Abstract
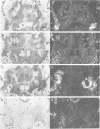
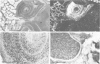
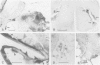
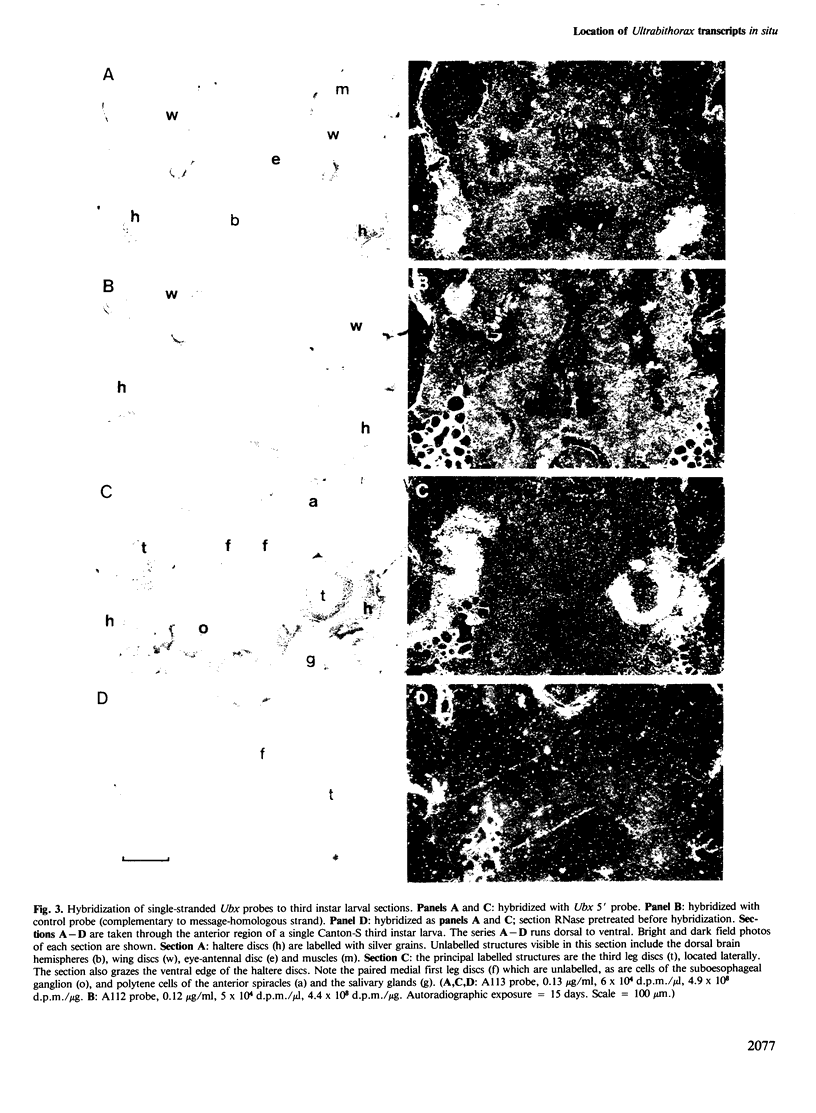
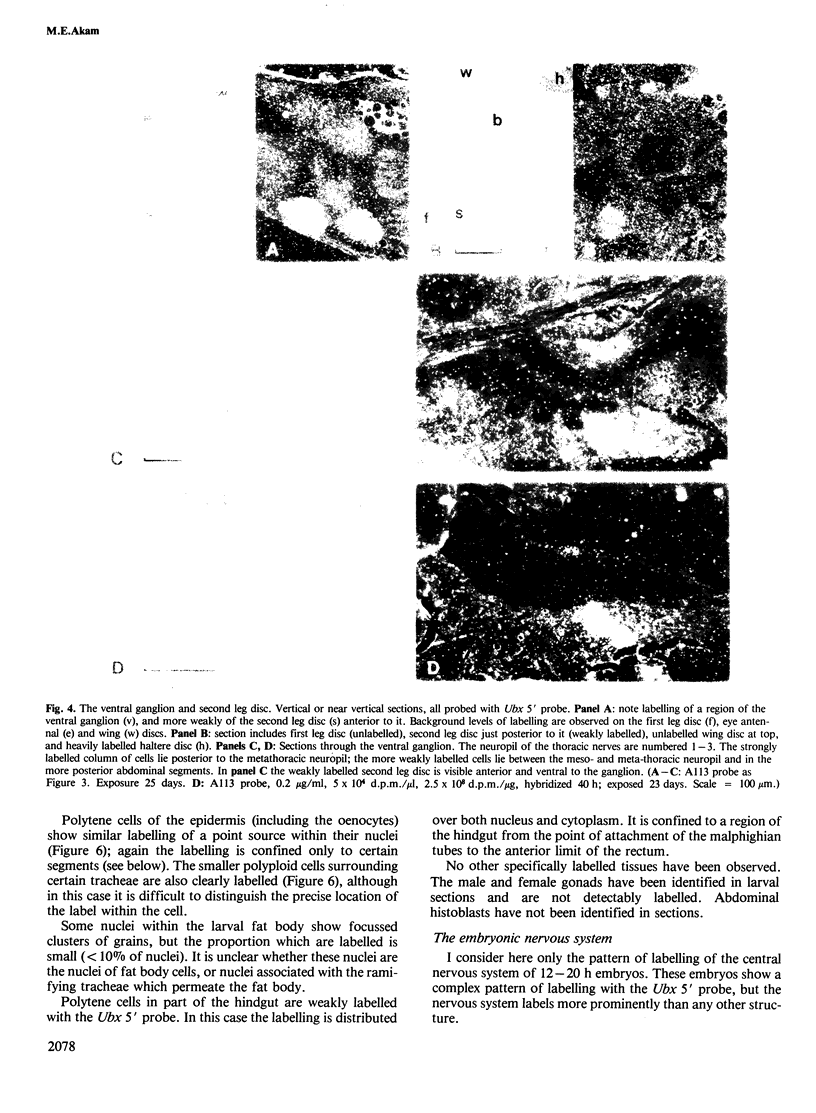
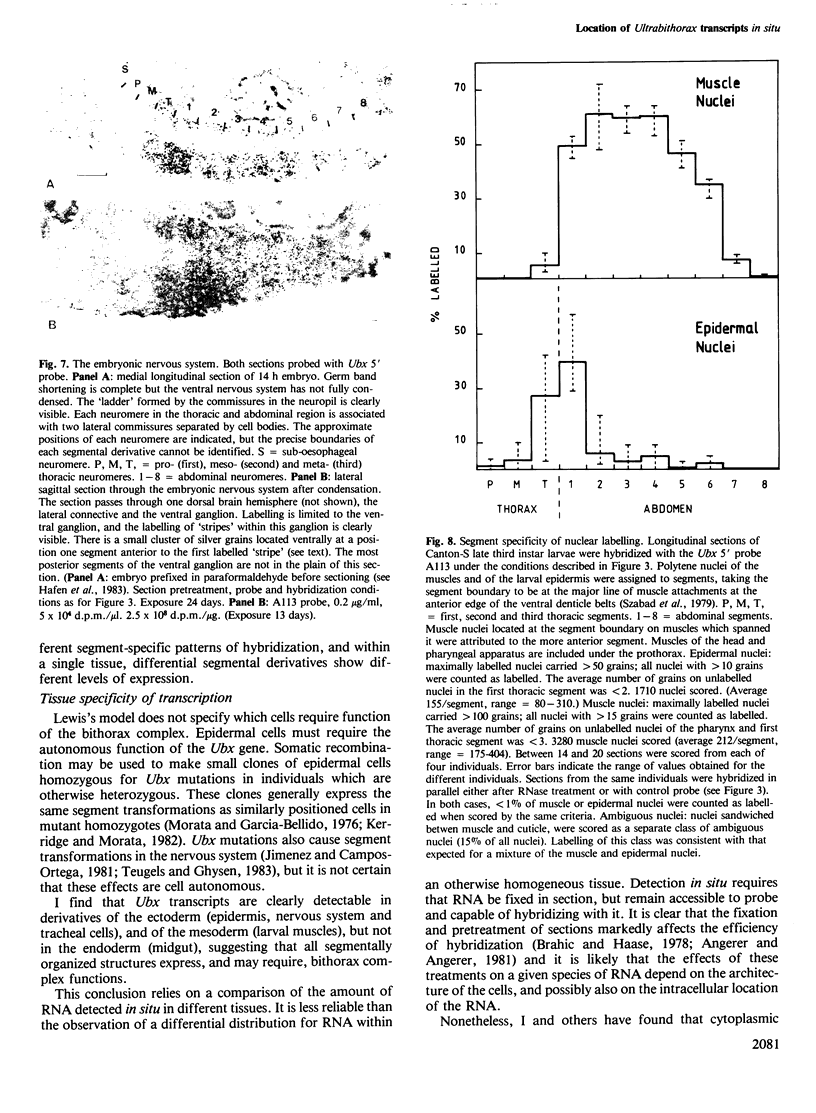
I have used in situ hybridization to map the distribution of transcripts of the Ultrabithorax unit in tissue sections of Drosophila embryos and larvae. The results confirm the prediction of Lewis that genes of the bithorax complex will show segmentally regulated patterns of expression. Transcripts of the Ultrabithorax unit are detected in derivatives of the ectoderm and in some derivatives of the mesoderm, but not in the endoderm. In each of these tissues the transcripts are found in the derivatives of some segments but not of others. In third instar larvae they are most abundant in the imaginal discs of the third thoracic segment, and in a region of the central nervous system that includes parts of the metathoracic and first abdominal neuromeres. They are also detected in the nuclei of polytene cells of the larval epidermis, principally in the third thoracic and first abdominal segments, and in the nuclei of larval muscles in the first six abdominal segments. In late stages of embryogenesis the central nervous system is the most prominently labelled tissue; within it transcripts are found only in the neuromeres of the thoracic and abdominal segments. They are most abundant in a region which includes parts of the neuromeres of the metathorax and the first abdominal segment.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angerer L. M., Angerer R. C. Detection of poly A+ RNA in sea urchin eggs and embryos by quantitative in situ hybridization. Nucleic Acids Res. 1981 Jun 25;9(12):2819–2840. doi: 10.1093/nar/9.12.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M., Spierer P., Lewis E. B., Hogness D. S. Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Science. 1983 Jul 1;221(4605):23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Brahic M., Haase A. T. Detection of viral sequences of low reiteration frequency by in situ hybridization. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6125–6129. doi: 10.1073/pnas.75.12.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafen E., Levine M., Garber R. L., Gehring W. J. An improved in situ hybridization method for the detection of cellular RNAs in Drosophila tissue sections and its application for localizing transcripts of the homeotic Antennapedia gene complex. EMBO J. 1983;2(4):617–623. doi: 10.1002/j.1460-2075.1983.tb01472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerridge S., Morata G. Developmental effects of some newly induced Ultrabithorax alleles of Drosophila. J Embryol Exp Morphol. 1982 Apr;68:211–234. [PubMed] [Google Scholar]
- Lewis E. B. A gene complex controlling segmentation in Drosophila. Nature. 1978 Dec 7;276(5688):565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Szabad J., Schüpbach T., Wieschaus E. Cell lineage and development in the larval epidermis of Drosophila melanogaster. Dev Biol. 1979 Dec;73(2):256–271. doi: 10.1016/0012-1606(79)90066-6. [DOI] [PubMed] [Google Scholar]