Abstract
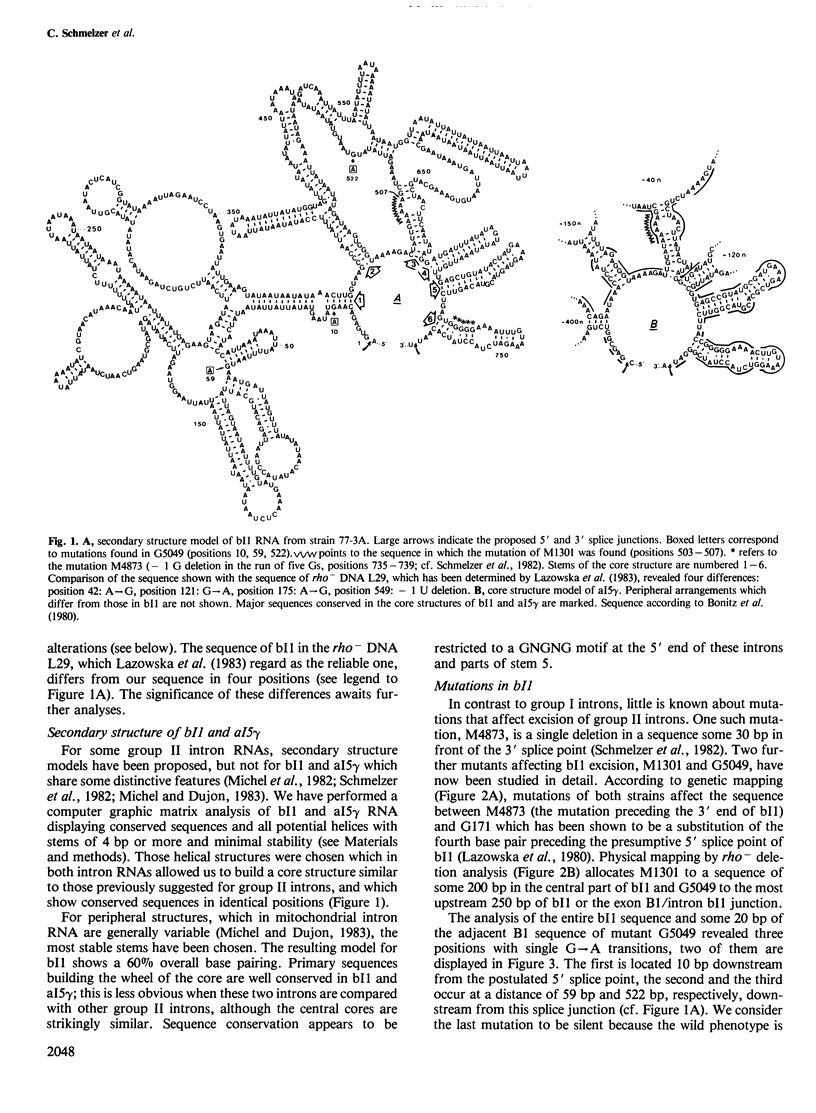
The sequence of intron 1 in the cob gene in mtDNA (bI1) of the yeast strain 777-3A has been determined. Furthermore, we have performed a systematic search for complementary sequence stretches within this intron RNA, and within the RNA of intron 5 gamma of the oxi3 gene (aI5 gamma) which shares distinctive sequences with bI1. Possible secondary structure models derived from this analysis show nearly identical core structures for bI1 and aI5 gamma RNA with conserved sequence stretches in prominent positions. These core structures are similar to those previously reported for RNAs of introns having very limited sequence homology with bI1 and aI5 gamma. In two mutants which are defective in bI1 excision from cob pre-mRNA, nucleotide sequence alterations in bI1 have been determined. One mutation (G5049) apparently affects the stability of a hybrid stretch in the proposed secondary structure of bI1 RNA whereas the other one (M1301), a deletion of one A in a run of five As, affects a sequence which is conserved in bI1 and aI5 gamma and is involved in the formation of a distinct secondary structure. Out of seven revertants of M1301, three were found to have restored the wild-type bI1 sequence AAAAA, three others had the related sequence AAAAG which is functionally indistinguishable from wild-type, whereas one revertant had a nuclear mutation which suppresses the splicing defect exerted by the mitochondrial mutation M1301. This nuclear suppressor (SUP-101) is allele specific and dominant. The possible role of the sequence affected by M1301 in terms of a recognition site for a nuclear gene product will be discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anziano P. Q., Hanson D. K., Mahler H. R., Perlman P. S. Functional domains in introns: trans-acting and cis-acting regions of intron 4 of the cob gene. Cell. 1982 Oct;30(3):925–932. doi: 10.1016/0092-8674(82)90297-5. [DOI] [PubMed] [Google Scholar]
- Arnberg A. C., Van Ommen G. J., Grivell L. A., Van Bruggen E. F., Borst P. Some yeast mitochondrial RNAs are circular. Cell. 1980 Feb;19(2):313–319. doi: 10.1016/0092-8674(80)90505-x. [DOI] [PubMed] [Google Scholar]
- Bechmann H., Haid A., Schweyen R. J., Mathews S., Kaudewitz F. Expression of the "split gene" COB in yeast mtDNA. Translation of intervening sequences in mutant strains. J Biol Chem. 1981 Apr 10;256(7):3525–3531. [PubMed] [Google Scholar]
- Bonitz S. G., Coruzzi G., Thalenfeld B. E., Tzagoloff A., Macino G. Assembly of the mitochondrial membrane system. Structure and nucleotide sequence of the gene coding for subunit 1 of yeast cytochrme oxidase. J Biol Chem. 1980 Dec 25;255(24):11927–11941. [PubMed] [Google Scholar]
- Breathnach R., Benoist C., O'Hare K., Gannon F., Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde J., Fink G. R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- De La Salle H., Jacq C., Slonimski P. P. Critical sequences within mitochondrial introns: pleiotropic mRNA maturase and cis-dominant signals of the box intron controlling reductase and oxidase. Cell. 1982 Apr;28(4):721–732. doi: 10.1016/0092-8674(82)90051-4. [DOI] [PubMed] [Google Scholar]
- Dujardin G., Pajot P., Groudinsky O., Slonimski P. P. Long range control circuits within mitochondria and between nucleus and mitochondria. I. Methodology and phenomenology of suppressors. Mol Gen Genet. 1980;179(3):469–482. doi: 10.1007/BF00271736. [DOI] [PubMed] [Google Scholar]
- Groudinsky O., Dujardin G., Slonimski P. P. Long range control circuits within mitochondria and between nucleus and mitochondria. II. Genetic and biochemical analyses of suppressors which selectively alleviate the mitochondrial intron mutations. Mol Gen Genet. 1981;184(3):493–503. doi: 10.1007/BF00352529. [DOI] [PubMed] [Google Scholar]
- Halbreich A., Pajot P., Foucher M., Grandchamp C., Slonimski P. A pathway of cytochrome b mRNA processing in yeast mitochondria: specific splicing steps and an intron-derived circular DNA. Cell. 1980 Feb;19(2):321–329. doi: 10.1016/0092-8674(80)90506-1. [DOI] [PubMed] [Google Scholar]
- Hensgens L. A., Bonen L., de Haan M., van der Horst G., Grivell L. A. Two intron sequences in yeast mitochondrial COX1 gene: homology among URF-containing introns and strain-dependent variation in flanking exons. Cell. 1983 Feb;32(2):379–389. doi: 10.1016/0092-8674(83)90457-9. [DOI] [PubMed] [Google Scholar]
- Klapholz S., Esposito R. E. A new mapping method employing a meiotic rec-mutant of yeast. Genetics. 1982 Mar;100(3):387–412. doi: 10.1093/genetics/100.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancashire W. E., Mattoon J. R. Cytoduction: a tool for mitochondrial genetic studies in yeast. Utilization of the nuclear-fusion mutation kar 1-1 for transfer of drug r and mit genomes in Saccharomyces cerevisiae. Mol Gen Genet. 1979 Mar 5;170(3):333–344. doi: 10.1007/BF00267067. [DOI] [PubMed] [Google Scholar]
- Lang B., Burger G., Doxiadis I., Thomas D. Y., Bandlow W., Kaudewitz F. A simple method for the large-scale preparation of mitochondria from microorganisms. Anal Biochem. 1977 Jan;77(1):110–121. doi: 10.1016/0003-2697(77)90295-0. [DOI] [PubMed] [Google Scholar]
- Lazowska J., Jacq C., Slonimski P. P. Sequence of introns and flanking exons in wild-type and box3 mutants of cytochrome b reveals an interlaced splicing protein coded by an intron. Cell. 1980 Nov;22(2 Pt 2):333–348. doi: 10.1016/0092-8674(80)90344-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Netter P., Jacq C., Carignani G., Slonimski P. P. Critical sequences within mitochondrial introns: cis-dominant mutations of the "cytochrome-b-like" intron of the oxidase gene. Cell. 1982 Apr;28(4):733–738. doi: 10.1016/0092-8674(82)90052-6. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Tinoco I., Jr, Jacobson A. B. Small changes in free energy assignments for unpaired bases do not affect predicted secondary structures in single stranded RNA. Nucleic Acids Res. 1982 Jan 11;10(1):341–349. doi: 10.1093/nar/10.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer C., Haid A., Grosch G., Schweyen R. J., Kaudewitz F. Pathways of transcript splicing in yeast mitochondria. Mutations in intervening sequences of the split gene COB reveal a requirement for intervening sequence-encoded products. J Biol Chem. 1981 Jul 25;256(14):7610–7619. [PubMed] [Google Scholar]
- Schmelzer C., Schmidt C., Schweyen R. J. Identification of splicing signals in introns of yeast mitochondrial split genes: mutational alterations in intron bI1 and secondary structures in related introns. Nucleic Acids Res. 1982 Nov 11;10(21):6797–6808. doi: 10.1093/nar/10.21.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Uhlenbeck O. C., Levine M. D. Estimation of secondary structure in ribonucleic acids. Nature. 1971 Apr 9;230(5293):362–367. doi: 10.1038/230362a0. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Scazzocchio C., Brown T. A., Davies R. W. Close relationship between certain nuclear and mitochondrial introns. Implications for the mechanism of RNA splicing. J Mol Biol. 1983 Jul 5;167(3):595–605. doi: 10.1016/s0022-2836(83)80100-4. [DOI] [PubMed] [Google Scholar]
- Weiss-Brummer B., Holl J., Schweyen R. J., Rödel G., Kaudewitz F. Processing of yeast mitochondrial RNA: involvement of intramolecular hybrids in splicing of cob intron 4 RNA by mutation and reversion. Cell. 1983 May;33(1):195–202. doi: 10.1016/0092-8674(83)90348-3. [DOI] [PubMed] [Google Scholar]
- Weiss-Brummer B., Rödel G., Schweyen R. J., Kaudewitz F. Expression of the split gene cob in yeast: evidence for a precursor of a "maturase" protein translated from intron 4 and preceding exons. Cell. 1982 Jun;29(2):527–536. doi: 10.1016/0092-8674(82)90169-6. [DOI] [PubMed] [Google Scholar]