Abstract
Gemfibrozil, a peroxisome proliferator-activated receptor α (PPARα) agonist, is widely used for hypertriglyceridemia and mixed hyperlipidemia. Drug-drug interaction of gemfibrozil and other PPARα agonists has been reported. But the role of PPARα in Cytochrome P450 (CYP) induction by fibrates is not well known. In this study, wild-type mice were first fed gemfibrozil-containing diets (0.375%, 0.75% and 1.5%) for 14 days to establish a dose-response relationship for CYP induction. Then wild-type mice and Pparα-null mice were treated with 0.75% gemfibrozil-containing diet for 7 days. CYP3a, CYP2b and CYP2c were induced in a dose-dependent manner by gemfibrozil. In Pparα-null mice, their mRNA level, protein level and activity were induced more than that in wild-type mice. So gemfibrozil induced CYP and this action was inhibited by activated PPARα. These data suggested that the induction potential of CYPs was suppressed by activated PPARα, showing a potential role of this receptor in drug-drug interactions and metabolic diseases treated with fibrates.
Keywords: PPARα, gemfibrozil, cytochrome P450, suppression
Introduction
Gemfibrozil belongs to a group of agents known as fibrate drugs which are prescribed primarily to treat hypertriglyceridemia and mixed hyperlipidemia associated with atherosclerosis [1]. Fibrate drugs promote the catabolism of triglyceride and very low-density lipoprotein, reduces the risk of cardiovascular events. Pharmacologically, they activate the peroxisome proliferator-activated receptor α (PPARα), alter adipokine release, and modulate glucose homeostasis [2, 3]. However, drug-drug interaction associated with gemfibrozil has raised many concerns in recent years.
Drug-drug interaction between gemfibrozil and statins was investigated to explore the mechanism of myotoxicity which led to the withdrawal of cerivastatin [4]. Gemfibrozil potently inhibits CYP2C9, and its metabolite gemfibrozil-1-O-glu is a mechanism-dependent inhibitor of CYP2C8 which is important for the oxidation of cerivastatin [5]. Additionally, CYP3A4, which mediates metabolism of cerivastatin, simvastatin and atorvastatin, could be inhibited by both gemfibrozil and gemfibrozil-1-O-glu [6]. Gemfibrozil also inhibits UDP-glucuronyltransferases (UGT) 1A1 and 1A3, which are responsible for the glucuronidation and elimination of statins [7]. Additionally, the uptake of cerivastatin by the hepatocyte transporter organic anion transporting polypeptide 1A4 was inhibited by both gemfibrozil and gemfibrozil-1-O-glu [6]. Based on the inhibitory actions, gemfibrozil was suggested as a CYP2C8 inhibitor by the U.S. Food and Drug Administration and the European Medicine Agency [8].
Besides inhibition of xenobiotic metabolism, the inductive potential of gemfibrozil and other PPARα agonists for CYPs and UGTs have been evaluated. In HepG2 cells, PPARα agonist bezafibrate induces CYP2C8 mRNA and protein levels in a time-dependent manner [9]. In Sprague-Dawley rats, CYP3A1 and CYP3A2, and CYP2C6 activities were induced (32–77%) by gemfibrozil when the exposure concentration was in the clinical range [10]. In primary human hepatocytes, gemfibrozil induced CYP3A4 and CYP2C8 without pregnane X-receptor (PXR) activation [11]. In humanized mice, induction of CYP3A4 mRNA and protein by WY14,643 was verified [12]. However, little is known about PPARα’s role for CYP induction potential by its agonists.
PPARα regulates genes involved in the metabolism of endogenous and exogenous substrates. PPARα also mediates the up-regulation of mRNA encoding CYP7A1 and CYP8B1 involved in bile acid synthesis and homeostasis [13, 14]. In the dextran sulfate sodium-induced mouse colitis model, activation of PPARα-UGTs axis accelerated the elimination of intestinal bile acids, resulting in suppression of farnesoid X receptor (FXR)-FGF15 signaling. The subsequent up-regulation of hepatic CYP7A1 increased de novo bile acid synthesis [15]. PPARα agonist perfluorodecanoic acid (PFDA), an environmental pollutant, was found to increase expression of Cyp2b10 (20-fold), 3a11 (2-fold), and 4a14 (32-fold) mRNAs in wild type but not Pparα-null mice [16]. Both the disruptive and protective action of PPARα in PFDA induced hepatotoxicity was revealed recently [17]. These data indicated pleiotropic role of PPARα in liver metabolism and function. But a suppressive effect of activated PPARα on CYP induction potential is not reported.
In the present study, the dose-dependent induction of CYPs by the PPARα agonist gemfibrozil was investigated. Then Pparα-null mice fed a 0.75% gemfibrozil-containing diet were used to reveal a suppressive role of PPARα for induction potential of CYPs by gemfibrozil.
Materials and Methods
Chemicals and reagents
Gemfibrozil was obtained from Hunan Qianjin Xiangjiang Pharmaceutical Co. Ltd (Zhuzhou, China). Trizol was obtained from Invitrogen (CA, USA). The reverse transcription kit was purchased from Thermo Fisher (PA, USA). The LightCycle 480 SYBR Green I Master Mix was purchased from Roche. Antibodies against CYP3A (sc-30621) and CYP2B1/2 (sc-73546) were products by Santa Cruz Biotechnologies (CA, USA). Antibody against GAPDH (ab181602), CYP2C (ab137015) were obtained from Abcam (Cambridge, UK). Midazolam, testosterone, pentoxyresorufin, diclofenac were purchased from Sigma-Aldrich (Sigma-Aldrich, St. Louis, MO, USA). 1′-hydroxy-midazolam (1-OH-M), 6β-hydroxyl-testosterone (6β-OH-T), resorufin (Reso), and 4-hydroxy-diclofenac (4-OH-D) were obtained from Toronto Research Chemical Inc. (Toronto, ON, Canada). Purified water was freshly prepared using a Millipore Elix (Bedford, USA). All the other chemicals were of the highest grade from commercial sources.
Animals and treatment
The Pparα-null mice used in this study were described earlier [18]. Wild-type and Pparα-null mice are on the 129/Sv genetic background. The animal protocols were reviewed and approved by the Institutional Laboratory Animal Care and Use Committee of the Medical School at Ningbo University and the National Cancer Institute Animal Care and Use Committee. Wild-type and Pparα-null mice were maintained under a standard 12 h light/12 h dark cycle with free access to water and a commercial diet. Male, 5- to 7-week mice were acclimated to a commercial purified diet for 7 days before the experiment. Three groups of wild-type mice (n = 5 in each group) were fed with 0.375%, 0.75%, and 1.5% gemfibrozil diet (g/g) respectively for 14 days. This dosage was designed based on published data with close relevance to clinical regime [13]. Considering the dose-response relationship observed in the wild-type mice, a new set of wild-type and Pparα-null mice with the same background as the above were fed a 0.75% gemfibrozil-containing diet only for 7 days. The control groups for both mouse lines were treated with purified diet only. At the end of each experiment, all mice were weighed and killed by CO2 overdose and blood was collected for analysis. Liver tissues harvested were flash frozen in liquid nitrogen and stored at −80°C pending analysis.
Quantitative polymerase chain reaction (Q-PCR) analysis
Total RNA from mouse hepatic tissues homogenized in Trizol reagent was determined by Multiskan Go (Thermofisher, USA). The reverse transcription system (20 μL) included the following items: 5 × Reaction buffer 4 μL, total mRNA 1 μg, Oligo dT18 1 μL, Random primer 1 μL, 10 mM dNTPs Mix 2 μL, 200 U RevertAid M-MuLV RT 1μL and 20 U Ribolock RNase Inhibitor 1 μL. The synthesized cDNA was stored at −20°C and subjected to analysis within 7 days. The primer sequences, listed in Table 1, were extracted from http://mouseprimerdepot.nci.nih.gov/. Each 10 μL reaction contained total cDNA 1 μL, LightCycle 480 SYBR Green I Master Mix (FastStartTaq DNA Polymerase, reaction buffer, dNTPmix, SYBR Green I dye, and MgCl2) 5 μL, forward primer 0.2 μL, reverse primer 0.2 μL, nuclease-free water 3.6 μL. Amplification was performed using 40 reaction cycles of 95°C 10 s, 60°C 10 s, and then 72°C 15 s. The fluorescence signal was detected at the end of each cycle. All mRNA expression levels were normalized by transcription of internal standard 18S rRNA and expressed as a ratio compared with that of the control group which was arbitrarily set as 1 in each experiment. The classical function 2−ΔΔCt was used throughout all the qPCR data analysis.
Table 1.
Sequences of the qPCR primers used in this study
| Genes | Forward primer | Reverse primer | Product(bp) |
|---|---|---|---|
| 18S rRNA | ATTGGAGCTGGAATTACCGC | CGGCTACCACATCCAAGGAA | 102 |
| Cyp3a11 | AGCAGGGATGGACCTGG | CGGTAGAGGAGCACCAA | 109 |
| Cyp2b10 | CACACAGCATAACCACAGGC | TGCAGATGGACAGAGGAGG | 108 |
| Cyp2c29 | TTTTTCAGCCATTGGAAAGC | TGGGCTCAAAGCCTACTGTC | 109 |
| Cyp2c37 | TCATGCAGCACTGATGACAG | AACAGCCTTCCCCATGAAGT | 101 |
| Cyp2c39 | AGCTCAGAATGAATGTGGGG | AAGGAACATTGAGGACCGTG | 101 |
| Cyp4a10 | GATGGACGCTCTTTACCCAAC | AAGGGTCAAACACCTCTGGA | 100 |
| Ehhadh | TATGATCCGCCTCTGCAA | TGGCTCTAACCGTATGGTCCGG | 108 |
| Acot1 | CTGGCGCATGCAGGATC | CACTTTTCTTGGATAGCTCC | 107 |
Western blot analysis
Liver tissues were homogenized with a MagNALyser (Roche, USA) using RIPA buffer (1:10, g/v) containing 1% PMSF (Shanghai, China). Tissue debris was removed by centrifugation at 10,000 g and 4°C for 5 min. Total protein was quantified using a BCA protein assay kit from Beyotime Biotech Co., Ltd (Nantong, China). An equivalent volume of 5 x SDS-PAGE sample loading buffer (Shanghai, China) was added to the tubes that were boiled for 5 min. The samples were loaded and separated on 10% SDS-polyacrylamide electrophoresis gels. The samples were transblotted onto PVDF membranes blocked with 5% fat-free milk at 37°C for 2 h. Membranes were incubated overnight with primary antibodies against CYP3A, CYP2B, CYP2C and GAPDH. After a second antibody incubation for 2 h, the blotted membranes were exposed to ECL substrates (Advansta, USA) and the signals were detected by Tanon 4200SF (Shanghai, China).
Assessment of induced CYP activities
Liver homogenate from 300 mg liver tissues were prepared in PB (0.1 M, 1 mM EDTA, 0.9% NaCl, pH 7.4) and centrifuged at 9, 000 g for 30 min at 4 °C. The supernatant was spun at 100, 000 g for 60 min at 4 °C and then discarded. The microsomal preparations were re-suspended in ice-coolled PB (0.1 M, 1 mM EDTA, 0.25M D-(+)-Sucrose, pH 7.4). Protein concentration was determined using a BCA protein assay kit. Midazolam 1′-hydroxylation, testosterone 6β-hydroxylation were used as the marker reactions of CYP3A. Diclofenac 4-hydroxylation and pentoxyresorufin O-deethylation were used as marker reactions for CYP2C and CYP2B activity respectively. To determine the maximal activity, the substrate concentrations were 4 Km based on human microsome data reported earlier [19]. The incubation was performed similarly as previously reported [10]. The reactions were terminated by the addition of 200 mL ice-cold acetonitrile with 0.04 mg/mL bezafibrate as the internal standard. The ion transitions m/z 342.1/324.6 for 1′-hydroxy-midazolam, m/z 305.3/269.2 for 6β-hydroxyl-testosterone, m/z 312.1/265.8 for 4-hydroxy-diclofenac and m/z 213.9/186.0 for resorufin were determined using an ABI 3000 triple quadrupole mass spectrometer (Applied Biosystems, USA) coupled with HPLC (SHIMADZU 10A, Japan) and an MPS3C autosampler (GERSTEL, Germany) controlled by Analyst 1.4.2 workstation software (Applied Biosystems Biosystems, Foster City, USA). Relative abundance of the produced metabolites compared with internal standard was used to evaluate the CYP activities.
Statistical analysis
All the experimental values were expressed as mean ± SD. Statistical analysis was performed using SPSS 13.0. One-way ANOVA, followed by Dunnett-t post hoc tests, was used for multiple comparisons of data from wild-type mice dosed with three different gemfibrozil diets. The results following treatment in both wild-type and Pparα-null mice were analyzed using a two-tailed, independent-sample t-test. The difference was considered significant when the P values were less than 0.05 (P< 0.05)
Results
Dose-response of CYP induction by gemfibrozil
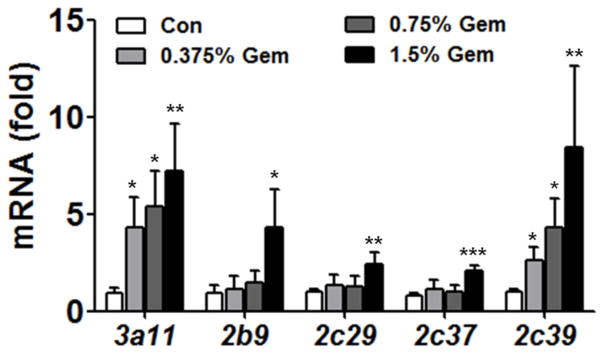
Messenger RNA, protein expression and metabolic activity were assayed to evaluate the dose-response relationship of the CYP induction by gemfibrozil. Cyp3a11 mRNA level was elevated 4.7-, 5.9- and 8-fold after 0.375%, 0.75% and 1.5% gemfibrozil respectively compared with control mice, showing Cyp3a11 mRNA induction was dose-dependent on gemfibrozil. A similar tendency was also observed for Cyp2c39 where the mRNA increased by 2.6-, 4.4- and 8.5-fold respectively after increasing doses of gemfibrozil. However, induction of Cyp2b10, Cyp2c29 and Cy2c37 was only observed in the 1.5% gemfibrozil group (4.5-, 2.4-, 2.5-fold respectively).
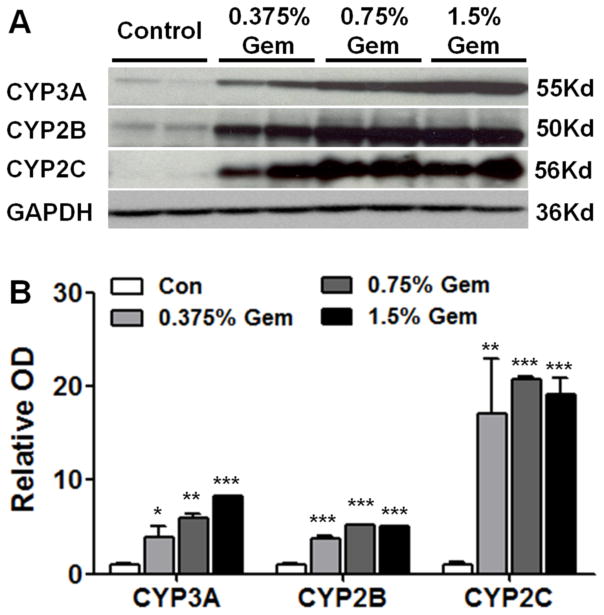
In accordance with the mRNA expression level of Cyp3a, Cyp2b and Cyp2c, western blot results showed that levels of CYP3A, CYP2B and CYP2C were higher in gemfibrozil treated than in control mice, and the tendency was also dose-dependent (Figure 2A). CYP3A, CYP2B and CYP2C were moderately induced in 0.375% the gemfibrozil group and strongly induced in 1.5% gemfibrozil group (Figure 2B).
Fig. 2.
Western-blot analysis of CYPs. (A) Protein levels of CYP3A, CYP2B and CYP2C. (B) Densitometry analysis of western-blot results. Specific band intensity was quantified, normalized to GAPDH. Data were from liver samples collected 14 days after gemfibrozil diet treatment and two were randomly selected for protein analysis. GAPDH was used as a loading control. Data are expressed as mean ± SD (Gem: gemfibrozil, n=5, *P<0.05, **P<0.01, ***P<0.001).
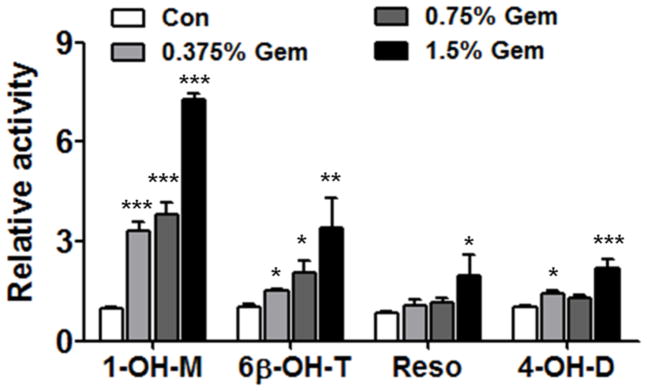
The activities of CYP3A, CYP2B and CYP2C were evaluated using an internal standard method. Similar to the mRNAs and proteins, CYP3A activity, according to 1-OH-M, increased by 3.4-, 3.9-, 7.4-fold respectively in 0.375%, 0.75% and 1.5% gemfibrozil groups and showed dose dependency (Figure 3). A much smaller dose-dependent increase of 6β-OH-T production as a result of CYP3A activity was also noted. Catalytic activity of CYP2B was only induced in the 1.5% gemfibrozil group by 2.3-fold according to the Resorufin metabolism. The activity of CYP2C showed a slight induction in the 0.375% gemfibrozil mice (1.4-fold) and moderately in the 1.5% gemfibrozil group (2.0-fold) according to the production of 4-OH-D. But the difference compared with that the control group was evident.
Fig. 3.
Relative activities of three CYPs after induction by different doses of gemfibrozil. Data are expressed as mean ± SD (Gem: gemfibrozil; 1-OH-M: 1′-hydroxy-midazolam; 6β-OH-T: 6β-hydroxyl-testosterone; Reso: resorufin; 4-OH-D: 4-hydroxy-diclofenac. n=5, *P<0.05, **P<0.01; ***P<0.001).
Comparison of CYP induction in wild-type and Pparα-null mice
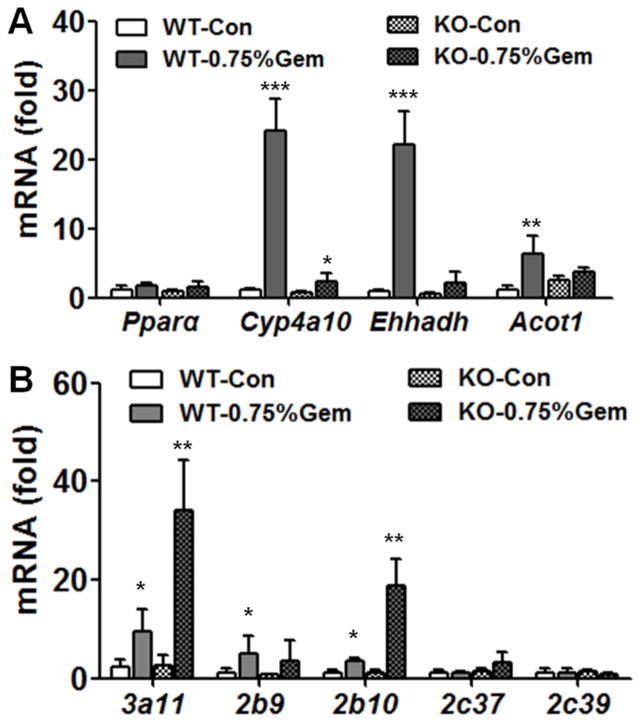
After gemfibrozil feeding for 7 days, mRNAs encoded by the PPARα target genes Cyp4a10, Ehhadh and Acot1, were highly induced in wild-type mice but not in Pparα-null mice (Figure 4A), which indicated that PPARα was differentially activated in the two mouse lines as expected. Expression of Cyp3a11 mRNA in gemfibrozil-fed mice was 4.3-fold higher compared with the control group in wild-type mice (Figure 4B). While in Pparα-null mice, the Cyp3a11 mRNA level was 13-fold higher than in the control group. Cyp2b10 mRNA was induced by 3.2-fold in wild-type mice and 17-fold in Pparα-null mice respectively. Induction of Cyp2c37 (2.5-fold) was observed only in Pparα-null mice but not in wild-type mice (Figure 3A). The mRNA results indicated that activated PPARα played a suppressive role in CYP induction by gemfibrozil, which was evident for Cyp3a11 and Cyp2b10, moderate for Cyp2c37 and negative Cyp2c39.
Fig. 4.
Comparison of Cyp mRNAs between wild-type and Pparα-null mice. (A) Levels of PPARα target gene mRNAs, indicating activation of PPARα in wild-type mice. (B) Hepatic Cyp mRNA involved in xenobiotic metabolism in wild-type and Pparα-null mice. Pparα-null mice were fed 0.75% gemfibrozil-containing diets for 14 days. Data are expressed as mean ± SD (WT: wild-type; KO: Pparα-null; Gem: gemfibrozil, n=5, *P<0.05, **P<0.01, ***P<0.001).
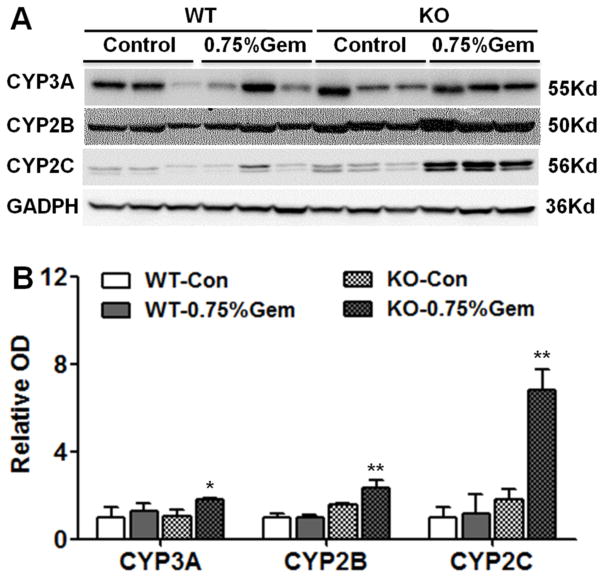
Induction of Cyp3a, Cyp2b and Cyp2c mRNAs in wild-type mice was not as significant as the above 14-day treatment, probably due to the shorter treatment (7-day). But the levels of Cyp3a, Cyp2b and Cyp2c mRNAs were significantly higher in Pparα-null mice than in wild-type mice and (Figure 5A). For CYP3A, the protein level observed were 1.4-fold in wild-type mice, a little lower than the 2-fold increase in Pparα-null mice. Notably, the protein level of CYP2B was higher in Pparα-null compared with wild-type mice after gemfibrozil treatment. CYP2C protein levels were robustly induced in Pparα-null mice treated with gemfibrozil, which is in accordance with above mRNA results (Figure 5B).
Fig. 5.
Western blot and densitometry analysis of CYPs in wild-type and Pparα-null mouse liver extracts. Liver samples were collected 14 days after 0.75% gemfibrozil-containing diet treatment. GAPDH was used as a loading control. The molecular weight was indicated at the right side of the respective band. (A) Western blot of CYP3A, CYP2B, and CYP2C in both mouse lines. (B) Densitometry analysis of CYP3A, CYP2B, CYP2C in liver extracts. Results of the control group were set as 1 and the data expressed as relative intensity respectively. Data are expressed as mean ± SD (WT: wild-type; KO: Pparα-null; Gem: gemfibrozil, n=3, *P<0.05, **P<0.01).
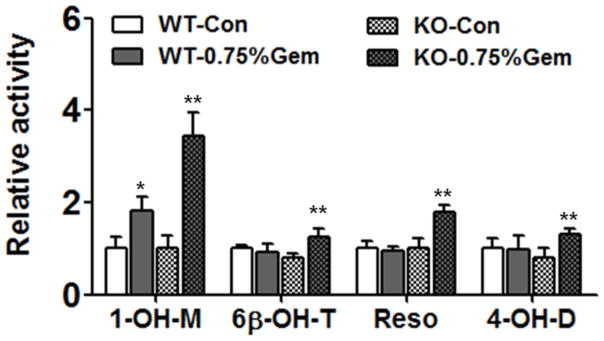
By in vitro activity analysis, the activity of CYP3A, CYP2C, and CYP2B were increased in both wild-type or Pparα-null mice compared with their control groups (Figure 6). The activity of CYP3A was increased 1.8-fold in wild-type mice and 3.5-fold in Pparα-null mice. For the 6β-OH-T, the increase was only found in Pparα-null mice (1.6-fold). The activity increase of CYP2B and CYP2C were only observed in Pparα-null mice by 1.8- and 1.6-fold respectively.
Fig. 6.
Relative enzyme activities of three CYPs after induction by gemfibrozil. Data are expressed as mean ± SD (WT: wild-type; KO: Pparα-null; Gem: gemfibrozil, 1-OH-M: 1′-hydroxy-midazolam; 6β-OH-T: 6β-hydroxyl-testosterone; Reso: resorufin; 4-OH-D: 4-hydroxy-diclofenac. n=5, *P<0.05, **P<0.01).
Discussion
Classically, PPARα is involved in modulating lipid homeostasis and also has a role in hepatic metabolism of glucose, amino acid and bile acid [20]. PPARα’s role in the induction of CYPs and UGTs was reported but not completely understood at the molecular level other than the known Cyp4a as PPARα target gene [16, 17]. PFDA, an activator of PPARα, induced Cyp2b10 (20-fold), Cyp3a11 (2-fold), and Cyp4a14 (32-fold) in wild-type but not in Pparα-null mice [16]. The above data indicated a role for PPARα in CYP induction by exogenous agonists. For gemfibrozil, besides the well-known inhibitory drug-drug interaction, its inductive potential was recently reported [10]. In this study, the Cyp induction level of 3 subfamilies by gemfibrozil was much higher in Pparα-null mice than that in wild-type mice, indicating a suppressive role of PPARα.
CYP3A4 is considered to be the most important CYP subfamily for metabolism of clinical medicines [21]. Nuclear receptors PPARα, PXR, constitutive androstane receptor (CAR) are involved in the regulation of CYP3A4 expression [22, 23]. CYP3A, CYP2B, CYP4A are target genes of PXR, CAR, PPARα respectively, with notable cross-talk among these receptors [24]. Three functional PPARα-binding regions (PBR-I, -II, and -III) within ~ 12 kb of the CYP3A4 upstream sequence were ever reported [12]. So there was a biological basis for CYP3A regulation by PPARα.
The DNA binding preference of CAR and PXR is quite similar, so it’s possible that these nuclear receptors may activate both CYP3A and CYP2B genes. Using CAR-, PXR-, and Pparα-null mouse models, PPARα and CAR were shown to play central roles in the induction of CYPs by PFDA. Cyp2b10 mRNA was induced higher in Pparα-null than that in wild-type mice by the PPARα activator PFDA, but not in Car-null mice [16]. These findings indicated a cross-talk between PPARα and CAR in the regulation of CYP induction by PPARα activators. The above finding also suggested PPARα activation decreased the induction of CYP2B in wild-type mice because of its cross-talk with CAR. The data of CYP2B induction by gemfibrozil in this study was quite similar as the above report. Probably when PPARα is absent, its cross-talk with CAR would be abolished, leading to enhanced induction of Cyp2b10 gene.
Similarly for the CYP2C subfamilies, they were induced much more in Pparα-null mice than that in wild-type mice. An early study reported peroxisome proliferators down-regulated liver-specific genes including CYP2C family members, which was dependent on PPARα [25]. In human primary hepatocytes, PPARα binds a peroxisome proliferator response element (DR1-B) in the promoter of CYP2C8 gene, suggesting the basis for direct action of PPARα on CYP2C gene expression [9]., Another group found direct binding of PPARα to a DR-1 motif located at positions −2762/−2775 bp upstream of the Cyp2C8 was involved in regulation of human CYP2C8 by PPARα [26]. PPARα activators induce the pantothenate kinase 1 gene and miR107, and the latter down-regulated CYP2C8 protein level in human liver [27]. Additionally, Cyp2b and Cyp2c subfamilies are target genes of the transcription factor signal transducer and activator of transcription 5B, which was regulated by growth hormone signaling and cross-talk among nuclear factors [28]. So diverse mechanism might be involved in regulation of CYP2C subfamily by PPARα agonists.
Gemfibrozil’s molecule weight is only 250.35 and its absorption is about 1.5 h, suggesting its diffusion and distribution in hepatic tissues is quite easy. After 14-day treatment, trough concentration of gemfibrozil did not differ between wild-type and Pparα-null mice [13]. CYP- and UGT-mediated depletion of gemfibrozil were 303 and 1607 nmol/min/mg in rat liver microsome versus 86 and 243 nmol/min/mg in human liver microsome, suggesting phase I metabolism contributed quite less for elimination of gemfibrozil [29]. These data indicated the chance was slim that self-induction of CYP by gemfibrozil or other pharmacokinetic variations might interfere with the pharmacodynamic actions.
There are 3 marker responses to evaluate CYP induction potential of compounds, namely mRNA level, protein level and activity level. And the CYP activity was considered as gold-standard to confirm the induction potential of given compounds. In this study, three marker responses and dose relationship were used to confirm the induction potential of gemfibrozil. The dose-response experiment in wild-type mice and suppression experiment in Pparα-null mice were performed separately. The weaker regulation of CYPs in the second experiment could be explained by shorter treatment time. Although the CYP2C mRNA was not strictly consistent with its protein and activity levels in Pparα-null mice, the CYP induction tendency was affirmative and the suppressive effect was clear. Thus the conclusion could be made by combination of the three responses mentioned above for three Cyp subfamilies.
Conclusively in this study, gemfibrozil induced CYP3A, CYP2B and CYP2C in a dose-dependent manner in wild-type mice, while induction of these CYPs was stronger in the Pparα-null mice. These data indicated induction potential of CYPs by gemfibrozil was inhibited by PPARα, suggesting a potential role of this receptor in drug-drug interactions as well as metabolic diseases treated with fibrates.
Fig. 1.
Cyp mRNA induction by gemfibrozil in mice. Fifteen wild-type mice were fed a gemfibrozil-containing diet (0.375%, 0.75% and 1.5%) for 14 days. Data were from liver samples collected 14 days after gemfibrozil treatment and expressed as mean ± SD (Gem: gemfibrozil, n=5, *P<0.05, **P<0.01, ***P<0.001).
Acknowledgments
This work was supported by National Natural Science Foundation of China [Grant 81273582, 81302848], the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health, and the K.C. Wong Magna Fund in Ningbo University.
Footnotes
Competing interest
The authors declare no conflict of interest.
References
- 1.Fievet C, Staels B. Combination therapy of statins and fibrates in the management of cardiovascular risk. Curr Opin Lipidol. 2009;20:505–511. doi: 10.1097/MOL.0b013e328332e9ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacobson TA, Zimmerman FH. Fibrates in combination with statins in the management of dyslipidemia. J Clin Hypertens (Greenwich) 2006;8:35–41. doi: 10.1111/j.1524-6175.2005.05278.x. quiz 42–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krysiak R, Labuzek K, Okopien B. Effect of atorvastatin and fenofibric acid on adipokine release from visceral and subcutaneous adipose tissue of patients with mixed dyslipidemia and normolipidemic subjects. Pharmacol Rep. 2009;61:1134–1145. doi: 10.1016/s1734-1140(09)70176-8. [DOI] [PubMed] [Google Scholar]
- 4.Farmer JA. Learning from the cerivastatin experience. Lancet. 2001;358:1383–1385. doi: 10.1016/S0140-6736(01)06489-3. [DOI] [PubMed] [Google Scholar]
- 5.Ogilvie BW, Zhang D, Li W, et al. Glucuronidation converts gemfibrozil to a potent, metabolism-dependent inhibitor of CYP2C8: implications for drug-drug interactions. Drug Metab Dispos. 2006;34:191–197. doi: 10.1124/dmd.105.007633. [DOI] [PubMed] [Google Scholar]
- 6.Shitara Y, Hirano M, Sato H, Sugiyama Y. Gemfibrozil and its glucuronide inhibit the organic anion transporting polypeptide 2 (OATP2/OATP1B1:SLC21A6)-mediated hepatic uptake and CYP2C8-mediated metabolism of cerivastatin: analysis of the mechanism of the clinically relevant drug-drug interaction between cerivastatin and gemfibrozil. J Pharmacol Exp Ther. 2004;311:228–236. doi: 10.1124/jpet.104.068536. [DOI] [PubMed] [Google Scholar]
- 7.Prueksaritanont T, Zhao JJ, Ma B, et al. Mechanistic studies on metabolic interactions between gemfibrozil and statins. J Pharmacol Exp Ther. 2002;301:1042–1051. doi: 10.1124/jpet.301.3.1042. [DOI] [PubMed] [Google Scholar]
- 8.Varma MV, Lin J, Bi YA, Kimoto E, Rodrigues AD. Quantitative Rationalization of Gemfibrozil Drug Interactions: Consideration of Transporters-Enzyme Interplay and the Role of Circulating Metabolite Gemfibrozil 1-O-beta-Glucuronide. Drug Metab Dispos. 2015;43:1108–1118. doi: 10.1124/dmd.115.064303. [DOI] [PubMed] [Google Scholar]
- 9.Makia NL, Goldstein JA. CYP2C8 Is a Novel Target of Peroxisome Proliferator-Activated Receptor alpha in Human Liver. Mol Pharmacol. 2016;89:154–164. doi: 10.1124/mol.115.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu A, Yang J, Zhao X, et al. Induction of P450 3A1/2 and 2C6 by gemfibrozil in Sprague Dawley rats. Pharmacol Rep. 2011;63:157–164. doi: 10.1016/s1734-1140(11)70410-8. [DOI] [PubMed] [Google Scholar]
- 11.Prueksaritanont T, Richards KM, Qiu Y, et al. Comparative effects of fibrates on drug metabolizing enzymes in human hepatocytes. Pharm Res. 2005;22:71–78. doi: 10.1007/s11095-004-9011-5. [DOI] [PubMed] [Google Scholar]
- 12.Thomas M, Burk O, Klumpp B, et al. Direct transcriptional regulation of human hepatic cytochrome P450 3A4 (CYP3A4) by peroxisome proliferator-activated receptor alpha (PPARalpha) Mol Pharmacol. 2013;83:709–718. doi: 10.1124/mol.112.082503. [DOI] [PubMed] [Google Scholar]
- 13.Liu A, Krausz KW, Fang ZZ, et al. Gemfibrozil disrupts lysophosphatidylcholine and bile acid homeostasis via PPARalpha and its relevance to hepatotoxicity. Arch Toxicol. 2014;88:983–996. doi: 10.1007/s00204-013-1188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li F, Patterson AD, Krausz KW, Tanaka N, Gonzalez FJ. Metabolomics reveals an essential role for peroxisome proliferator-activated receptor alpha in bile acid homeostasis. J Lipid Res. 2012;53:1625–1635. doi: 10.1194/jlr.M027433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X, Cao L, Jiang C, et al. PPARalpha-UGT axis activation represses intestinal FXR-FGF15 feedback signalling and exacerbates experimental colitis. Nat Commun. 2014;5:4573. doi: 10.1038/ncomms5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng X, Klaassen CD. Perfluorocarboxylic acids induce cytochrome P450 enzymes in mouse liver through activation of PPAR-alpha and CAR transcription factors. Toxicol Sci. 2008;106:29–36. doi: 10.1093/toxsci/kfn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo M, Tan Z, Dai M, et al. Dual action of peroxisome proliferator-activated receptor alpha in perfluorodecanoic acid-induced hepatotoxicity. Arch Toxicol. 2016 doi: 10.1007/s00204-016-1779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SS, Pineau T, Drago J, et al. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao M, Zhu M, Sinz MW, et al. Development and full validation of six inhibition assays for five major cytochrome P450 enzymes in human liver microsomes using an automated 96-well microplate incubation format and LC-MS/MS analysis. J Pharm Biomed Anal. 2007;44:211–223. doi: 10.1016/j.jpba.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 20.Peters JM, Cheung C, Gonzalez FJ. Peroxisome proliferator-activated receptor-alpha and liver cancer: where do we stand? J Mol Med (Berl) 2005;83:774–785. doi: 10.1007/s00109-005-0678-9. [DOI] [PubMed] [Google Scholar]
- 21.Daly AK. Significance of the minor cytochrome P450 3A isoforms. Clin Pharmacokinet. 2006;45:13–31. doi: 10.2165/00003088-200645010-00002. [DOI] [PubMed] [Google Scholar]
- 22.Betts S, Bjorkhem-Bergman L, Rane A, Ekstrom L. Expression of CYP3A4 and CYP3A7 in Human Foetal Tissues and its Correlation with Nuclear Receptors. Basic Clin Pharmacol Toxicol. 2015;117:261–266. doi: 10.1111/bcpt.12392. [DOI] [PubMed] [Google Scholar]
- 23.Lindh JD, Andersson ML, Eliasson E, Bjorkhem-Bergman L. Seasonal variation in blood drug concentrations and a potential relationship to vitamin D. Drug Metab Dispos. 2011;39:933–937. doi: 10.1124/dmd.111.038125. [DOI] [PubMed] [Google Scholar]
- 24.Honkakoski P, Negishi M. Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochem J. 2000;347:321–337. doi: 10.1042/0264-6021:3470321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ripp SL, Falkner KC, Pendleton ML, Tamasi V, Prough RA. Regulation of CYP2C11 by dehydroepiandrosterone and peroxisome proliferators: identification of the negative regulatory region of the gene. Mol Pharmacol. 2003;64:113–122. doi: 10.1124/mol.64.1.113. [DOI] [PubMed] [Google Scholar]
- 26.Thomas M, Winter S, Klumpp B, et al. Peroxisome proliferator-activated receptor alpha, PPARα, directly regulates transcription of cytochrome P450 CYP2C8. Frontiers in Pharmacology. 2015:6. doi: 10.3389/fphar.2015.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang SY, Surapureddi S, Coulter S, Ferguson SS, Goldstein JA. Human CYP2C8 is post-transcriptionally regulated by microRNAs 103 and 107 in human liver. Mol Pharmacol. 2012;82:529–540. doi: 10.1124/mol.112.078386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshida K, Waxman DJ, Corton JC. Chemical and Hormonal Effects on STAT5b-Dependent Sexual Dimorphism of the Liver Transcriptome. PLoS One. 2016;11:e0150284. doi: 10.1371/journal.pone.0150284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Liu A, Xie H, Cheng GQ, Dai R. Metabolism of fibrates by cytochrome P450s and UDP-glycosyltransferases in rat and human liver microsomes. Chinese Science Bulletin. 2012;57:1142–1149. [Google Scholar]