;21 Suppl 2:S72–4. [PubMed] [Google Scholar]
- 11.Ljungman P, Perez-Bercoff L, Jonsson J, Avetisyan G, Sparrelid E, Aschan J, et al. Risk factors for the development of cytomegalovirus disease after allogeneic stem cell transplantation. Haematologica 2006. January;91(1):78–83. [PubMed] [Google Scholar]
- 12.Cantoni N, Hirsch HH, Khanna N, Gerull S, Buser A, Bucher C, et al. Evidence for a bidirectional relationship between cytomegalovirus replication and acute graft-versus-host disease. Biol Blood Marrow Transplant 2010. September;16(9):1309–1314. doi: 10.1016/j.bbmt.2010.03.020 [DOI] [PubMed] [Google Scholar]
- 13.Gimenez E, Solano C, Pinana JL, Amat P, Vinuesa V, Navarro D. Re-examining the relationship between active cytomegalovirus (CMV) infection and acute graft-versus-host disease in allogeneic stem cell transplant recipients in the era of real-time PCR CMV assays. Transpl Int 2016. January;29(1):126–128. doi: 10.1111/tri.12689 [DOI] [PubMed] [Google Scholar]
- 14.Sampson JK, Sheth NU, Koparde VN, Scalora AF, Serrano MG, Lee V, et al. Whole exome sequencing to estimate alloreactivity potential between donors and recipients in stem cell transplantation. Br J Haematol 2014. August;166(4):566–570. doi: 10.1111/bjh.12898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jameson-Lee M, Koparde V, Griffith P, Scalora AF, Sampson JK, Khalid H, et al. In silico Derivation of HLA-Specific Alloreactivity Potential from Whole Exome Sequencing of Stem-Cell Transplant Donors and Recipients: Understanding the Quantitative Immunobiology of Allogeneic Transplantation. Front Immunol 2014. November 6;5:529 doi: 10.3389/fimmu.2014.00529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mason D. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol Today 1998. September;19(9):395–404. [DOI] [PubMed] [Google Scholar]
- 17.Regner M. Cross-reactivity in T-cell antigen recognition. Immunol Cell Biol 2001. April;79(2):91–100. doi: 10.1046/j.1440-1711.2001.00994.x [DOI] [PubMed] [Google Scholar]
- 18.Hiemstra HS, Schloot NC, van Veelen PA, Willemen SJ, Franken KL, van Rood JJ, et al. Cytomegalovirus in autoimmunity: T cell crossreactivity to viral antigen and autoantigen glutamic acid decarboxylase. Proc Natl Acad Sci U S A 2001. March 27;98(7):3988–3991. doi: 10.1073/pnas.071050898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sewell AK. Why must T cells be cross-reactive? Nat Rev Immunol 2012. September;12(9):669–677. doi: 10.1038/nri3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen TH, Rowntree LC, Pellicci DG, Bird NL, Handel A, Kjer-Nielsen L, et al. Recognition of distinct cross-reactive virus-specific CD8+ T cells reveals a unique TCR signature in a clinical setting. J Immunol 2014. June 1;192(11):5039–5049. doi: 10.4049/jimmunol.1303147 [DOI] [PubMed] [Google Scholar]
- 21.Tong CY, Bakran A, Peiris JS, Muir P, Herrington CS. The association of viral infection and chronic allograft nephropathy with graft dysfunction after renal transplantation. Transplantation 2002. August 27;74(4):576–578. [DOI] [PubMed] [Google Scholar]
- 22.Snyder LD, Finlen-Copeland CA, Turbyfill WJ, Howell D, Willner DA, Palmer SM. Cytomegalovirus pneumonitis is a risk for bronchiolitis obliterans syndrome in lung transplantation. Am J Respir Crit Care Med 2010. June 15;181(12):1391–1396. doi: 10.1164/rccm.200911-1786OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheper W, van Dorp S, Kersting S, Pietersma F, Lindemans C, Hol S, et al. gammadeltaT cells elicited by CMV reactivation after allo-SCT cross-recognize CMV and leukemia. Leukemia 2013. June;27(6):1328–1338. [DOI] [PubMed] [Google Scholar]
- 24.Halenius A, Hengel H. Human cytomegalovirus and autoimmune disease. Biomed Res Int 2014;2014:472978 doi: 10.1155/2014/472978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elmaagacli AH, Steckel NK, Koldehoff M, Hegerfeldt Y, Trenschel R, Ditschkowski M, et al. Early human cytomegalovirus replication after transplantation is associated with a decreased relapse risk: evidence for a putative virus-versus-leukemia effect in acute myeloid leukemia patients. Blood 2011. August 4;118(5):1402–1412. doi: 10.1182/blood-2010-08-304121 [DOI] [PubMed] [Google Scholar]
- 26.Barrett AJ. CMV: when bad viruses turn good. Blood 2011. August 4;118(5):1193–1194. doi: 10.1182/blood-2011-06-354340 [DOI] [PubMed] [Google Scholar]
- 27.Crough T, Beagley L, Smith C, Jones L, Walker DG, Khanna R. Ex vivo functional analysis, expansion and adoptive transfer of cytomegalovirus-specific T-cells in patients with glioblastoma multiforme. Immunol Cell Biol 2012. October;90(9):872–880. doi: 10.1038/icb.2012.19 [DOI] [PubMed] [Google Scholar]
- 28.Schuessler A, Smith C, Beagley L, Boyle GM, Rehan S, Matthews K, et al. Autologous T-cell therapy for cytomegalovirus as a consolidative treatment for recurrent glioblastoma. Cancer Res 2014. July 1;74(13):3466–3476. doi: 10.1158/0008-5472.CAN-14-0296 [DOI] [PubMed] [Google Scholar]
- 29.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010. September;38(16):e164 doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.IEDB Analysis Resource. MHC-I binding predictions—Download. 4 APR 2016; http://tools.immuneepitope.org/analyze/html_mhcibinding20090901B/download_mhc_I_binding.html.
- 31.Center for Biological Sequence Analysis, Technical University of Denmark. CBS Software Download—Amino Acid Sequences—Immunological Features—NetMHCpan. 2016; http://www.cbs.dtu.dk/services/NetMHCpan/. Accessed July 15, 2016.
- 32.Armistead PM, Liang S, Li H, Lu S, Van Bergen CA, Alatrash G, et al. Common minor histocompatibility antigen discovery based upon patient clinical outcomes and genomic data. PLoS One 2011;6(8):e23217 doi: 10.1371/journal.pone.0023217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat Biotechnol 2006. July;24(7):817–819. doi: 10.1038/nbt1215 [DOI] [PubMed] [Google Scholar]
- 34.National Center for Biotechnology Information, U.S. National Library of Medicine. Human Cytomegalovirus. http://www.ncbi.nlm.nih.gov/protein/?term=Human+Herpesvirus+5+and+HCMV. Accessed July 15, 2016.
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990. October 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 36.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med 2005. September 5;202(5):673–685. doi: 10.1084/jem.20050882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.GTEx Consortium. Gene Expression. 16 March 2016; http://gtexportal.org/home/gene/. Accessed July 15, 2016.
- 38.Toor AA, Sabo RT, Roberts CH, Moore BL, Salman SR, Scalora AF, et al. Dynamical System Modeling of Immune Reconstitution after Allogeneic Stem Cell Transplantation Identifies Patients at Risk for Adverse Outcomes. Biol Blood Marrow Transplant 2015. July;21(7):1237–1245. doi: 10.1016/j.bbmt.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdul Razzaq B, Scalora A, Koparde VN, Meier J, Mahmood M, Salman S, et al. Dynamical System Modeling to Simulate Donor T Cell Response to Whole Exome Sequencing-Derived Recipient Peptides Demonstrates Different Alloreactivity Potential in HLA-Matched and -Mismatched Donor-Recipient Pairs. Biol Blood Marrow Transplant 2016. May;22(5):850–861. doi: 10.1016/j.bbmt.2015.11.1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Motozono C, Miles JJ, Hasan Z, Gatanaga H, Meribe SC, Price DA, et al. CD8(+) T Cell Cross-Reactivity Profiles and HIV-1 Immune Escape towards an HLA-B35-Restricted Immunodominant Nef Epitope. PLoS One 2013;8(6):e66152 doi: 10.1371/journal.pone.0066152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Motozono C, Yokoyama M, Sato H, Ueno T. Cross-reactivity analysis of T cell receptors specific for overlapping HIV-1 Nef epitopes of different lengths. Microbes Infect 2014. April;16(4):320–327. doi: 10.1016/j.micinf.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 42.Derre L, Bruyninx M, Baumgaertner P, Ferber M, Schmid D, Leimgruber A, et al. Distinct sets of alphabeta TCRs confer similar recognition of tumor antigen NY-ESO-1157-165 by interacting with its central Met/Trp residues. Proc Natl Acad Sci U S A 2008. September 30;105(39):15010–15015. doi: 10.1073/pnas.0807954105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borbulevych OY, Insaidoo FK, Baxter TK, Powell DJ Jr, Johnson LA, Restifo NP, et al. Structures of MART-126/27-35 Peptide/HLA-A2 complexes reveal a remarkable disconnect between antigen structural homology and T cell recognition. J Mol Biol 2007. October 5;372(5):1123–1136. doi: 10.1016/j.jmb.2007.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hemmer B, Vergelli M, Gran B, Ling N, Conlon P, Pinilla C, et al. Predictable TCR antigen recognition based on peptide scans leads to the identification of agonist ligands with no sequence homology. J Immunol 1998. April 15;160(8):3631–3636. [PubMed] [Google Scholar]
- 45.Hiemstra HS, van Veelen PA, Schloot NC, Geluk A, van Meijgaarden KE, Willemen SJ, et al. Definition of natural T cell antigens with mimicry epitopes obtained from dedicated synthetic peptide libraries. J Immunol 1998. October 15;161(8):4078–4082. [PubMed] [Google Scholar]
- 46.Hiemstra HS, van Veelen PA, Willemen SJ, Benckhuijsen WE, Geluk A, de Vries RR, et al. Quantitative determination of TCR cross-reactivity using peptide libraries and protein databases. Eur J Immunol 1999. August;29(8):2385–2391. [DOI] [PubMed] [Google Scholar]
- 47.Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell 2014. May 22;157(5):1073–1087. doi: 10.1016/j.cell.2014.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peters HL, Tripathi SC, Kerros C, Katayama H, Garber HR, St John LS, et al. Serine Proteases Enhance Immunogenic Antigen Presentation on Lung Cancer Cells. Cancer Immunol Res 2017. March 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abelin JG, Keskin DB, Sarkizova S, Hartigan CR, Zhang W, Sidney J, et al. Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-allelic Cells Enables More Accurate Epitope Prediction. Immunity 2017. February 21;46(2):315–326. doi: 10.1016/j.immuni.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ, Mann M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol Cell Proteomics 2015. March;14(3):658–673. doi: 10.1074/mcp.M114.042812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amir AL, van der Steen DM, Hagedoorn RS, Kester MG, van Bergen CA, Drijfhout JW, et al. Allo-HLA-reactive T cells inducing graft-versus-host disease are single peptide specific. Blood 2011. December 22;118(26):6733–6742. doi: 10.1182/blood-2011-05-354787 [DOI] [PubMed] [Google Scholar]
- 52.Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet 2009. May 2;373(9674):1550–1561. doi: 10.1016/S0140-6736(09)60237-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackson SE, Mason GM, Wills MR. Human cytomegalovirus immunity and immune evasion. Virus Res 2011. May;157(2):151–160. doi: 10.1016/j.virusres.2010.10.031 [DOI] [PubMed] [Google Scholar]
- 54.Krenger W, Blazar BR, Hollander GA. Thymic T-cell development in allogeneic stem cell transplantation. Blood 2011. June 23;117(25):6768–6776. doi: 10.1182/blood-2011-02-334623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suessmuth Y, Mukherjee R, Watkins B, Koura DT, Finstermeier K, Desmarais C, et al. CMV reactivation drives posttransplant T-cell reconstitution and results in defects in the underlying TCRbeta repertoire. Blood 2015. June 18;125(25):3835–3850. doi: 10.1182/blood-2015-03-631853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paine A, Oelke M, Tischer S, Heuft HG, Blasczyk R, Eiz-Vesper B. Soluble recombinant CMVpp65 spanning multiple HLA alleles for reconstitution of antiviral CD4+ and CD8+ T-cell responses after allogeneic stem cell transplantation. J Immunother 2010. January;33(1):60–72. doi: 10.1097/CJI.0b013e3181b56dcc [DOI] [PubMed] [Google Scholar]
- 57.Delogu LG, Deidda S, Delitala G, Manetti R. Infectious diseases and autoimmunity. J Infect Dev Ctries 2011. October 13;5(10):679–687. [DOI] [PubMed] [Google Scholar]
- 58.Vanheusden M, Stinissen P, 't Hart BA, Hellings N. Cytomegalovirus: a culprit or protector in multiple sclerosis? Trends Mol Med 2015. January;21(1):16–23. doi: 10.1016/j.molmed.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 59.Sijmons S, Van Ranst M, Maes P. Genomic and functional characteristics of human cytomegalovirus revealed by next-generation sequencing. Viruses 2014. March 5;6(3):1049–1072. doi: 10.3390/v6031049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scrivano L, Sinzger C, Nitschko H, Koszinowski UH, Adler B. HCMV spread and cell tropism are determined by distinct virus populations. PLoS Pathog 2011. January 13;7(1):e1001256 doi: 10.1371/journal.ppat.1001256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elkington R, Walker S, Crough T, Menzies M, Tellam J, Bharadwaj M, et al. Ex vivo profiling of CD8+-T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers. J Virol 2003. May;77(9):5226–5240. doi: 10.1128/JVI.77.9.5226-5240.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.George B, Kerridge IH, Gilroy N, Huang G, Hertzberg MS, Bradstock KF, et al. A risk score for early cytomegalovirus reactivation after allogeneic stem cell transplantation identifies low-, intermediate-, and high-risk groups: reactivation risk is increased by graft-versus-host disease only in the intermediate-risk group. Transpl Infect Dis 2012. April;14(2):141–148. doi: 10.1111/j.1399-3062.2011.00706.x [DOI] [PubMed] [Google Scholar]
- 63.Manjappa S, Bhamidipati PK, Stokerl-Goldstein KE, DiPersio JF, Uy GL, Westervelt P, et al. Protective effect of cytomegalovirus reactivation on relapse after allogeneic hematopoietic cell transplantation in acute myeloid leukemia patients is influenced by conditioning regimen. Biol Blood Marrow Transplant 2014. January;20(1):46–52. doi: 10.1016/j.bbmt.2013.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bao X, Zhu Q, Xue S, Hu X, Ma X, Chen F, et al. Cytomegalovirus induces strong antileukemic effect in acute myeloid leukemia patients following sibling HSCT without ATG-containing regimen. Am J Transl Res 2016. February 15;8(2):653–661. [PMC free article] [PubMed] [Google Scholar]
- 65.Takenaka K, Nishida T, Asano-Mori Y, Oshima K, Ohashi K, Mori T, et al. Cytomegalovirus Reactivation after Allogeneic Hematopoietic Stem Cell Transplantation is Associated with a Reduced Risk of Relapse in Patients with Acute Myeloid Leukemia Who Survived to Day 100 after Transplantation: The Japan Society for Hematopoietic Cell Transplantation Transplantation-related Complication Working Group. Biol Blood Marrow Transplant 2015. November;21(11):2008–2016. doi: 10.1016/j.bbmt.2015.07.019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CMV CROSS database tools excel spreadsheet.
- Sheet 1. CMV proteome—NCBI accessions: hCMV/HHV5
- Sheet 2. CMV immunogenic protein—CD8+ list from Ref. #36.
(XLSX)
Match analysis excel spreadsheet.
- Sheet 1. Raw CMV match algorithm results—pre-IC50 screening
- Sheet 2. Match analysis summary—total peptide # by patient
- Sheet 3. Match analysis peptides—screened, sorted pt. peptides
- Sheet 4. All CMV case studies, tables, and figures
- Sheet 5. Peptide-HLA binding patterns sorted by CMV+GVH
- Sheet 6. Exome dataset—retrospective, de-identified info.
(XLSX)
Total number of Human—CMV sequence homologous peptides without binding affinity filtering by IC50 values for TCR relevance. Range of IC50 values: 0.01–50000 nM. Averaged number of total matches for MRD DRP (Left, Blue) versus MUD DRP (Right, Green) were displayed above the graph to indicate the observed difference of CMV peptide matches to alloreactive peptides prior to screening by donor type.
(TIFF)
Average number of HLA bound human—CMV sequence homologous peptides ordered by number of peptides presented by HLA locus. Descending order of average matches per HLA (>5 matches per HLA as threshold). Error bars indicated where HLA molecules were shared by more than one DRP and matches averaged.
(TIF)
Binding affinity differences indicated by inverse IC50 values from matched human and CMV peptides bound to the same HLA. Each data point represents the intersection of a matched peptide bound to an HLA class I molecule (CMV→Human). These peptides may be cross reactive, with varying degrees of T cell cross reactivity potential for alloreactivity trigger to ensue (towards the origin on the human peptide axis being the greatest).
(TIF)
GVHD-independent effects of CMV post SCT in CMV seropositive DRP (n = 46).
(TIF)
Cumulative effects of CMV viremia + GVHD post SCT in CMV-seropositive DRP (n = 46).
(TIF)
(Top) Lymphoid Recovery Patterns by ALC value achieved by Day 45 post SCT. Pattern A showed early lymphoid recovery (Patient 2), Pattern B showed a medium time to lymphoid recovery (Patient 48) and Pattern C showed late or very low lymphoid recovery (Patient 16). Trendlines indicated the fit to the logistic pattern previously observed. (Bottom) Lymphoid Recovery Patterns by CMV+GVHD Status (n = 77): CMV Reactivation triggers Lymphocytosis that may influence GVHD onset. Fisher’s Exact Chi Square compared CMV + GVHD groups with lymphoid recovery patterns as shown.
(TIF)
CMV reactivation with or without GVHD onset affects the survival negatively compared to the protective GVHD effect.
(TIFF)
(A) CMV-reactivation course with acute GVHD onset and progression: Patients 47 and 68. Patient 47, grade IV GVHD of the GI tract; patient 68,steroid refractory grade IV GI tract GVHD, skin and liver. Both patients showed signs of CMV reactivation and bursts of lymphocytosis prior to GVHD onset during stable immunosuppression (TAC, Tacrolimus; SIR, Sirolimus; Cyclosporin) as measured by serum levels or following taper. (B) CMV-reactivation with chronic GVHD onset: Patients 10 and 27. Patient 10 had CMV reactivation prior to a gradual lymphocyte proliferation during stable immunosuppression levels (TAC, Tacrolimus) and eventually developed relapsed malignancy. Patient 27 exhibited a CMV reactivation event prior to lymphocytosis at stable tacrolimus levels, and had mainly skin and oral GVHD. (C) CMV reactivation/de novo infection in patients with aGVHD + cGVHD: Patients 71 (new, continuous infection for 200+ days) and 84 (reactivation). Patient 71 showed poor lymphoid recovery, continuous CMV viremia and eventual relapsed malignancy, also developed aGVHD of the skin grade I and also developed mild cGVHD of skin. Patient 84 exhibited lymphocytosis following CMV reactivation events and eventually developed acute recurrent gut and skin grade IV GVHD. This patient also developed moderate cGVHD of gut, skin and liver. Both patients received tacrolimus.
(TIF)
(TIF)
Data Availability Statement
Human CMV accessions from the NCBI protein database (with full accession numbers and sequences) used to determine amino acid sequence homology with patient alloreactive peptide libraries (Previously published with VCU MCC-14012 Whole Exome Sequencing Study) for cross-reactivity potential determination were listed in S1 File. All raw patient data necessary to analyze the resulting peptide matches between CMV & patient peptide libraries were de-identified and compiled into S2 File with sorting based on the screening parameters set for the HLA-specific peptide binding affinity, IC50<500 nM, using netMHCpan version 2.8. These binding data were acquired for use in the bioinformatics pipeline from IEDB as previously described. GVHD tissue-specific peptide expression data was obtained from GTEx through third-party public download for CMV-reactivation cross-sectional analyses. The predictive binding affinity classification along with the data from the Gene-Tissue Expression Database (GTEx) were used to sort patient data by GVHD-tissue for statistical correlation and precluded patient alloreactive peptide library publication in the data repositories, dbSNP/ClinVar, per their own specifications. All other previously published alloreactivity data from the earlier VCU Whole Exome Sequencing Study MCC-14012 may be available upon request by contacting: amir.toor@vcuhealth.org.