Abstract
To compare protein expression levels, gene mutation and survival among Right-Sided Colon Cancer (RSCC), Left-Sided Colon Cancer (LSCC) and rectal cancer patients, 57 cases of RSCC, 87 LSCC and 145 rectal cancer patients were included retrospectively. Our results demonstrated significant differences existed among RSCC, LSCC and rectal cancer regarding tumor diameter, differentiation, invasion depth and TNM stage. No significant difference was identified in expression levels of MLH1, MSH2, MSH6, PMS2, β-Tubulin III, P53, Ki67 and TOPIIα, and gene mutation of KRAS and BRAF among three groups. Progression Free Survival (PFS) of RSCC was significantly lower than that of LRCC and rectal cancer. In univariate analyses, RSCC, preoperative chemoradiotherapy, poor differentiation, advanced TNM stage, elevated serum CEA and CA19-9 level, tumor deposit, perineural and vascular invasion were found to be predictive factors of shorter PFS. In multivariate analyses, only differentiation and TNM stages were found to be independent predictors of PFS. In conclusion, compared with LSCC and rectal cancer, RSCC has larger tumor size, poor differentiation, advanced TNM stage and shorter survival. The shorter survival in RSCC might be attributed to the advanced tumor stage caused by its inherent position feature of proximal colon rather than genetic difference.
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer globally, accounting for 10.0% of all new cancer cases. An estimated 746,300 of new CRC cases and 614,300 CRC deaths occurred in 2012 worldwide. It is also the fourth common cause of cancer-related deaths in men and the third in women worldwide1. According to the tumor position, CRCs are usually classified into three types: Right-Sided Colon Cancer (RSCC), Left-Sided Colon Cancer (LSCC) and Rectal Cancer, and each type approximately accounts for 30%2, 3. Colon cancers consist of RSCC and LSCC, divided at the splenic flexure. Rectal cancers are referred to lesions located within 12 cm from the anal verge. The issue whether these three types should be considered as a single entity or three distinct entities is still controversial2.
It is reported that rectal cancer is different from colon cancer in aetiology, genetics, anatomy, clinical manifestation, biological feature, treatment response and clinical outcomes3–6. Lifestyle factors such as diet, smoking and physical activity have different effects in colon cancer than in rectal cancer7. The treatments for rectal and colon cancer are different, depending on the TNM stage. For stage I and IV, rectal and colon cancers are commonly regarded as one entity and treated alike3. For stage II–III CRC, neoadjuvant radiochemotherapy is recommended for rectal cancer patients, but not for colon cancer patients. Neoadjuvant radiochemotherapy resulted in decrease of local recurrence rate, but no increase in overall survival (OS) compared to surgery alone8. Furthermore, a study which includes 372,130 patients from the SEER database with a median follow-up of 32 months, showed that there was no difference in OS between colon and rectal cancer9. Frattini’s study showed that significant differences existed in KRAS mutation and APC mutation between colon cancers and rectal cancers2. However, the Cancer Genome Atlas Network conducted a genome-scale analysis of 276 samples, analyzing exome sequence, DNA copy number, promoter methylation, mRNA and microRNA expression, and concluded that colon and rectal cancers had similar patterns of genomic alteration, and gene mutations of APC, TP53, SMAD4, PIK3CA and KRAS10. So, whether colon and rectal cancers have different gene expression and prognosis is still in debate.
The distinction between RSCC and LSCC has received increasing attention in recent years. Some suggested that they were two distinct categories of colon cancer11. Many publications reported that there were significant differences regarding epidemiology, clinical presentation, pathology, genetic mutations and survival between RSCC and LSCC11. RSCC had been reported to be older, more often female and more often poorly differentiated tumors, and have more advanced stages, increased tumor sizes and different molecular features11–14. Data regarding prognosis in RSCC versus LSCC are conflicting, and it remains a matter of great debate whether tumor location itself has a significant prognostic impact12. Most studies demonstrated a poorer survival in RSCC compared to LSCC15–17. In contrast, several scholars found no difference in OS between RSCC and LSCC after adjusting for various variables11, 18, 19. Warschkow et al.12 carried out a study including 91,416 patients, and found that RSCC patients had worse OS compared to LSCC patients; but the prognosis of RSCC was better than LSCC after matching clinical features. In addition, whether molecular features differ between LSCC and RSCC remains unclear20. Kuramochi et al.21 had detected mRNA expression levels of 14 signal transduction genes in 52 cases of CRC, but only identified significant differences in PTEN mRNA expression level.
Furthermore, some authors suggested that LSCC and rectal cancer shared multiple common characteristics and were different from RSCC, which was supported by several histological, genetic and methylation findings3, 22, 23. And a new term, Left-sided Colorectal Cancer (LCRC) which included LSCC and rectal cancer, was created24. There are three main types of (epi)genetic instability in CRC: (1) chromosomal instability (CIN) caused by KRAS mutations; (2) microsatellite instability (MSI) resulted from deficient DNA mismatch repair (MMR); (3) CpG island methylator phenotype (CIMP) epigenetic instability3. The mutational profiles (KRAS, MMR, CIMP) of LSCC and rectal cancer were similar, but were different from that of RSCC. This result was attributed to their differing origins: RSCC originated from midgut, while LCRC originated from hindgut3. BRAF was preferentially mutated in RSCC, and EGFR (epidermal growth factor receptor) was prevalently amplified in LCRC3. Class III beta-tubulin (β-Tubulin III) had been reported to express at the invasive margin of CRC, and its expression level was correlated with tumor differentiation and lymphatic metastasis25. The mutation incidence of p53 gene was reported to be as high as 42.4% in CRC26. P53 plays an important role in the transformation from colorectal normal mucosa to carcinoma through adenoma27. Several studies reported that gene mutation and protein expression of P53 differed significantly between RSCC and LSCRC26, 28, 29, but others showed that no significant association was identified between p53 protein expression and tumor site30. Ki67 is a marker of cell proliferation, and it plays a vital role in the development of CRC31. High Ki67 labeling index had been reported to be an independent prognostic biomarker in TNM III and IV CRC32. In addition, our previous study showed that topoisomerase II alpha (TOPIIα) expression was related with T stage, N stage, recurrence and prognosis33.
In the light of the aforementioned considerations, it is urgent for us to explore the possible differences of gene expression level and prognosis among RSCC, LSCC and rectal cancers. Knowledge of the molecular differences would help us to improve the diagnosis and treatment strategy in clinical practice. Thus, we investigated the protein expression levels of MLH1, MSH2, MSH6, PMS2, β-Tubulin III, P53, Ki67 and TOPIIα, and the gene mutation of KRAS and BRAF in 289 specimens of sporadic CRC patients, and their implications on survival were also investigated.
Methods
Patients
From January 2015 to December 2016, 289 cases of sporadic CRC patients were recruited from the Department of Colorectal Surgery of Changhai Hospital, Second Military Medical University, Shanghai, China. All patients received radical resection of the primary tumor. The clinicopathological characteristics were extracted from the electronic medical records. All patients were followed up every 3 months, with a median follow up period of 10 months, ranging from 3 to 23 months. Informed consent had been obtained from all patients and the project had been approved by the Ethics Committee of Changhai Hospital, Second Military Medical University. All methods were performed in accordance with the relevant guidelines and regulations.
Immunohistochemistry
Paraffin-embedded tumor tissues were examined for MLH1, MSH2, MSH6, PMS2, β-tubulin III, P53, Ki67 and TopIIα expression, using the Envision method following the manufacturer’s instruction. After deparaffinization and re-hydration, antigen retrieval was done with Citrate buffer (0.01 mol/L, pH = 6.0) by pressure cooker. Primary antibodies (Table 1) were incubated on the slides for 2 hours at room temperature in a hydrated chamber. The sections were stained with DAB and counterstained with Mayer’s hematoxylin, washed again, dehydrated in alcohol, cleared in xylene, mounted with Pertex mounting medium, and coverslipped. All sections were scored blindly by two pathologists (Xu Y & Bai CG) under microscope, by randomly selecting 10 high-power (×400) view fields in each sample and scoring the protein expression in tumor cells.
Table 1.
The primary antibody of MLH1, MSH2, MSH6, PMS2, β-Tubulin III, P53, Ki67 and TOPIIα used in this study.
| Antibody | Corporation | Manufacturer City | Product code | Dilution | Clonality |
|---|---|---|---|---|---|
| MLH1 | Maixin Biotech | Fuzhou, China | MAB-0642 | 1:50 | G168-15 |
| MSH2 | Maixin Biotech | Fuzhou, China | MAB-0291 | 1:50 | 25D12 |
| MSH6 | Maixin Biotech | Fuzhou, China | MAB-0643 | 1:50 | 44 |
| PMS2 | Maixin Biotech | Fuzhou, China | MAB-0656 | 1:50 | MOR4G |
| β-Tubulin III | Maixin Biotech | Fuzhou, China | MAB-0636 | 1:100 | TUJ1 |
| P53 | Maixin Biotech | Fuzhou, China | Kit-0010 | 1:100 | Do-7 |
| Ki-67 | Maixin Biotech | Fuzhou, China | Kit-0005 | 1:100 | MIB-1 |
| TOPIIα | Maixin Biotech | Fuzhou, China | MAB-0588 | 1:100 | 3F6 |
Expression defects of MLH1, MSH2, MSH6 and PMS2 proteins were defined as complete absence of detectable nuclear staining in tumor cells. Intact nuclear staining of the colorectal crypts of the peritumoral normal mucosa, stromal cells and lymphocytes served as internal positive control and was required for adequate evaluation34.
To measure the expression level of β-tubulin III, P53, Ki67 and TopIIα, each sample was scored according to the intensity of the nucleic or cytoplasmic staining (0, no staining; 1, weak staining; 2, moderate staining; and 3, strong staining) and the extent of stained cells (0%, 0; 1–25%, 1; 26–50%, 2; 51–75%, 3; and 76–100%, 4). The multiplication of the intensity and extent score was used as the final staining scores (0 to 12). Tumors having final staining scores of 0, 1~4, 5~8 and 9~12 were considered to be negative (−), slightly positive (+), moderately positive (++) and strongly positive (+++), respectively35. There was a close agreement on staining intensity (91%) and staining extent (93%) between the two pathologists. Disagreements were resolved by consensus. Immunohistochemical labeling index was defined as the percentage of positive nuclei in relation to the whole tumor area.
Sample preparation, DNA extraction, amplification of KRAS and BRAF genes
Surgically resected primary CRC tumor tissue specimens were fixed in formalin and preserved in paraffin blocks for histological examination. After evaluating the standard hematoxylin/eosin-stained slides from each specimen, appropriate samples were specifically chosen by a pathologist to include predominantly tumor cells without significant necrosis or inflammation36. Eight 10 μm-thick formalin-fixed paraffin-embedded (FFPE) sections were used for this study, and placed in 2-mL sterile Eppendorf tubes. AmoyDx FFPE DNA Kit (AmoyDx, Xiamen, China) was used for DNA extraction. Human genomic DNA was amplified for KRAS in exons 2, 3 and 4 (codons 12, 13, 59, 61, 117 and 146), and BRAF in exon 15 (codon 600) by using AmoyDx gene mutation real-time PCR kits (AmoyDx, Xiamen, China). All the experiments are performed following manufacturer’s instructions. 5 μL DNA was used for PCR amplification in each reaction, the PCR cycling conditions were: 5 min incubation at 95 °C, followed by 15 cycles of 95 °C for 25 sec, 64 °C for 20 sec, 72 °C for 20 sec and then 31 cycles of 93 °C for 25 sec, 60 °C for 35 sec, 72 °C for 20 sec. Fluorescent signal was collected from FAM and HEX channels. Each PCR run must contain one negative control and one positive control. KRAS and BRAF mutation status were determined according to the Ct value as indicated in the manufacturer’s instructions.
Statistical analysis
Associations between tumor position and categorical clinicopathological variables were analyzed by χ2 test, using Fisher Exact test if one or more expects in the cross table is less than 5. Numerical data which were consistent with normal distribution were presented as the mean ± standard deviation, and comparisons were performed with the student’s t-test or one-way ANOVA analysis. Numerical data which were inconsistent with normal distribution were presented as median (minimum-maximum), and comparisons were performed with nonparametric test. The Progression Free Survival (PFS) was estimated by the Kaplan-Meier method and survival differences were analyzed with the log-rank test. The Cox proportional hazards model was used for multivariate analysis of prognostic factors. P < 0.05 was considered statistically significant (two-sided). All statistical analyses were conducted using SPSS 17.0 statistical software (SPSS, Inc., Chicago, IL, USA).
Results
A total of 289 cases of CRC patients were included, consisting of 57 cases of RSCC, 87 LSCC and 145 rectal cancers (Table 2). Of the 289 CRC patients, 186 were males and 103 were females, with a mean age of 59.6 years. Thirty-three (33/289, 11.4%) patients received preoperative chemoradiotherapy, 156 (156/289, 54.0%) received postoperative chemoradiotherapy; 82 were resected laparoscopically, and the remaining 207 were removed by open surgery. All of the primary tumors were resected radically, and combined resections of metastatic lesions were performed in 29 patients. There were 60, 82, 89 and 58 cases of stage I, II, III and IV according to the UICC-AJCC TNM stage classification system (7th edition).
Table 2.
Relationship between Tumor Position and Clinicopathological Parameters in 289 Colorectal Cancer Patients.
| Clinicopathological Parameters | RSCC# (n = 57) | LSCC▴ (n = 87) | Rectal cancer (n = 145) | Total (n = 289) | P1* | P2* | P3* | P4* | P5* |
|---|---|---|---|---|---|---|---|---|---|
| Gender | 0.869 | 0.615 | 0.648 | 0.915 | 0.868 | ||||
| Male | 35 | 57 | 94 | 186 | |||||
| Female | 22 | 30 | 51 | 103 | |||||
| Age (year) | 58.6 ± 12.9 | 60.0 ± 11.7 | 59.8 ± 10.1 | 59.6 ± 11.2 | 0.743 | 0.511 | 0.484 | 0.910 | 0.775 |
| Preoperative chemotherapy | 0.845 | 0.949 | 0.691 | 0.591 | 0.565 | ||||
| No | 50 | 76 | 130 | 256 | |||||
| Yes | 7 | 11 | 15 | 33 | |||||
| Open or Laparoscopic | 0.004 | 0.003 | 0.001 | 0.885 | 0.073 | ||||
| Open | 51 | 59 | 97 | 207 | |||||
| Laparoscopic | 6 | 28 | 48 | 82 | |||||
| Gross Type | 0.598 | 0.567 | 0.318 | 0.648 | 0.394 | ||||
| Protruding | 9 | 17 | 32 | 58 | |||||
| Ulcerative & Infiltratie | 48 | 70 | 113 | 231 | |||||
| Diameter (cm) | 5.4 ± 2.3 | 4.3 ± 1.5 | 4.1 ± 1.6 | 4.4 ± 1.8 | 0.000 | 0.002 | 0.000 | 0.415 | 0.004 |
| Differentiation | 0.000 | 0.000 | 0.000 | 0.363 | 0.066 | ||||
| Well & Moderate | 33 | 78 | 124 | 235 | |||||
| Poor☆ | 24 | 9 | 21 | 54 | |||||
| T | 0.000 | 0.110 | 0.000 | 0.015 | 0.000 | ||||
| T1-T2 | 5 | 16 | 48 | 69 | |||||
| T3-T4 | 52 | 71 | 97 | 220 | |||||
| N | 0.708 | 0.437 | 0.466 | 0.891 | 0.772 | ||||
| N0 | 29 | 50 | 82 | 161 | |||||
| N1-N2 | 28 | 37 | 63 | 128 | |||||
| M | 0.667 | 0.980 | 0.502 | 0.423 | 0.368 | ||||
| M0 | 44 | 67 | 118 | 229 | |||||
| M1 | 13 | 20 | 27 | 60 | |||||
| TNM | 0.011 | 0.711 | 0.008 | 0.042 | 0.001 | ||||
| 1 | 5 | 12 | 43 | 60 | |||||
| 2 | 22 | 27 | 33 | 82 | |||||
| 3 | 17 | 28 | 44 | 89 | |||||
| 4 | 13 | 20 | 25 | 58 | |||||
| CEA (ng/mL) | 0.590 | 0.951 | 0.473 | 0.273 | 0.259 | ||||
| Unknown | 3 | 5 | 3 | 11 | |||||
| <5 | 31 | 45 | 84 | 160 | |||||
| >=5 | 23 | 37 | 58 | 118 | |||||
| CA199 (U/mL) | 0.298 | 0.439 | 0.142 | 0.297 | 0.200 | ||||
| Unknown | 4 | 5 | 3 | 12 | |||||
| <37 | 40 | 69 | 116 | 225 | |||||
| >=37 | 13 | 13 | 26 | 52 | |||||
| Tumor Deposit | 0.207 | 0.677 | 0.311 | 0.094 | 0.083 | ||||
| No | 47 | 74 | 110 | 231 | |||||
| Yes | 10 | 13 | 35 | 58 | |||||
| Perineural Invasion | 0.643 | 0.967 | 0.505 | 0.410 | 0.348 | ||||
| No | 49 | 75 | 119 | 243 | |||||
| Yes | 8 | 12 | 26 | 46 | |||||
| Vascular Invasion | 0.454 | 0.493 | 0.757 | 0.213 | 0.305 | ||||
| No | 49 | 71 | 127 | 247 | |||||
| Yes | 8 | 16 | 18 | 42 |
#RSCC: Right-sided colon cancer; ▴LSCC: Left-sided colon cancer; *P1: comparison of three group; P2: RSCC vs. LSCC; P3: RSCC vs. rectal; P4: LSCC vs. rectal; P5:colon vs. rectal; ☆Poorly differentiated, Mucinous adenocarcinoma, signet ring cell carcinoma.
Comparisons of clinicopathological parameters among RSCC, LSCC and rectal cancer patients
As demonstrated in Table 2, significant differences were observed among the RSCC, LSCC and rectal cancer patients regarding surgical procedure (Open/Laparoscopic), tumor diameter, differentiation, invasion depth (T) and TNM stage (all P < 0.05). No significant difference was observed in other characteristics, including gender, age, preoperative chemoradiotherapy, gross type, lymph node metastasis(N), distant metastasis(M), serum CEA level, serum CA 19-9 level, tumor deposit, perineural invasion and vascular invasion (Table 2, all P > 0.05). Of the 58 cases of TNM stage IV patients, 38 had received postoperative chemoradiotherapy, 12 had received postoperative biological therapy, and 8 had received second line chemoradiotherapy after recurrence. No significant difference was identified in postoperative chemoradiotherapy, biological therapy and second line chemoradiotherapy among the three groups (Table 3, all P > 0.05).
Table 3.
Postoperative chemoradiotherapy and Biological therapy in TNM stage IV Colorectal Cancer Patients.
| Postoperative treatment | Right-sided colon cancer (n = 13) | Left-sided colon cancer (n = 20) | Rectal cancer (n = 25) | Total (n = 58) | P |
|---|---|---|---|---|---|
| Postoperative chemoradiotherapy | 0.386 | ||||
| No | 3 | 1 | 7 | 11 | |
| Yes | 8 | 16 | 14 | 38 | |
| Unknown | 2 | 3 | 4 | 9 | |
| Postoperative biological therapy | 0.551 | ||||
| No | 9 | 8 | 15 | 32 | |
| Anti-EGFR | 1 | 4 | 2 | 7 | |
| Anti-VEGF | 1 | 3 | 1 | 5 | |
| Unknown | 2 | 5 | 7 | 14 | |
| Postoperative second line chemoradiotherapy | 0.089 | ||||
| No | 7 | 13 | 21 | 41 | |
| Yes | 4 | 4 | 0 | 8 | |
| Unknown | 2 | 3 | 4 | 9 |
Comparisons of clinicopathological parameters between each two groups
Compared with LSCC, RSCC was associated with less laparoscopic resection (P = 0.003), larger tumor size (P = 0.002) and poor differentiation (P < 0.001) (Table 2). Compared with rectal cancer, RSCC was associated with less laparoscopic resection (P = 0.001), larger tumor size (P < 0.001), poor differentiation (P < 0.001), advanced T stage (P < 0.001) and advanced TNM stage (P = 0.008) (Table 2). Compared with rectal cancer, LSCC was associated with advanced T stage (P = 0.015) and advanced TNM stage (P = 0.042) (Table 2). Compared with rectal cancer, colon cancer was associated with larger tumor size (P = 0.004), advanced T stage (P < 0.001) and advanced TNM stage (P = 0.001) (Table 2).
Comparisons of protein expression levels, KRAS and BRAF mutation among RSCC, LSCC and rectal cancer patients
No significant difference was observed among RSCC, LSCC and rectal cancer, regarding protein expression levels of MLH1, MSH2, MSH6, PMS2, β-tubulin III, P53, Ki67 and TopIIα (Figs 1 and 2 & Table 4). Similarly, no significant difference was identified in gene mutation incidences of KRAS and BRAF (Fig. 3, Table 4). Immunohistochemical labeling index of TOPIIα in RSCC was significantly lower than that in LSCC (P = 0.007) and that in rectal cancer (P = 0.010) (Table 4).
Figure 1.
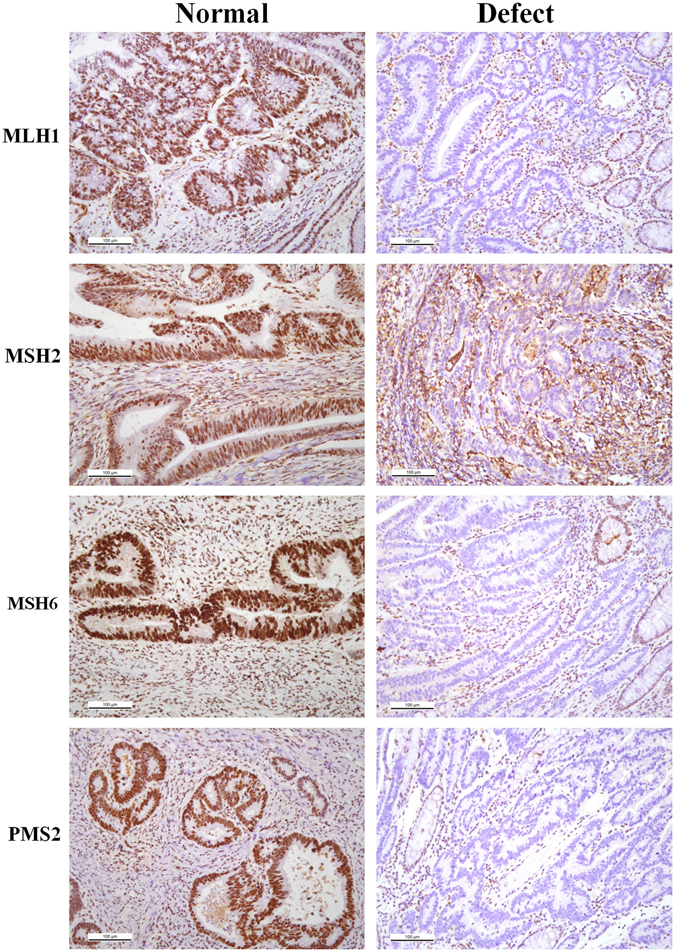
Representative images of MLH1, MSH2, MSH6 and PMS2 in colorectal cancer tissue.
Figure 2.
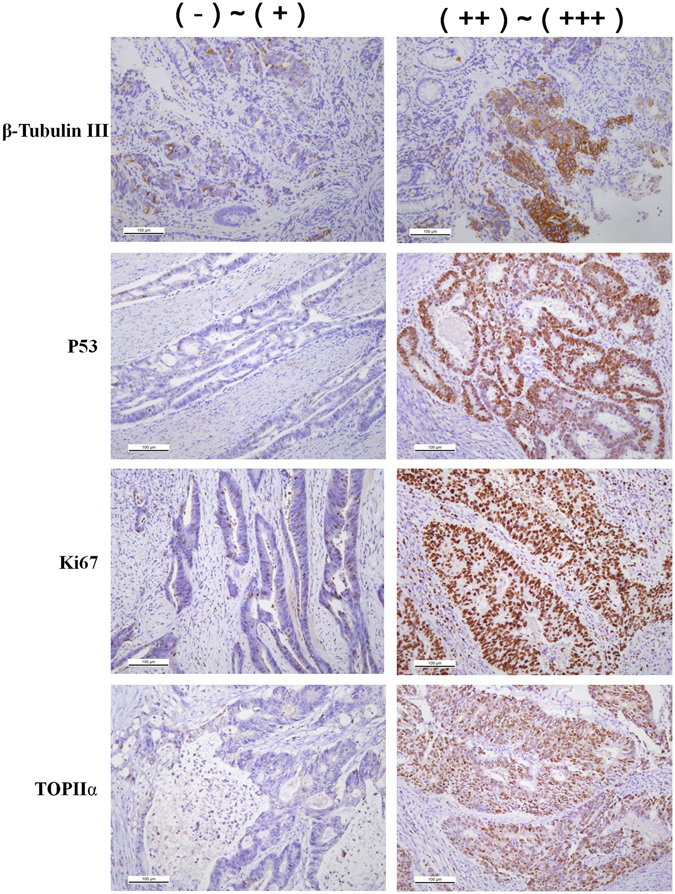
Representative images of different expression level of β-Tubublin III, P53, Ki67 and TOPIIα in colorectal cancer (×200).
Table 4.
Relationship between Tumor Position and Gene Mutation, Protein Expression Levels in 289 Colorectal Cancer Patients.
| Gene Mutation & Expression Level | Right-sided colon cancer (n = 57) | Left-sided colon cancer (n = 87) | Rectal cancer (n = 145) | Total (n = 289) | P1* | P2* | P3* | P4* | P5* |
|---|---|---|---|---|---|---|---|---|---|
| KRAS | 0.685 | 0.637 | 0.392 | 0.673 | 0.464 | ||||
| Wild | 27 | 42 | 59 | 128 | |||||
| Mutant | 16 | 30 | 48 | 94 | |||||
| Unknown | 14 | 15 | 38 | 67 | |||||
| BRAF | 0.074 | 0.144 | 0.068 | 0.769 | 0.202 | ||||
| Wild | 39 | 70 | 106 | 215 | |||||
| Mutant | 3 | 1 | 1 | 5 | |||||
| Unknown | 15 | 16 | 38 | 69 | |||||
| MLH1 | 0.735 | 0.614 | 0.860 | 0.509 | 0.701 | ||||
| Defect | 6 | 13 | 17 | 36 | |||||
| Normal | 44 | 67 | 114 | 225 | |||||
| Unknown☆ | 7 | 7 | 14 | 28 | |||||
| MSH2 | 0.898 | 0.641 | 0.786 | 0.792 | 0.974 | ||||
| Defect | 2 | 2 | 4 | 8 | |||||
| Normal | 50 | 80 | 127 | 257 | |||||
| Unknown☆ | 5 | 5 | 14 | 24 | |||||
| MSH6 | 0.989 | 0.886 | 0.945 | 0.920 | 0.972 | ||||
| Defect | 4 | 6 | 10 | 20 | |||||
| Normal | 46 | 76 | 120 | 242 | |||||
| Unknown☆ | 7 | 5 | 15 | 27 | |||||
| PMS2 | 0.497 | 0.388 | 0.828 | 0.268 | 0.566 | ||||
| Defect | 1 | 0 | 2 | 3 | |||||
| Normal | 49 | 79 | 128 | 256 | |||||
| Unknown☆ | 7 | 8 | 15 | 30 | |||||
| β-Tubulin III | 0.452 | 0.280 | 0.221 | 0.957 | 0.563 | ||||
| (−)~(+) | 8 | 27 | 52 | 87 | |||||
| (++)~(+++) | 5 | 8 | 15 | 28 | |||||
| Unknown☆ | 44 | 52 | 78 | 174 | |||||
| TOPIIα | 0.223 | 0.088 | 0.317 | 0.308 | 0.818 | ||||
| (−)~(+) | 41 | 54 | 96 | 191 | |||||
| (++)~(+++) | 11 | 29 | 38 | 78 | |||||
| Unknown☆ | 5 | 4 | 11 | 20 | |||||
| P53 | 0.660 | 0.366 | 0.607 | 0.585 | 0.902 | ||||
| (−)~(+) | 26 | 36 | 63 | 125 | |||||
| (++)~(+++) | 24 | 46 | 69 | 139 | |||||
| Unknown☆ | 7 | 5 | 13 | 25 | |||||
| Ki67 | 0.875 | 0.922 | 0.746 | 0.619 | 0.612 | ||||
| (−)~(+) | 3 | 5 | 6 | 14 | |||||
| (++)~(+++) | 51 | 79 | 129 | 259 | |||||
| Unknown☆ | 3 | 3 | 10 | 16 | |||||
| TOPIIα index (%) | 19 (0–70) | 27 (0–90) | 25 (0–90) | 20 (0–90) | 0.015 | 0.007 | 0.010 | 0.635 | 0.322 |
| P53 index (%) | 38 (0–95) | 46 (0–95) | 42 (0–95) | 45 (0–95) | 0.311 | 0.138 | 0.430 | 0.323 | 0.765 |
| Ki67 index (%) | 68 (5–90) | 69 (0–100) | 69 (10–90) | 70 (0–100) | 0.651 | 0.439 | 0.878 | 0.412 | 0.622 |
#RSCC: Right-sided colon cancer; ▴LSCC: Left-sided colon cancer; *P1: comparison of three group; P2: RSCC vs. LSCC; P3: RSCC vs. rectal; P4: LSCC vs. rectal; P5:colon vs. rectal; ☆All of the “Unknown” groups were not taken into analysis in this table.
Figure 3.
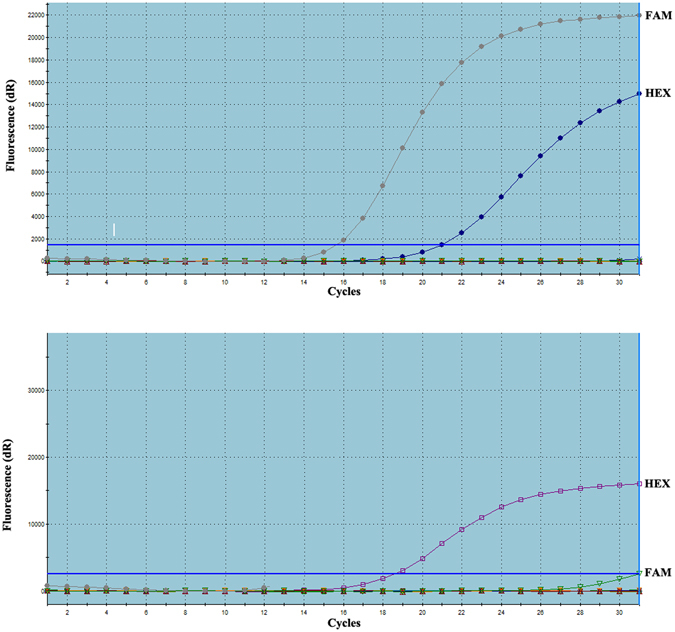
Graphic representations of gene mutation detection of KRAS (A, FAM(+)HEX(+), mutant type) and BRAF (B, FAM(−)HEX(+), wild type). (HEX: Internal reference sample, FAM: Test sample).
Influencing factors of Progression Free Survival (PFS)
Our results showed that tumor location was related with PFS (Fig. 4, P = 0.002). The PFS of RSCC was significantly shorter than that of LRCC (P = 0.003) and rectal cancer (P = 0.004). No significant difference was identified in PFS between LSCC and rectal cancer patients (P = 0.392).
Figure 4.
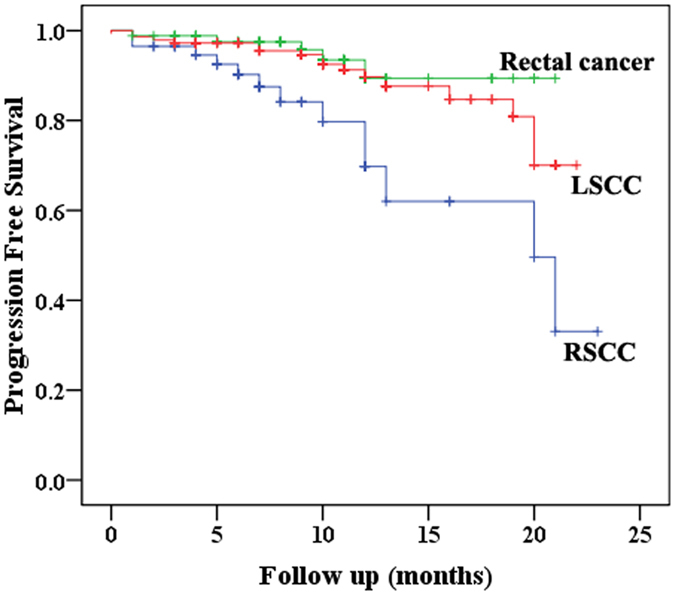
The relationship between tumor location and Progression Free Survival (PFS, P = 0.002) in 289 cases of Stage I~IV colorectal cancer patients (RSCC: Right-Sided Colon Cancer; LSCC: Left-Sided Colon Cancer).
By univariate analysis, RSCC (P = 0.018), preoperative chemoradiotherapy (P = 0.010), poor differentiation (P = 0.001), advanced T stage (P = 0.018), lymph node metastasis (P = 0.001), distant metastasis (P < 0.001), advanced TNM stage (P < 0.001), elevated serum CEA level (P = 0.035), elevated serum CA19-9 level (P = 0.025), tumor deposit (P = 0.001), perineural invasion (P = 0.003) and vascular invasion (P = 0.001) were all found to be associated with shorter PFS (Table 5). Multivariate analyses only identified poor differentiation (P = 0.048) and advanced TNM stage (P < 0.001) as predictor of shorter PFS (Table 6).
Table 5.
Univariate Analyses of Factors Associated With Progression Free Survival (n = 289).
| Variables | 95%CI | P |
|---|---|---|
| Tumor Position (Right vs. Left&Rectum)# | 0.612 (0.407–0.919) | 0.018 |
| Gender (Male vs. Female) | 1.428 (0.720–2.833) | 0.308 |
| Age (<65 y vs. >=65 y) | 1.098 (0.549–2.196) | 0.791 |
| Preoperative chemotherapy (No vs. Yes) | 3.295 (1.332–8.153) | 0.010 |
| Preoperative radiation (No vs. Yes) | 0.048 (0–1858.733) | 0.573 |
| Combined Resection (No vs. Yes) | 2.087 (0.863–5.046) | 0.102 |
| Open or Laparoscopic (Open vs. Laparoscopic) | 0.611 (0.275–1.357) | 0.226 |
| Gross Type (Protruding vs. (Ulcerative, Infiltratie)) | 1.919 (0.581–6.342) | 0.285 |
| Circumference (<1/2 vs. >=1/2) | 1.037 (0.925–1.162) | 0.538 |
| Differentiation (Well&Moderate vs. Poor) | 3.161 (1.560–6.407) | 0.001 |
| T stage ((T1-T2) vs. (T3-T4)) | 5.586 (1.338–23.317) | 0.018 |
| N (N0 vs. (N1-N2)) | 3.586 (1.673–7.686) | 0.001 |
| M (M0 vs. M1) | 4.356 (2.211–8.579) | 0.000 |
| TNM ((1–2) vs. (3–4)) | 2.673 (1.775–4.026) | 0.000 |
| CEA (<5ng/mL vs. >=5ng/mL) | 2.007 (1.051–3.833) | 0.035 |
| CA199 (<37 U/mL vs. >=37 U/mL) | 2.225 (1.106–4.473) | 0.025 |
| KRAS (Wild vs. Mutant) | 0.945 (0.842–1.061) | 0.340 |
| BRAF (Wild vs. Mutant) | 0.954 (0.856–1.064) | 0.397 |
| Tumor Deposit (No vs. Yes) | 3.270 (1.659–6.445) | 0.001 |
| Perineural Invasion (No vs. Yes) | 2.862 (1.414–5.789) | 0.003 |
| Vascular Invasion (No vs. Yes) | 3.166 (1.556–6.444) | 0.001 |
| MLH1 (Defect vs. Normal) | 0.966 (0.837–1.114) | 0.619 |
| MSH2 (Defect vs. Normal) | 0.934 (0.785–1.111) | 0.438 |
| MSH6 (Defect vs. Normal) | 0.905 (0.760–1.078) | 0.262 |
| PMS2 (Defect vs. Normal) | 0.907 (0.755–1.090) | 0.298 |
| β-Tubulin III ((−)~(+) vs. (++)~(+++)) | 1.016 (0.928–1.111) | 0.735 |
| TOPIIα ((−)~(+) vs. (++)~(+++)) | 0.930 (0.748–1.158) | 0.518 |
| P53 ((−)~(+) vs. (++)~(+++)) | 0.938 (0.774–1.137) | 0.514 |
| Ki67 ((−)~(+) vs. (++)~(+++)) | 1.001 (0.811–1.237) | 0.991 |
#Right-sided colon cancer vs. Left -sided colon cancer & Rectal cancer.
Table 6.
Multivariate Analyses of Factors Associated With Progression Free Survival (n = 289).
| Variables | 95%CI | P |
|---|---|---|
| Tumor Position (Right vs. Left &Rectum)# | 0.646 (0.415–1.006) | 0.053 |
| Preoperative chemotherapy (No vs. Yes) | 1.881 (0.553–6.397) | 0.312 |
| Differentiation (Well& Moderate vs. Poor) | 2.066 (1.005–4.247) | 0.048 |
| T stage ((T1-T2) vs. (T3-T4)) | 0.622 (0.105–3.692) | 0.601 |
| N (N0 vs. (N1-N2)) | 0.470 (0.124–1.789) | 0.268 |
| M (M0 vs. M1) | 0.287 (0.052–1.591) | 0.153 |
| TNM ((1–2) vs. (3–4)) | 2.484 (1.640–3.763) | 0.000 |
| CEA (<5 ng/mL vs. >=5 ng/mL) | 1.709 (0.846–3.450) | 0.135 |
| CA199 (<37 U/mL vs. >=37 U/mL) | 1.183 (0.552–2.535) | 0.665 |
| Tumor Deposit (No vs. Yes) | 2.098 (0.870–5.058) | 0.099 |
| Perineural Invasion (No vs. Yes) | 1.484 (0.700–3.144) | 0.303 |
| Vascular Invasion (No vs. Yes) | 1.474 (0.653–3.331) | 0.351 |
#Right-sided colon cancer vs. Left -sided colon cancer & Rectal cancer.
Discussion
The difference among RSCC, LSCC and rectal cancer patients has been remaining a serious debate for a long time. Our comparative study showed that these three groups had similar baseline in most clinicopathological characteristics. But there were still significant differences in surgical procedure (Open/Laparoscopic), tumor diameter, differentiation, invasion depth (T) and TNM stage among the three groups. Compared with LCRC, RSCC had less laparoscopic resection, larger tumor size and poor differentiation. As far as T stage and TNM stage were concerned, RSCC was also found to have similar tumor stage compared with LSCC in our study, which might have something to do with the small sample size. Our results were consistent with that in the published literature, which reported that RSCC had more advanced tumor stages, increased tumor sizes and poorly differentiation3, 11–14. Due to the larger bowel lumen, RSCC usually becomes symptomatic later than LSCC, which in turn leads to later diagnosis, larger tumor size and advanced tumor stage37, 38. Secondly, RSCC is located far away from the anal verge, so it is more difficult to be discovered by digit rectal examination and sigmoidoscopy. Hugen et al.39 reported that the frequency of mucinous and signet-ring cell tumors was higher in RSCC (45%) than in that in LCRC (20%). It was consistent with our results, but the underlying cause for poorer differentiation in RSCC is still unknown. Some oncologists hypothesized that it could be attributed to different underlying genetic and biological features20, 40. The reported differences in age, gender, perineural invasion and vascular invasion by other authors37 was not found in our study, which may be attributed to the relatively small sample size. In our study, RSCC was less likely to undergo laparoscopic resection, which may have something to do with the relatively higher difficulty of laparoscopic right hemicolectomy.
It had been hypothesized that there were significant differences in molecular features between RSCC and LSCC, which might serve as the cause of clinicopathological differences40. RSCC was reported to have a higher frequency of KRAS mutation than LSCC (57.3% vs 40.4%; P < 0.0001)41, and a higher incidence of BRAF mutation with 18.4–22.4% in RSCC and 1.3–7.8% in LCRC42. But RSCC had also been reported to be associated with more mutant KRAS and more wild-type BRAF tumors19. But no significant difference was found in KRAS and BRAF mutation in our study. Similarly, except for the TOPIIα immunostaining index, no significant difference was found in protein expression levels of MLH1, MSH2, MSH6, PMS2, β-tubulin III, P53, Ki67 and TopIIα between RSCC and LSCC. Our results were consistent with that of Zhu et al.20 and Cancer Genome Atlas Network43. Zhu et al. compared gene expression profiling of RSCC and LSCC using the Human Genome Array gene chip in 100 cases of patients, but only 11 genes were identified to be differentially expressed between RSCC and LSCC20. Cancer Genome Atlas Network conducted a genome-scale analysis of 276 samples, analyzing exome sequence, DNA copy number, promoter methylation, mRNA and microRNA expression, and concluded colon and rectal cancers had similar patterns of genomic alteration43. No significant relationship was found between p53 protein expression and tumor position in our study, which was consistent with the results of Ghavam-Nasiri30. The reported prognostic role of P53, Ki67 and TopIIα expression in the literature is conflicting44–46. It confirmed our results, which showed none of β-tubulin III, P53, Ki67 and TopIIα expression level was not prognostic factor in CRC.
Our study demonstrated that RSCC patients had shorter PFS compared with LCRC patient. Our result was consistent with that in most literature. Petrelli et al.47 conducted a systematic review including 66 studies and 1,437,846 patients, and found that RSCC had shorter OS than LSCC. And Lee’s systematic review showed that RSCC had shorter OS than LCRC24. He also suggested that CRC should be classified into two types: the RSCC and the LCRC24.
Since the follow-up is relatively short and few patients died in this period, so it is unreasonable for us to compare OS. In addition, since combined resections of metastatic lesions were performed in only 29 of the 60 stage IV patients with distant metastasis, so the Disease Free Survival (DFS) can’t be applied in the other 31 patients with residual tumor. For these reasons, we chose PFS to measure survival in our study. We proved that RSCC patients had longer PFS than LCRC patients, but no difference between LSCC and rectal cancer. In univariate analysis, tumor position (RSCC vs. LSCC&Rectal), preoperative chemoradiotherapy, poor differentiation, TNM stage, serum CEA and CA19-9 level, tumor deposit, perineural and vascular invasion were all found to be predictor of PFS. Petrelli et al. summarized that TNM stage, differentiation and vascular invasion were well-recognized risk factors for CRC47. Mutation of KRAS and BRAF had also been reported to predict poor survival in CRC patients13, 48, 49, but no similar finding was identified in our study which may be related with relatively small sample size and short follow up period. NRAS mutations (codons 12, 13 and 61) were reported to occur in 3–5% of colorectal cancer48, 49. NRAS mutation was a predictive factor of response to anti-EGFR biological therapy and OS49. In our study, only 9 of the included 289 colorectal cancer patients had undergone the NRAS tests. It is difficult to reach statistical significance with such a small sample size, so NRAS gene mutation status was not analyzed in this study. A meta-analysis of FIRE-3, SWOG 80405 and PEAK trials indicated that RAS wild LSCC patients benefited more from anti-EGFR treatment (P < 0.001), while RSCC patients benefit more from anti-VEGF treatment (P > 0.05)50. Since no significant difference was identified in postoperative biological therapy in our study, it might not be a major contributing factor to survivals of our patients.
In the following multivariate analysis, only differentiation and TNM stage were found to be independent predictor of PFS. It indicated that tumor position might not be an independent influencing factor of PFS. Many authors also found that no difference existed in survival between RSCC and LSCC after adjusting for various variables11, 18, 19. In terms of these phenomena, we suggest the survival difference between RSCC and LCRC would be caused by differences in tumor stage. The RSCC’s larger lumen and longer distance from anal verge makes it less symptomatic and difficult to be discovered, which then leads to advanced tumor stage and poor prognosis38.
In conclusion, RSCC patients have larger tumor size, poor differentiation, advanced TNM stage and poor survival, compared with LCRC patients. Except for TOPIIα immunostaining index, no significant difference was identified in the expression levels of MLH1, MSH2, MSH6, PMS2, β-tubulin III, P53, Ki67 and TopIIα, and gene mutation of KRAS and BRAF. The shorter survival in RSCC might be attributed to the advanced tumor stage caused by its inherent position feature of proximal colon.
Acknowledgements
This study was supported in part by grants from the National Natural Science Foundation of China (#81572358, #81572332, and #81201936).
Author Contributions
Gao X.H., Yu G.Y., Gong H.F., Liu L.J., Hao L.Q., Liu P. and Liu Z.H. collected the data. Xu Y. and Bai C.G. evaluated the expression level. Gao X.H. performed statistical analyses. Gao X.H., Yu G.Y. and Gong H.F. drafted the manuscript. Zhang W. and Bai C.G. conceived and designed the study.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Xian Hua Gao, Guan Yu Yu and Hai Feng Gong contributed equally to this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chen Guang Bai, Email: bcg709@126.com.
Wei Zhang, Email: weizhang2000cn@163.com.
References
- 1.Torre LA, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Frattini M, et al. Different genetic features associated with colon and rectal carcinogenesis. Clin Cancer Res. 2004;10:4015–4021. doi: 10.1158/1078-0432.CCR-04-0031. [DOI] [PubMed] [Google Scholar]
- 3.Tamas K, et al. Rectal and colon cancer: Not just a different anatomic site. Cancer Treat Rev. 2015;41:671–679. doi: 10.1016/j.ctrv.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 4.van der Sijp MP, et al. Differences between colon and rectal cancer in complications, short-term survival and recurrences. Int J Colorectal Dis. 2016;31:1683–1691. doi: 10.1007/s00384-016-2633-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goto S, et al. Differences in surgical site infection between laparoscopic colon and rectal surgeries: sub-analysis of a multicenter randomized controlled trial (Japan-Multinational Trial Organization PREV 07-01) Int J Colorectal Dis. 2016;31:1775–1784. doi: 10.1007/s00384-016-2643-1. [DOI] [PubMed] [Google Scholar]
- 6.Doumouras AG, Tsao MW, Saleh F, Hong D. A population-based comparison of 30-day readmission after surgery for colon and rectal cancer: How are they different? J Surg Oncol. 2016;114:354–360. doi: 10.1002/jso.24334. [DOI] [PubMed] [Google Scholar]
- 7.Harriss DJ, et al. Lifestyle factors and colorectal cancer risk (2): a systematic review and meta-analysis of associations with leisure-time physical activity. Colorectal Dis. 2009;11:689–701. doi: 10.1111/j.1463-1318.2009.01767.x. [DOI] [PubMed] [Google Scholar]
- 8.Bosset JF, et al. Chemotherapy with preoperative radiotherapy in rectal cancer. N Engl J Med. 2006;355:1114–1123. doi: 10.1056/NEJMoa060829. [DOI] [PubMed] [Google Scholar]
- 9.Lee YC, Lee YL, Chuang JP, Lee JC. Differences in survival between colon and rectal cancer from SEER data. PLoS One. 2013;8:e78709. doi: 10.1371/journal.pone.0078709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature487, 330–337 (2012). [DOI] [PMC free article] [PubMed]
- 11.Hansen IO, Jess P. Possible better long-term survival in left versus right-sided colon cancer - a systematic review. Dan Med J. 2012;59:A4444. [PubMed] [Google Scholar]
- 12.Warschkow R, et al. Better survival in right-sided versus left-sided stage I - III colon cancer patients. BMC Cancer. 2016;16:554. doi: 10.1186/s12885-016-2412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinicrope FA, et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology. 2015;148:88–99. doi: 10.1053/j.gastro.2014.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nawa T, et al. Differences between right- and left-sided colon cancer in patient characteristics, cancer morphology and histology. J Gastroenterol Hepatol. 2008;23:418–423. doi: 10.1111/j.1440-1746.2007.04923.x. [DOI] [PubMed] [Google Scholar]
- 15.Yahagi M, Okabayashi K, Hasegawa H, Tsuruta M, Kitagawa Y. The Worse Prognosis of Right-Sided Compared with Left-Sided Colon Cancers: a Systematic Review and Meta-analysis. J Gastrointest Surg. 2016;20:648–655. doi: 10.1007/s11605-015-3026-6. [DOI] [PubMed] [Google Scholar]
- 16.Benedix F, et al. Comparison of 17,641 patients with right- and left-sided colon cancer: differences in epidemiology, perioperative course, histology, and survival. Dis Colon Rectum. 2010;53:57–64. doi: 10.1007/DCR.0b013e3181c703a4. [DOI] [PubMed] [Google Scholar]
- 17.Suttie SA, et al. Outcome of right- and left-sided colonic and rectal cancer following surgical resection. Colorectal Dis. 2011;13:884–889. doi: 10.1111/j.1463-1318.2010.02356.x. [DOI] [PubMed] [Google Scholar]
- 18.Weiss JM, et al. Mortality by stage for right- versus left-sided colon cancer: analysis of surveillance, epidemiology, and end results–Medicare data. J Clin Oncol. 2011;29:4401–4409. doi: 10.1200/JCO.2011.36.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brule SY, et al. Location of colon cancer (right-sided versus left-sided) as a prognostic factor and a predictor of benefit from cetuximab in NCIC CO.17. Eur J Cancer. 2015;51:1405–1414. doi: 10.1016/j.ejca.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Zhu H, et al. Screening for differentially expressed genes between left- and right-sided colon carcinoma by microarray analysis. Oncol Lett. 2013;6:353–358. doi: 10.3892/ol.2013.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuramochi H, et al. PTEN mRNA expression is less pronounced in left- than right-sided colon cancer: a retrospective observational study. BMC Cancer. 2016;16:366. doi: 10.1186/s12885-016-2400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Engeland M, Derks S, Smits KM, Meijer GA, Herman JG. Colorectal cancer epigenetics: complex simplicity. J Clin Oncol. 2011;29:1382–1391. doi: 10.1200/JCO.2010.28.2319. [DOI] [PubMed] [Google Scholar]
- 23.Minoo P, Zlobec I, Peterson M, Terracciano L, Lugli A. Characterization of rectal, proximal and distal colon cancers based on clinicopathological, molecular and protein profiles. Int J Oncol. 2010;37:707–718. doi: 10.3892/ijo_00000720. [DOI] [PubMed] [Google Scholar]
- 24.Lee GH, et al. Is right-sided colon cancer different to left-sided colorectal cancer? - A systematic review. Eur J Surg Oncol. 2015;41:300–308. doi: 10.1016/j.ejso.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X, et al. Class III beta-Tubulin in Colorectal Cancer: Tissue Distribution and Clinical Analysis of Chinese Patients. Med Sci Monit. 2016;22:3915–3924. doi: 10.12659/MSM.901252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lleonart ME, et al. Microsatellite instability and p53 mutations in sporadic right and left colon carcinoma: different clinical and molecular implications. Cancer. 1998;83:889–895. doi: 10.1002/(SICI)1097-0142(19980901)83:5<889::AID-CNCR13>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 27.Xi HQ, et al. Expression and clinicopathologic significance of TUFM and p53 for the normal-adenoma-carcinoma sequence in colorectal epithelia. World J Surg Oncol. 2017;15:90. doi: 10.1186/s12957-017-1111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rambau PF, Odida M, Wabinga H. p53 expression in colorectal carcinoma in relation to histopathological features in Ugandan patients. Afr Health Sci. 2008;8:234–238. [PMC free article] [PubMed] [Google Scholar]
- 29.Paluszkiewicz P, Berbec H, Pawlowska-Wakowicz B, Cybulski M, Paszkowska A. p53 protein accumulation in colorectal cancer tissue has prognostic value only in left-sided colon tumours. Cancer Detect Prev. 2004;28:252–259. doi: 10.1016/j.cdp.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Ghavam-Nasiri MR, Rezaei E, Ghafarzadegan K, Seilanian-Toosi M, Malekifard H. Expression of p53 in colorectal carcinoma: correlation with clinicopathologic features. Arch Iran Med. 2007;10:38–42. [PubMed] [Google Scholar]
- 31.Zhang L, et al. Partial Oxygen Pressure Affects the Expression of Prognostic Biomarkers HIF-1 Alpha, Ki67, and CK20 in the Microenvironment of Colorectal Cancer Tissue. Oxid Med Cell Longev. 2016;2016:1204715. doi: 10.1155/2016/1204715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li P, et al. Association between Ki67 Index and Clinicopathological Features in Colorectal Cancer. Oncol Res Treat. 2016;39:696–702. doi: 10.1159/000450623. [DOI] [PubMed] [Google Scholar]
- 33.Gao XH, et al. DNA topoisomerase II alpha: a favorable prognostic factor in colorectal caner. Int J Colorectal Dis. 2012;27:429–435. doi: 10.1007/s00384-011-1346-x. [DOI] [PubMed] [Google Scholar]
- 34.Paraf F, et al. MLH1 and MSH2 protein immunohistochemistry is useful for detection of hereditary non-polyposis colorectal cancer in young patients. Histopathology. 2001;39:250–258. doi: 10.1046/j.1365-2559.2001.01203.x. [DOI] [PubMed] [Google Scholar]
- 35.Gao XH, et al. Expression of ZNF148 in different developing stages of colorectal cancer and its prognostic value: a large Chinese study based on tissue microarray. Cancer. 2013;119:2212–2222. doi: 10.1002/cncr.28052. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, et al. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: analysis of 1,110 cases. Sci Rep. 2015;5:18678. doi: 10.1038/srep18678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hussain M, et al. Right-Sided and Left-Sided Colon Cancers are Two Distinct Disease Entities: an Analysis of 200 Cases in Pakistan. Asian Pac J Cancer Prev. 2016;17:2545–2548. [PubMed] [Google Scholar]
- 38.Christodoulidis G, et al. Clinicopathological differences between right- and left-sided colonic tumors and impact upon survival. Tech Coloproctol. 2010;14(Suppl 1):S45–47. doi: 10.1007/s10151-010-0622-z. [DOI] [PubMed] [Google Scholar]
- 39.Hugen N, van de Velde CJ, de Wilt JH, Nagtegaal ID. Metastatic pattern in colorectal cancer is strongly influenced by histological subtype. Ann Oncol. 2014;25:651–657. doi: 10.1093/annonc/mdt591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen H, et al. Different treatment strategies and molecular features between right-sided and left-sided colon cancers. World J Gastroenterol. 2015;21:6470–6478. doi: 10.3748/wjg.v21.i21.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tong JH, et al. Characterization of rare transforming KRAS mutations in sporadic colorectal cancer. Cancer Biol Ther. 2014;15:768–776. doi: 10.4161/cbt.28550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishida H, et al. Clinical significant of semiquantificating DNA topoisomerase- I mRNA in colorectal cancer. Gan To Kagaku Ryoho. 2005;32:1295–1299. [PubMed] [Google Scholar]
- 43.Horvath B, et al. Overexpression of p53 predicts colorectal neoplasia risk in patients with inflammatory bowel disease and mucosa changes indefinite for dysplasia. Gastroenterol Rep (Oxf) 2015;3:344–349. doi: 10.1093/gastro/gov022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang P, et al. The prognostic value of p53 positive in colorectal cancer: A retrospective cohort study. Tumour Biol. 2017;39:1010428317703651. doi: 10.1177/1010428317703651. [DOI] [PubMed] [Google Scholar]
- 45.Coss A, et al. Increased topoisomerase IIalpha expression in colorectal cancer is associated with advanced disease and chemotherapeutic resistance via inhibition of apoptosis. Cancer Lett. 2009;276:228–238. doi: 10.1016/j.canlet.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 46.Melling N, et al. High Ki67 expression is an independent good prognostic marker in colorectal cancer. J Clin Pathol. 2016;69:209–214. doi: 10.1136/jclinpath-2015-202985. [DOI] [PubMed] [Google Scholar]
- 47.Petrelli, F. et al. Prognostic Survival Associated With Left-Sided vs Right-Sided Colon Cancer: A Systematic Review and Meta-analysis. JAMA Oncol (2016). [DOI] [PubMed]
- 48.Foltran L, et al. Prognostic role of KRAS, NRAS, BRAF and PIK3CA mutations in advanced colorectal cancer. Future Oncol. 2015;11:629–640. doi: 10.2217/fon.14.279. [DOI] [PubMed] [Google Scholar]
- 49.Schirripa M, et al. Role of NRAS mutations as prognostic and predictive markers in metastatic colorectal cancer. Int J Cancer. 2015;136:83–90. doi: 10.1002/ijc.28955. [DOI] [PubMed] [Google Scholar]
- 50.Holch JW, Ricard I, Stintzing S, Modest DP, Heinemann V. The relevance of primary tumour location in patients with metastatic colorectal cancer: A meta-analysis of first-line clinical trials. Eur J Cancer. 2017;70:87–98. doi: 10.1016/j.ejca.2016.10.007. [DOI] [PubMed] [Google Scholar]


