Abstract
Sheath blight, caused by the necrotrophic fungal pathogen Rhizoctonia solani, is a serious and destructive disease of the rice. In order to improve sheath blight resistance, we developed three different kinds of transgenic rice lines. The first transgenic line overexpresses the rice chitinase gene (OsCHI11); the second contains the Arabidopsis NPR1 (AtNPR1) gene and, the third has pyramided constructs with both the genes (OsCHI11 and AtNPR1). This is a comparative study between the single-gene transgenic lines and the double gene transgenic in terms of their ability to activate the plant defense system. Rice plants of each individual construct were screened via PCR, Southern hybridization, activity assays, and expression analysis. The best transgenic lines of each construct were chosen for comparative study. The fold change in qRT-PCR and activity assays revealed that the pyramided transgenic rice plants show a significant upregulation of defense-related genes, PR genes, and antioxidant marker genes as compared to the single transgene. Simultaneous co-expression of both the genes was found to be more efficient in tolerating oxidative stress. In R. solani (RS) toxin assay, mycelial agar disc bioassay, and in vivo plant bioassay, pyramided transgenic plant lines were more competent at restricting the pathogen development and enhancing sheath blight tolerance as compared to single gene transformants.
Introduction
Rice (Oryza sativa L.), being the most important staple food crop, is a major nutritional food supplement for more than half of the global population. One of the major constraints of rice productivity is the frequent occurrence of the diseases caused by fungi, bacteria, and viruses. The three major diseases that affect the rice crop are blast (causative agent: Magnaporthe grisea), sheath blight (causative agent: Rhizoctonia solani), and bacterial leaf blight (Xanthomonas oryzae pv. Oryzae)1. When considered in combination, these three diseases can account for an annual loss of as much as 50% of rice productivity2.
Sheath blight, caused by the fungus Rhizoctonia solani AGI-IA, is the second most devastating disease of rice3. This disease causes an annual yield loss of around 10–25%4. The damage caused by the sheath blight fungus includes a rapid decrease in chlorophyll content and the loss of photosynthetically active area. This occurs due to the development of lesions on both leaf blade as well as sheaths5. In addition, the array of lytic enzymes and toxins secreted by this necrotrophic fungal pathogen causes an alteration in the photosynthetic and respiratory processes of the green tissues6. The management of rice sheath blight is difficult due to the absence of the desired level of resistance in cultivated and wild rice. Resistance is conferred to rice solely by the presence of the non-race-specific resistance QTL factor; however, no R genes, corresponding to R. solani have been discovered to date7. Hence, it is imperative to develop alternative strategies for the development of durable and broad-spectrum resistance to sheath blight. Genetic engineering is considered as a promising alternative tool for enhancing the tolerance of rice plants toward sheath blight disease.
Systemic Acquired Resistance (SAR) is an innate plant defense strategy, where the expression of the defense genes increases; this enhances resistance toward pathogens8. SAR has been characterized in the model plant Arabidopsis thaliana, and it has been documented that the accumulation of pathogenesis-related (PR) proteins confers broad-spectrum protection against a wide variety of microorganisms9. Arabidopsis NPR1 gene encodes a key SAR regulatory component10. The over-expression of NPR1 in Arabidopsis showed that the NPR1 protein accumulates in the cytosol as an inactive multimeric protein complex11. The changes in the redox level during SA (Salicylic Acid) treatment or pathogen infection causes this protein complex to break into a monomeric form that is then transported to the nucleus where it interacts with the TGA family of basic leucine zipper transcription factors12. This interaction induces the expression of various PR genes13. In several crop plants such as tomato, tobacco, and rice14–18, AtNPR1 or its orthologs have been found to be associated with the phenomenon of conferring enhanced resistance against a wide variety of pathogens. These observations strongly suggest that AtNPR1 and its homologs could serve as important candidates for providing resistance against rice sheath blight.
Chitin, a ß-(1, 4) linked unit of amino sugar, constitutes about 3–60% of the cell wall in various fungi19. Chitinase catalyzes the hydrolysis of the ß-(1, 4) linkages of chitin. The inhibitory action of this enzyme is directly linked to a balloon-like swelling followed by the thinning of the growing hyphal tips leading to bursting of fungal hyphae. In addition, the fungal cell wall degradation products, which are generated in the process, especially the monomers and oligomers, act as potent elicitors of plant defense20. Among the various plant chitinase genes used in genetic transformation, rice chitinases have been most extensively studied. The resistance to a wide range of plant pathogenic fungi by the constitutive expression of rice chitinase gene has been well documented in various economically important plants such as rice21, grapevine22, and cucumber23. On the basis of aforementioned studies, the inhibition of chitin metabolism in fungi such as R. solani, via the expression of rice chitinase, is a promising strategy for controlling the disease like rice sheath blight.
In the present investigation, our objectives were to (i) generate rice plant lines harboring single as well as the pyramided construct of OsCHI11 and AtNPR1 genes under two different green tissue-specific promoters along with molecular evaluation of the transgenic plants; (ii) to identify and validate the best transgenic line for each construct; (iii) to compare single gene derived transgenic lines with the double gene construct derived transgenic lines using gene expression, histochemical, and biochemical analyses subsequent to the sheath blight infection; (iv) in vitro and whole plant bioassay of transgenic plants for measuring the tolerance level against sheath blight disease; and (v) to evaluate the agronomic and phenotypical parameters of different transgenic lines.
Results
Generation, molecular evaluation, and selection of best transgenic line of individual construct
Fifty-three independent T0 transgenic lines with a total of 140 plants were developed from Agrobacterium-mediated transformation with three different constructs. Hygromycin (50 mg/L) selected putative T0 plants were chosen to advance for T1 generation. On the basis of the phenotype, hygromycin response, and molecular characterization, ten plants containing each construct (i.e., CN, C, and N) independently were further advanced to T2 generation. PCR analysis of the T2 transgenic lines, using partial gene specific primers of OsCHI11 and AtNPR1 genes, revealed amplification of 490 bp and 1.7 kbp fragments, respectively, while no amplification was observed in non-transformed plants [Fig. 1b(I),(II),(III),(IV)]. The integration and transgene copy numbers were further examined by Southern hybridization and the transgenic lines containing a single copy of the transgene for each of the constructs were selected for further evaluation and comparison [Fig. 1c(I),(II),(III)]. The expression and transcript levels of the OsCHI11 and AtNPR1 genes were examined in different transgenic lines as also in non-transformed wild-type control (WT) at 2DPI (days post infection). The qRT-PCR results revealed that the expression levels of OsCHI11 and AtNPR1 genes increased substantially in the CN5-2-1 line (2.67 fold and 7.5 fold, respectively) with respect to the wild type control but the other two lines, i.e., CN4-2-2 (1.21 fold and 4.09 fold respectively) and CN2-5-3 (1.27 fold and 4.70 fold respectively), showed comparatively lower elevation (Fig. 2g and 2h). The differential expression in the transgenic and non-transformed plants were statistically significant (F = 25.60; P = 0.0002 for OsCHI11 and F = 21.90; P = 0.0003 for OsCHI11). Similarly, as compared to WT, the maximum expression of AtNPR1 gene was observed in N4-3-2 (7.76 fold) line whereas the lines N7-9-4 (6.62 fold) and N1-8-7 (4.46 fold) showed relatively lower expression (F = 28.55; P = 0.0001) (Fig. 2d). OsCHI11 gene in the C8-9-1 (2.52 fold) line showed maximum elevation with respect to non-transformed WT but the other two transgenic lines, viz., C5-9-8 (1.87 fold) and C1-2-4 (1.49 fold), showed a comparatively lower expression (F = 7.700; P = 0.0096) (Fig. 2b).
Figure 1.

Diagrammatic representation of gene constructs and molecular evaluation of transgenic rice lines. (a) (I) Schematic representation of T-DNA construct harboring OsCHI11 gene under the green tissue-specific promoter P D54O-544 used for rice transformation. (II) Diagrammatic representation of T-DNA construct AtNPR1 gene placed under the control of maize green tissue-specific PEPC promoter. (III) Diagrammatic representation of the T-DNA construct used to transform mature embryogenic calli of jaldi-13 rice variety. OsCHI11 and AtNPR1 genes under the control of the rice green tissue-specific promoter (P D54O–544) and maize green tissue-specific promoter (PEPC), respectively. (b) (I) PCR analysis of T2 transgenic rice plants with partial gene-specific (OsCHI11) primers which amplified 490 bp product. (II) PCR analysis of T2 transgenic rice plants with partial gene-specific (AtNPR1) primers which amplified 1.7Kbp product. (III) PCR based screening of T2 transgenic and non-transformed WT performed with partial gene specific primer (OsCHI11) which amplified 490 bp product. (IV) PCR analysis performed with partial gene specific primer (AtNPR1) showing amplification of 1.7Kbp product. PC-Positive control and NC- Negative control. (c) (I) Southern blot analysis of T2 transgenic plants (C): genomic DNA digested with SalI restriction enzyme and probed with 1.1 kbp HPT gene fragment. (II) Southern blot analysis T2 transgenic plants (N): genomic DNA digested by EcoRI restriction enzyme and probed with 1.1 kbp HPT gene fragment. (III) Southern hybridization analysis of T2 transgenic plants (C-N): genomic DNA was digested with SalI restriction enzyme and probed with 1.1 kbp HPT (hygromycin phosphotransferase) gene fragment. WT represents wild type.
Figure 2.
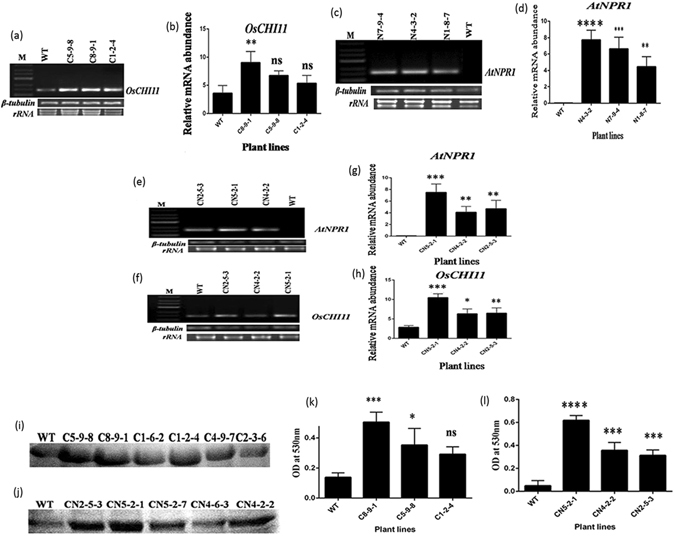
Expression analysis and activity assay of transgenic and wild type plants. (a) Semi quantitative RT-PCR analysis of selected T2 transgenic OsCHI11 overexpressing rice lines using gene specific primers taking β-tubulin as reference control.(b) Relative quantity of rice OsCHI11 mRNA in leaves of T2 (C8-9-1, C5-9-8, C1-2-4) overexpressing and WT rice lines as determined by RT-PCR (Real time PCR). (c) Semi quantitative RT-PCR analysis of selected T2 transgenic AtNPR1 rice lines using gene-specific primers taking β-tubulin as internal control. (d) Relative quantity of Arabidopsis NPR1 mRNA in leaves of T2 (N4-3-2, N7-9-4, C1-8-7) overexpressing and WT rice lines as determined by RT-PCR (Real time PCR). (e) Semi quantitative RT-PCR analysis of selected T2 transgenic OsCHI11-AtNPR1 rice lines using gene-specific (AtNPR1) primer with β-tubulin as internal control. (f) Semi quantitative RT-PCR analysis of selected T2 transgenic OsCHI11-AtNPR1 rice lines using gene-specific (OsCHI11) primer where β-tubulin acts as internal control. (g) Relative abundance of AtNPR1 mRNA in T2 transgenic (CN5-2-1, CN4-2-2, CN2-5-3) pyramided lines and untransformed WT was determined by real-time PCR using SYBR green.(h) Relative quantity of OsCHI11 mRNA in T2 transgenic (CN5-2-1, CN4-2-2, CN2-5-3) rice lines and non-transformed WT was determined real -time PCR using SYBR green. Values represent the mean ± SE of three independent experiments. (i) In-gel chitinase activity assay of T2 transgenic and non-transformed wild type (WT) plants. (j) In-gel chitinase activity assay of T2 pyramided transgenic and non-transformed wild type (WT) plants. (k) Activity assay (‘In-solution’) of chitinase in T2 transgenic and WT plants by dinitrosalycylic acid (DNSA) method. Activity was measured by spectrophotometer with an optical density of 530 nm (OD530). (l) ‘In-solution’ activity assay of chitinase in T2 transgenic and WT plants by dinitrosalycylic acid (DNSA) method. Chitinase activity was measured by spectrophotometer with an optical density of 530 nm (OD530). Values are presented as mean ± SE (n = 3).
With respect to chitinase activity, the C8-9-1 line was the best performer among the lines expressing the C-cassette, while the CN5-2-1 line showed the highest level of chitinase activity among the CN expressing lines (Fig. 2k and 2l). The difference between WT and transgenic lines was statistically significant (p < 0.005 in the case of C8-9-1 and p < 0.0001 in the case of CN5-2-1). A similar outcome was also obtained from the chitinase in-gel activity assay where the C8-9-1 and CN5-2-1 lines exhibited relatively strong lytic activity as compared to the other lines (Fig. 2i,j). The best T2 transgenic lines (C8-9-1, CN5-2-1, and N4-3-2) from each corresponding construct (C, CN, and N) were selected to validate the results further.
Elevated expression of PR genes, SA and JA pathway genes after R. solani infection in transgenic lines
In qRT-PCR analysis, the expression levels of different PR genes [OsPR10a, OsPR1b, RC24 (PR3) and OsPR5], the markers for plant defense responses, were investigated in the CN5-2-1, C8-9-1 and N4-3-2 transgenic lines with respect to the non-transformed control at 24 and 48 h post-inoculation (Fig. 3a,b,c,d). The three JA and SA-dependent signalling pathway genes, phenylalanine ammonia-lyase (OsPAL), allene oxide synthase (AOS) and a rice homolog of Arabidopsis NPRl (OsNHI), along with a chitin-induced phytoalexins producing gene, OsMAPK6, were also investigated. The strongest induction of all these biotic stress marker genes, except for OsPAL, was observed in the CN5-2-1 transgenic line whereas the lines, N4-3-2 and C8-9-1 showed moderate and lower level of induction, respectively (Fig. 3e,f,g,h). The comparison between the different transgenic lines and WT at 24 and 48 h with respect to the relative fold change in qRT-PCR products for different biotic stress marker genes is given in Supplementary Table 3.
Figure 3.
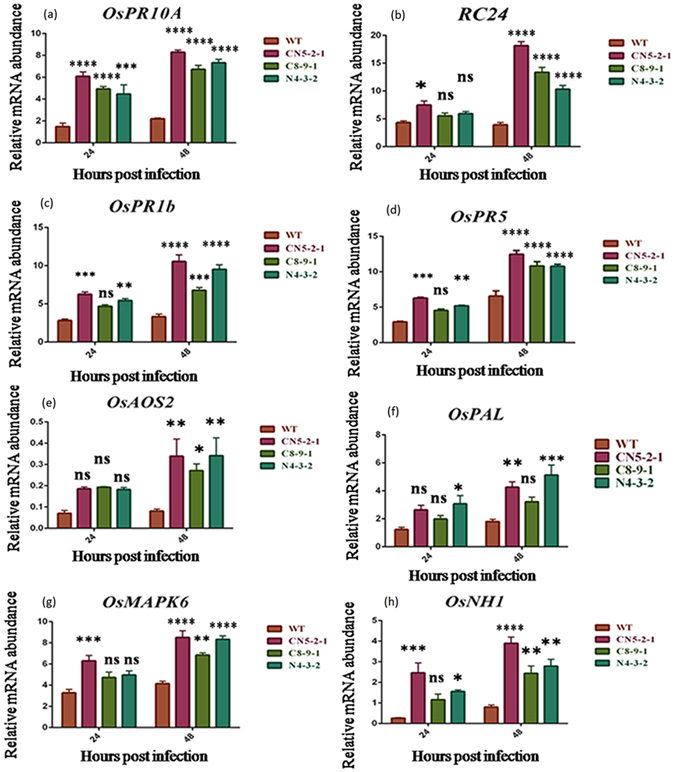
Real-time PCR analysis of some differentially expressed PR, SA and JA pathway genes in single transgene containing as well as pyramided rice lines. Total RNA isolated from leaves of both transgenic T2 and non-transformed WT plant, inoculated with sheath blight fungus R. solani, and harvested at 24 and 48 hpi (hours post infection). Experiment performed by SYBR green-based quantitative real-time PCR, using β-tubulin as internal control. Expression of (a) PR10A, (b) RC24, (c) OsPR1b, (d) OsPR5, (e) OsAOS2, (f) OsPAL, (g) OsMAPK6 and (h) OsNH1 gene in transgenic and wild type plants. Each bar represents the mean ± SE of three independent experiments.
Enhanced expression of different oxidative stress metabolizing enzymes upon R. solani infection in different transgenic lines
Plants execute a complex antioxidative defense system in order to maintain a stable ROS-scavenging system for the maintenance of intracellular ROS homeostasis during stress conditions. Some antioxidant markers, such as SOD, POD, CAT, and APX activity levels, were studied in order to examine the defense responses and biochemical changes triggered by R. solani in the WT and transgenic lines. In contrast to WT, the pathogen inoculation significantly (F = 53.83; P < 0.0001) increased the activity of SOD in all three tested T2 transgenic lines: CN5-2-1, C8-9-1, and N4-3-2 lines (Fig. 4e). Similarly, CAT activity was elevated in all T2 transgenic lines, viz., CN5-2-1, C8-9-1, and N4-3-2 lines with respect to WT (F = 12.86; P = 0.0020) (Fig. 4c). The APX activity under stress increased significantly (F = 17.97; P = 0.0006,) in CN5-2-1, C8-9-1 and N4-3-2 lines when compared with non-transgenic plants (Fig. 4b). A substantial increase in POD activity was also observed in the transgenic lines CN5-2-1, C8-9-1 and N4-3-2 as compared to the infected WT plants (F = 42.15; P < 0.0001) (Fig. 4d). These results indicate that the expression of the corresponding transgene in transgenic rice plants is likely to enhance the activity of the antioxidant enzyme.
Figure 4.

Oxidative damage and activities of different antioxidant enzymes in WT and T2 transgenic rice plants after sheath blight infection. (a) (I) In situ detection of O2− by NBT staining in WT and T2 transgenic lines. (II) In situ detection of H2O2 by DAB staining in WT and T2 transgenic rice lines. (b) APX activity; (c) CAT activity; (d) POD activity; (e) SOD activity; (f) Determination of lipid hydroperoxide content in both transgenic and WT plants; (g) Determination of MDA accumulation in leaves of WT and T2 transgenic rice plant lines. Leaves were collected from transgenic and wild type (WT) plants after 2 days post infection (dpi). Data represent means ± SE calculated from three replicates. Three independent experiments performed.
Transgenic rice plants showed reduced ROS accumulation upon sheath blight infection
The two main sources of ROS, H2O2 and O2 −, were detected in DAB and NBT staining, respectively, in a post-infection scenario. In DAB staining, two days after the inoculation, dark-brown polymeric oxidation products resulting from H2O2 were observed to accumulate in both WT as well as transgenic leaves; however, the leaves of WT exhibited significantly stronger staining as compared to the transgenic lines [Fig. 4a(II)]. Similarly, the blue polymerization product in NBT staining resulting from O2 − accumulation was observed to be substantially higher in WT in comparison to the transgenic lines [Fig. 4a(I)]. Among the transgenic lines, C8-9-1 exhibited the maximum accumulation of ROS and stained deeply with both NBT and DAB whereas the line CN5-2-1 was observed to accumulate the least amount of oxidation product.
Malondialdehyde (MDA), the final product of cellular membrane lipid peroxidation, is considered as a key parameter for measuring the extent of membrane damage in plants. The level of MDA in transgenic plants due to R. solani infection was found to be significantly lower in the CN5-2-1 and N4-3-2 lines as compared to WT at 2 DPI (Fig. 4g). However, in the case of the transgenic line C8-9-1, the difference in MDA content when compared to a non-transgenic control was not statistically significant. The accumulation of lipid hydroperoxide, another marker for membrane damage, was found to be significantly lower (F = 28.75; P = 0.0001) in the CN5-2-1, C8-9-1 and N4-3-2 transgenic lines at 2 DPI as compared to the WT (Fig. 4f).
AtNPR1-OsCHI11 pyramided line exhibits lower sheath blight symptom as compared to that of single-gene transgenic lines
In order to evaluate the level of disease resistance of transgenic lines, we performed both in vitro and in vivo plant bioassay by infecting the plants with sheath blight fungus R. solani. In the detached leaf bioassay conducted using the R. solani mycelial agar disc, all transgenic lines (CN5-2-1, C8-9-1 and N4-3-2) showed significantly (F = 43.96; P < 0.0001) lower disease response when compared with that of wild-type control plant. After three days of inoculation, a stereomicroscope was used to assess disease severity by counting the number of infection cushions (served as an indicator of disease incidence) developed per leaf. The number of infection cushions in the CN5-2-1 line (15) was much lower than that those in the C8-9-1 (30) and N4-3-2 (26) lines; while the maximum number of cushions was found in the wild type (38.66) (Fig. 5a). The size of the lesions on the transgenic leaves was found to be less than on the control plant leaves; this also confirms an effective restriction of pathogen invasion (Fig. 5b). The non-transformed plants showed large yellow colored lesions, which subsequently turned brown after 72 h of the infection. The detached leaf bioassay, using partially purified R. solani toxin, revealed that the transgenic plant lines (CN5-2-1, C8-9-1 and N4-3-2) were able to significantly reduce (F = 141.3; P < 0.0001) the disease severity as can be deduced from the lower percentage of infected leaf area. After five days of incubation, it was noted that the transgenic plant CN5-2-1 line showed lowest percentage of affected area (29.51%) whereas C8-9-1 (62.91%) and N4-3-2 (50.84%) lines displayed moderate level of affected area when compared to the WT control (86.43%) (Fig. 5c).
Figure 5.
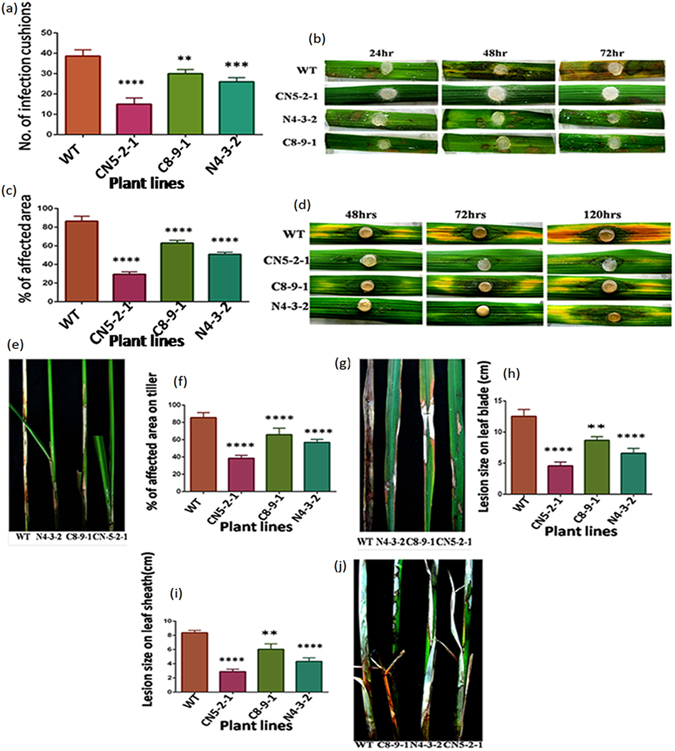
Evaluation of T2 transgenic lines against sheath blight disease along with non-transformed control through In vitro and In vivo plant bioassay. (a) Mycellial agar disc bioassay showing reduced infection cushion formation in T2 transgenic lines (CN5-2-1, N4-3-2, C8-9-1) than in the wild-type (WT). Experiments replicated three times. (b) Representative images of reduced lesion formation in transgenic leaves (CN5-2-1, N4-3-2, and C8-9-1) relative to WT in mycellial agar disc bioassay. (c) Bar diagram showing percentage of affected area after 72 hrs post infection in transgenic leaf samples than wild type (WT) in RS -toxin bioassay. (d) Images showing less affected area in transgenic leaves (CN5-2-1, C8-9-1, and N4-3-2) compared to WT control in the toxin bioassay. (e) Image showing reduced sheath blight symptoms development in rice tillers in T2 transgenic plants, than that in WT control after 21 dpi (Days post infection). (f) Percentage of infected area on tillers on T2 transgenic (CN5-2-1, C8-9-1, and N4-3-2) and WT plants after 21 dpi. (g) Images showing sheath blight symptoms on leaf blade of transgenic (CN5-2-1, C8-9-1, and N4-3-2) and non-transformed control. (h) Lesion size on leaf blade of WT and transgenic lines after 21 dpi. (i) Lesion size on leaf sheath of both WT and transgenic lines (CN5-2-1, C8-9-1, and N4-3-2) after 21 dpi. Values are presented as mean of 10 replicas ± SE. (j) Representative images showing reduction in symptoms development on rice sheath of transgenic lines as compared to WT after 21 dpi.
The highly virulent R. solani AG1-1A (Hyderabad isolate) strain was used for whole plant bioassay of transgenic and wild-type control plants. When compared with the wild type control plants, the highest protection against sheath blight disease was exhibited by the CN5-2-1 transgenic plant line as demonstrated by the percent disease index (PDI), which was the lowest among the three transgenic lines. Prominent lesions appeared within 5–7 days on the wild type control as well as on transgenic plants. We assessed the disease severity by measuring the lesion size on the leaf blade (Fig. 5g) and leaf sheath (Fig. 5j), and by measuring the affected area on tillers (Fig. 5e) at 14 DPI (days post infection). The lesion size on leaf blade (Fig. 5h), sheath (Fig. 5i), and affected area on tillers (Fig. 5f) was observed to be smallest in the CN5-2-1 transgenic line (4.56, 2.89 and 38.53% respectively) as compared to WT control (12.53, 8.37 and 85.54%), whereas other transgenic lines C8-9-1 and N4-3-2 exhibited a moderate level of resistance. In all three cases, the mean differences between transgenic and non-transformed control plants were statistically significant (P < 0.0001). The mean disease scores of transgenic lines were significantly lower (P < 0.0001) than the wild-type control lines at 7, 14, and 21 days post-inoculation (DPI). At 7, 14 and 21 DPI, the CN5-2-1 line displayed the highest level of protection (PDI-11.18%, 27.55% and 34.03%, respectively) among the wild type control (PDI-26.88%, 59.35% and 86.04%) and other transgenic plant lines studied (Fig. 6a,b).
Figure 6.
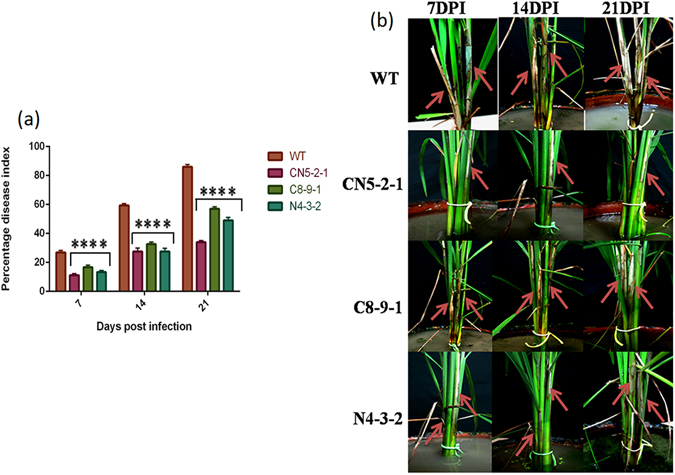
Whole plant bioassay of T2 transgenic and wild type (WT) plants with sheath blight fungus, R. solani. (a) Percent Disease Index (PDI) value in T2 transgenic plants with respect to wild type control at 7, 14 and 21 dpi (days post inoculation). The values represent as the mean ± SE (n = 15). (b) Representative images showing typical sheath blight symptoms development on rice tillers of T2 transgenic (CN5-2-1, C8-9-1, and N4-3-2) and non transformed wild type control. Pictures were taken at 7, 14 and 21 dpi. Red arrows indicate sheath blight symptoms.
Surviving tiller number and grain yield in transgenic plants after sheath blight infection
In order to analyze the physiological and yield related components in transgenic and WT plants after sheath blight infection, the percentage of surviving fertile green tillers and the number of grains per panicle were measured at 30 DPI. The transgenic line CN5-2-1 demonstrated the maximum surviving green tillers (66.83%) as compared to the non-transformed control (18.25%) while the other two lines, C8-9-1 (38.54%) and N4-3-2 (51.21%), displayed comparatively lower tiller count than that of CN5-2-1 (F = 68.59, P < 0.0001) (Fig. 7a,b). Furthermore, all the three transgenic rice lines, i.e., CN5-2-1(80.04%), C8-9-1 (37.45%) and N4-3-2 (53.14%), exhibited significantly higher grain count per panicle [Fig. 7c] (F = 116, P < 0.0001) when compared to the non-transformed control (22.28%), which had the maximum number of unfilled panicles (Fig. 7d). These results clearly demonstrated that the transgenic rice lines provide a significant and durable protection against the sheath blight fungus and enhance the ability to overcome the damage caused by the infection.
Figure 7.
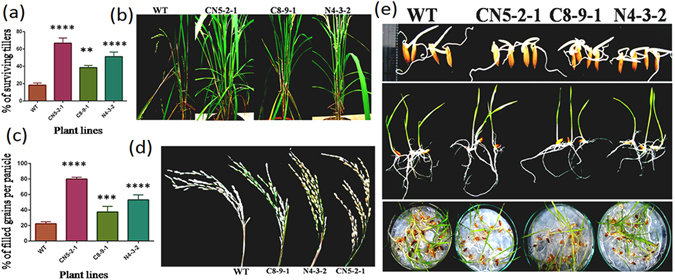
Enhanced resistance of transgenic lines to highly virulent isolates of R. solani and seed germination analysis. (a) Bar diagram showing percentage of survival tillers in T2 transgenic lines (CN5-2-1, C8-9-1, and N4-3-2) compared to wild type (WT) after 21 dpi. Bar represents mean ± SE of three independent experiments. (b) Images showing more number of surviving tillers in T2 transgenic lines (CN5-2-1, C8-9-1, and C4-3-2) compared to WT after 21 dpi. (c) Percentage of filled grains per panicle on transgenic and non-transformed control after 21 dpi. Bar represents mean ± SE of five independent experiments. (d) Images showing more grain count per panicle in transgenic plants after sheath blight infection with respect to WT at 21 dpi. (e) Images showing seed germination and phenotypic evaluation of T3 transgenic and wild type (WT) control seeds at juvenile stage.
Transgene insertion does not alter seed germination or any other agronomically important traits in transgenic rice
The agronomic performance of all three transgenic plants was compared with that of the non-transgenic control. All transgenic and non-transformed plants were observed to grow normally; no significant difference was observed in terms of the number of tillers, plant height, and panicles (Table 1). In addition, the weight of 1000 dry seeds obtained from the transgenic plants was not significantly different from that obtained from the non-transgenic control. We also analyzed the seed germination potentials of T3 transgenic line and WT plants by performing the control germination test (CGT). The transgenic seeds, compared to the non-transformed controls, germinated normally without any morphological or phenotypic alterations (Fig. 7e).
Table 1.
Agronomical data evaluation of transgenic lines.
| Group | Plant height (cm) | Number of tillers | Panicle length (cm) | Panicle per plant | 1000 seed weight (gm) |
|---|---|---|---|---|---|
| WT | 55.43 ± 3.06 | 12.43 ± 1.99 | 16.44 ± 1.03 | 10.25 ± 0.60 | 19.22 ± 0.11 |
| CN5-2-1 | 54.82 ± 5.33 | 17.32 ± 2.31 | 18.65 ± 1.11 | 12.09 ± 0.42 | 20.56 ± 0.12 |
| C8-9-1 | 53.12 ± 3.33 | 16.83 ± 1.11 | 15.03 ± 2.87 | 11.09 ± 0.46 | 18.56 ± 0.19 |
| N4-3-2 | 54.42. ± 4.90 | 13.83 ± 2.98 | 17.11 ± 2.00 | 10.10 ± 0.66 | 20.23 ± 0.17 |
All the essential parameters were measured for the transgenic plants with their wild type (WT) counterpart, grown in same condition in green house. The values represent the mean of progeny (T3) 10 plants from each line ± SE.
Discussion
Sheath blight disease is one of the major threats to rice production. As the food security of approximately half of the world’s population is directly dependent upon rice, improving the resistance in this crop against sheath blight is imperative for sustainable rice production. Breeding for sheath blight resistance has not yet become a realistic option largely because of our inability to identify potent resistant germplasm, major resistant QTL or any other major resistance/defense-related gene. Thus improving host resistance by over-expressing different defense genes derived from rice and non-rice sources is an attractive alternate strategy for combating the pathogen. For developing host plant resistance, single defense gene expression and pyramiding of more than one gene are the common strategies. Pyramiding defense genes is considered superior to monogenic resistance in achieving durable resistance against pathogen concerned. Several studies have reported successful defense gene pyramiding in rice24–27. In the current study, we attempted to enhance sheath blight tolerance by developing transgenic rice lines expressing either the chitinase (OsCHI11) gene [Fig. 1a(I)] or NPR1 (AtNPR1) [Fig. 1a(II)] gene alone and a line containing both the genes (OsCHI11 and AtNPR1) [Fig. 1a(III)]. Chitinase is a well-known antifungal gene; its product confers improved resistance by breaking the fungal cell wall component chitin28. Arabidopsis NPR1 is a master regulator of plant immunity that is known to provide broad spectrum resistance29. The expression of both the genes individually (OsCHI11 and AtNPR1) in rice has been demonstrated to confer enhanced tolerance to sheath blight disease17, 25, 30. The primary objective of our study was to compare transgenic lines expressing either of single genes (OsCHI11 or AtNPR1) with the line that contains both the genes. Three different types of transgenic lines were chosen based on the criteria that they should possess a single copy of their respective construct. C8-9-1 was selected for inclusion in the comparative study on the basis of CHI11 gene expression (Fig. 2a,b) and chitinase enzyme activity (Fig. 2i,k), whereas N4-3-2 was selected on the basis of AtNPR1 gene expression (Fig. 2c,d); the pyramid line CN5-2-1 was chosen on account of exhibiting the highest level of gene expression (Fig. 2e,f,g,h) as well as chitinase enzyme activity (Fig. 2j,l). A variety of parameters were used to compare these three lines so as to evaluate their efficacy in imparting tolerance towards the sheath blight pathogen Rhizoctonia solani.
The expressions of pathogenesis-related (PR) genes in response to invasion by pathogens are one of the most studied defense responses in case of plants31. In comparison to their wild-type counterparts, we observed an elevated level of expression of four different PR genes, viz., PR1b, PR5, PR10a and RC24 in all three transgenic lines at 24 and 48 DPI (Fig. 3a,b,c,d). The expression of PR1b and PR10a was found to be higher in the AtNPR1 expressed line as compared to the CHI11 line while the reverse was true in the case of RC24 expression. Earlier studies have also reported the induction of expression of PR1b, PR10A and PR5 genes in AtNPR1 transgenic rice plants upon pathogen infection17, 32. Upon infection, the over-expressed chitinase is hypothesized to act on the fungal cell wall to generate chitin oligomers which in turn intensifies the infection signaling and defense gene expression33. In all cases, the pyramided line CN5-2-1 was observed to exhibit a faster and stronger activation of endogenous PR genes in response to R. solani infection as compared to N4-3-2 and C8-9-1. The stronger activation of these defense genes in the pyramided line is likely to be an outcome of the synergistic effect of NPR1 and chitinase. The expressions of four other endogenous defense-related genes, viz., PAL, AOS2, NH1, MAPK6 were also found to be more enhanced upon pathogen infection in transgenic lines than that in their wild-type counterpart. Among the transgenic lines, the pyramided line showed the highest accumulation of transcripts of all the above-mentioned genes except for PAL (Fig. 3e,f,g,h). The expression of the AOS2 gene, a key enzyme in JA biosynthetic pathway, was also observed to be induced upon fungal infection. The over-expression of this gene has been linked to an enhanced PR gene expression and increased pathogen resistance34. R. solani is a necrotrophic fungus and it has been well established that JA-dependent defense pathways are conventionally activated in response of necrotroph35. Fungal MAMPs (microbial-associated molecular patterns), such as chitin elicitor, have been found to activate OsMPK6 so as to produce phytoalexin momilactone and phytocassane36. In our study, we found significant up-regulation of the OsMPK6 gene at 48 dpi. The transcripts of the PAL genes, which are related to the SAR pathway induction in the plant, were found to be accumulated more in N4-3-2 than the other two lines. Our result is in concurrence with an earlier study where PAL expression was reported to be enhanced in transgenic rice in response to R. solani infection3.
An increased accumulation of reactive oxygen species (ROS) is a common phenomenon in stress-related conditions37. The elevated activity of antioxidant enzymes is one of the major means by which plants curtail ROS-related damage. For neutralizing superoxide (O2 ·−) and hydrogen peroxide (H2O2), SOD, POD, CAT and APX are the key enzymes responsible. SOD acts as the first line of defense against membrane lipid peroxidation. It dismutases the superoxide radical into less damaging species like O2 or H2O2. POD, CAT and APX act as the next line of defense by detoxifying peroxides into H2O. In the present study, the activity of these antioxidant enzymes was observed to be induced upon R. solani inoculation (Fig. 4a,b,c and d). The antioxidant enzyme activities were similar or higher in the pyramided line compared to the other two lines after sheath blight infection. Our results are in accordance with a previous study which reported higher SOD and POD activity levels in hybrid rice plants infected with R. solani 38. Another study has also presented data that support a significant increase in SOD, CAT, APX, and POD activity in rice leaf sheath in response to R. solani infection39. Lignin or suberin mediated physical barrier formation also plays an important role in limiting pathogen invasion. Peroxidase enzyme helps in cross-linking lignin monomers and increasing plant cell wall rigidity40. As evidenced from DAB and NBT staining, transgenic plants showed less accumulation of ROS than the wild type plants. The pyramided line performed better in terms of lower accumulation of ROS as compared to the other two lines [Fig. 4a(I),(II)]. This result could be correlated with the higher activity of antioxidant enzymes in the pyramided line. Malondialdehyde (MDA) and lipid hydroperoxide are the two products generated by lipid peroxidation in plants. These two products are commonly correlated with the level of membrane damage caused by peroxidation. Interestingly, the pyramided line showed the lowest level of MDA and lipid hydroperoxide accumulation compared with the other two lines as well as wild type control (Fig. 4f,g). These observation concords well with the antioxidant enzyme activity and ROS accumulation data, as higher enzyme activity and lower ROS accumulation, can be directly correlated with lesser MDA and lipid hydroperoxide formation. This result indicates that the pyramided line that co-expresses both genes is better equipped than transgenic lines expressing either AtNPR1 or OsCHI11 to cope with the oxidative stress generated due to pathogen infection. It also has more potential to maintain homeostasis than the two lines studied.
As the next step toward evaluating the three transgenic lines, whether the elevated levels of defense gene expression and ROS homeostasis in the transgenic lines are capable of being translated into increased sheath blight disease resistance was studied. Three independent R. solani infection experiments, two detached leaf bioassays (Fig. 5b,d), and one whole plant bioassay (Fig. 6b) were performed on transgenic plants as well as on the wild-type control plants in order to assess their tolerance level. The bioassays using mycelial agar discs revealed that compared to the wild type, there was a significant reduction in the number of infection cushions in all transgenic lines, where the pyramided line (CN5-2-1) showed the highest level (61.2%) of reduction. Infection cushions develop from the aggregation of convoluted hyphae, which play an important role in direct cuticular penetration3. Therefore, a reduction in the number of infection cushions can be directly co-related to the level of tolerance. In a similar fashion, when RS toxin was applied on pricked leaves of transgenic plants, the pyramided line displayed smallest area of necrosis in contrast to other lines. However, the NPR1 expressing line performed better than the CHI11 expressing line in both the detached leaf bioassays. RS toxin, a host-specific toxin secreted by R. solani, is capable of producing all symptoms of the sheath blight disease41. Sheath blight sensitivity of rice cultivars has been demonstrated to be well correlated with the sensitivity to phytotoxin from R. solani 42. In the same way, whole plant bioassay comprising of analysis of percent disease index, lesion size on leaves, lesion size on sheath and affected area on tiller, established that the pyramided line had a stronger and more durable tolerance toward sheath blight as compared to the single-gene transgenic line (Figs 5g,j,e and 6a). The superior performance of the pyramided line can be attributed to the faster and stronger activation of rice defense genes during pathogen infection. The combination of NPR1 and chitinase in pyramided line was able to instigate the plant defense system to a higher level than either gene could do alone. This fact can also be visualized by the stronger activation of PR genes and other defense-related genes in the pyramided line as compared to the single-gene transgenic lines. An additional contribution to the increased sheath blight tolerance of transgenic lines may derive from the higher expression of MAPK6, which might have contributed to increased accumulation of rice phytoalexins36.
The constitutive expression of defense genes, especially those associated with the regulatory systems, for induced resistance may lead to different types of phenotypic shortcomings43. The expression of AtNPR1 that is under the control of a constitutive promoter has been shown to affect adversely rice plant phenotype32, 44. Although pyramiding of alfalfa β-1,3-glucanase gene (AGLU1) and rice chitinase gene (RCH10) under the control of constitutive promoter conferred tolerance to R. solani, but it had a negative impact on seed germination45. Recently, it has been demonstrated that the tissue-specific expression of AtNPR1 can bypass phenotypic costs while imparting tolerance to sheath blight17. In the present study, two genes of interest under the control of two different green tissue-specific promoters, viz., P D54O-544 3 and P PEPC 25 have been over-expressed in order to avoid any possible phenotypic cost. The analysis of several phenotypic and morphological parameters revealed no significant phenotypic abnormalities in any of the three different transgenic lines (Fig. 7e).
The current study indicates that the synergistic effect of AtNPR1 and OsCHI11 genes in transgenic rice plants leads to a better and improved performance against sheath blight than that of a single individual gene. It is evident from this study that combining a regulatory gene (NPR1) with a functional PR gene (CHI11) leads to the formation of a synergistic effect in rice with superior sheath blight tolerance properties. Stronger defense gene activation, higher activity of antioxidative enzymes, lower ROS accumulation and lesser membrane peroxidation account for the superior performance of pyramided line against sheath blight tolerance. However, for a better understanding, global transcriptome analysis of the transgenic lines is imperative. The strategy used in this study has the potential to be extrapolated to other economically important crop species for improving tolerance against fungal infection levels.
Methods
Cloning and vector construction
The maize PEPC (1.2 kb) (Ac no: x15642.1) promoter was taken out from a pUC19-P PEPC -OsOXO4-NOS vector25 by HindIII-BamHI digestion and inserted at the MCS of pCAMBIA1301 (Gus free)46, to generate pCAMBIA1301-P PEPC. NPR1-NOS fragment was released from pBI-PNN plasmid17 construct by digestion with BamHI-EcoRI and sub-cloned downstream of P PEPC in the pCAMBIA1301-PEPC vector to generate pCAMBIA1301-P PEPC -NPR1-NOS. This vector construct is designated as N.
Another OsCHI11 gene construct was liberated from the vector pUC19-P D54O–544-OsCHI11-NOS 25 by HindIII-EcoRI digestion and inserted at the MCS of pCAMBIA1301 so as to generate pCAMBIA1301-P D54O–544 -OsCHI11-NOS. The name of this vector construct is abbreviated as C.
The vector pUC19-P PEPC -OsOXO4-NOS 25 was digested by BamHI-EcoRI to eliminate the OsOXO4-NOS fragment and an NPR1-NOS fragment released from the pBI-PNN 17 plasmid was subcloned downstream of pUC19-P PEPC to generate pUC19-P PEPC -NPR1-NOS. This construct was again digested with EcoRI and the cassette was inserted into the EcoRI site of pCAMBIA1301-P D54O–544 -OsCHI11-NOS in order to generate pCAMBIA1301-P D54O–544 -OsCHI11-NOS- P PEPC -NPR1-NOS. This construct is abbreviated as C-N.
Rice transformation and analysis of transgenic plants
Rice (Oryza sativa L. subspecies indica cv. Jaldi-13) was transformed through the Agrobacterium-mediated method30 using 20–25 days old embryogenic calli as explants. The transformants were selected on Murashige-Skoog plates47 supplemented with 50 mg/L hygromycin B. The plants were grown in a growth chamber. PCR screening was performed using partial gene-specific primers for the OsCHI11 and AtNPR1 genes (Supplementary Table 1).
Southern hybridization
Total genomic DNA was extracted from greenhouse grown T2 transgenics as well as WT rice plants as per the methodology previously described48. Southern blot analysis was performed as per standard procedures49. A minimum of 15 µg of genomic DNA was digested with SalI (C and C-N constructs) or EcoRI (N construct) by incubating the reactions overnight at 37 °C. Digested DNA was electrophoresed on a 1% agarose gel at 50 V. The gel was denatured, neutralized, and blotted onto nylon membranes (Amersham Pharmacia, USA). Southern blots were hybridized with the partial HPT (hygromycin phosphotransferase) gene fragment, which acted as the probe. This gene fragment was labeled with (α-32P) dCTP using a Deca LabelTMDNA Labeling Kit (Roche) as per standard procedures prescribed by the manufacturer.
RNA isolation and semiquantitative and quantitative reverse transcription (qRT)-PCR
Total RNA was isolated from 100 mg of leaf tissues using a commercially available RNeasy Plant Mini Kit (Qiagen, USA). This was followed by DNaseI (Fermentas, Canada) treatment. Of RNA, 5 µg was reverse-transcribed into cDNA using a Maxima First Strand cDNA Synthesis Kit (Fermentas, Canada) as per the manufacturer’s instructions. qRT-PCR was performed in CFX-96 real-time PCR system (Bio-Rad) using a SYBR Green Kit (Fermentas, Canada). Rice β-tubulin gene was used as an endogenous control for qRT-PCR. A list of primers used in the procedure is given in Supplementary Table 2. Quantitative variation was evaluated by the 2−ΔΔCT method50. The specificity of PCR amplification was verified using a two-pronged strategy: analysis with a heat dissociation curve (65–95 °C) after the PCR procedure as well as by running the amplified products on a 2% agarose gel.
Each experiment was performed in triplicates using three independent tissue samples. Semi-quantitative RT-PCR was performed using 1 µL of cDNA as template. High fidelity Taq polymerase (Fermentas, Canada) was used in the procedure.
Chitinase activity assay
In-solution chitinase activity was determined according to a previously described protocol51 with modifications. The preparation of colloidal chitin has been described earlier52. The incubation of colloidal chitin with total protein was performed in a water bath at 45 °C for 1 h. HCl (100 µL, 1 N) was used to terminate the reaction. After 10 min of incubation on ice, the mixture was centrifuged at 13,000 rpm for 10 min in order to precipitate undigested substrate. The amount of reducing sugar released was measured in a spectrophotometer at 530 nm using dinitrosalicylic acid (DNS) reagent53. ‘In-gel’ activity assay for chitinase was performed as previously described in literature54. Total protein profile was resolved on a 12% discontinuous SDS-PAGE containing 0.05% (w/v) colloidal chitin. After electrophoresis, the gel was transferred into a 200 mM sodium acetate buffer (pH 5) solution containing 1% (v/v) deionized Triton X-100 and incubated in a shaking water bath for 2 h at 37 °C. The gel was stained for 5 min with Calcofluor white M-2R (0.01%) (Fluka, Sigma, USA) dissolved in 50 mM Tris-HCl (pH 8.0) and then washed several times with distilled water. Lytic zones were observed and photographed using a gel documentation system (BioRad, Hercules, California, USA).
Histochemical staining
The generation of superoxide (O2 −) radicals and hydrogen peroxide (H2O2) after sheath blight infection was analyzed through histochemical staining55. For O2 − detection, the leaves from both infected wild type and transgenic plants were floated in 50 mM sodium phosphate buffer (pH7.5) containing 0.2% fresh NBT solution at ambient temperature until the appearance of insoluble dark blue color. For detection of H2O2, the leaves were immersed in fresh DAB solution (1 mg mL−1 pH 3.8) prepared in 10 mM phosphate buffer (pH 7.8), and incubated overnight in light until brown spots were observed. The stained leaves were bleached in 95% ethanol in a boiling water bath and stored in 70% ethanol.
Activity assays of different reactive oxygen species (ROS) scavenging enzymes
The activity assays for various antioxidant enzymes were performed using total protein obtained from leaf samples (WT and transgenic) that were collected at different time intervals after post-infection39.
Superoxide dismutase
The SOD activity assay was performed as per a method previously described56. The inhibition of the photochemical reduction of NBT was measured at 530 nm in a spectrophotometer. One unit of SOD activity was defined as the amount of enzyme required to cause 50% inhibition of the reduction of NBT.
Ascorbate peroxidase (APX)
APX activity was measured as previously described57. The decrease in absorbance was recorded at 290 nm for 180 seconds. Enzyme activity is expressed as units per milligram of protein. Calculation was done using the extinction coefficient of Ascorbate (E): 2.8 mM−1cm−1.
Catalase (CAT)
CAT activity was measured following the procedure described previously58. Enzyme activity was estimated by monitoring the decrease in absorbance of H2O2 at 240 nm for 180 seconds. Activity is expressed as units per milligram of protein.
Peroxidase (POD)
Peroxidase (POD) activity was measured using the protocol designed59 with modifications. The increase in absorbance at 470 nm was recorded within 120 seconds after addition of enzyme extract; enzyme activity is expressed as mM H2O2 decomposed per milliliter of total soluble protein.
Lipid peroxidation assay
Lipid peroxidation was estimated by determining the MDA (malondialdehyde) content as previously described60 with some modifications. The MDA concentration was calculated using the extinction coefficient 155 mM−1 cm−1 and is expressed as nmole/g.
Lipid hydroperoxide assay by ferrous xylenol orange
Lipid hydroperoxide content was spectrophotometrically determined using the xylenol orange assay61. Absorbance was recorded at 560 nm and the values are expressed as H2O2 equivalent µmol/g.
Bioassay
Detached leaf bioassay with mycelial agar disc
Detached leaf bioassay using mycelial agar disc was performed in accordance with previous report3, 17. Sterilized Petri plates were lined with sterile cotton pads and sterile distilled water was sprayed to maintain moisture. Detached leaves, after surface sterilization, were placed on sterile glass slides so that both ends were inserted into the slits of moistened Whatman 3 mm filter paper. A mycelial block (5 mm in diameter), grown on a 5 day old PDA culture of R. solani, was placed on the middle of the leaf surface. The lid was sealed with parafilm and kept at 28 °C for 3 days.
RS-toxin isolation and detached leaf toxin bioassay
RS-toxin isolation and bioassay were done following previously described method41. Briefly, 50 μL toxin solution was used to inoculate the plant material. An injury was inflicted at the middle of each detached leaf piece with the help of a sterile needle. The filter paper disks of 5 mm diameter, soaked with the toxin sample, were placed on the injured leaf section. This set up was incubated at 28 °C (12 h of light and 12 h of darkness) for 5 days. After 5 days, symptom development was assessed.
Bioassay with whole plant
Whole plant bioassay of transgenic and WT plants were performed as described previously62. The rice hull-rice grain inoculum (1 g) of R. solani was placed in between the stem and the basal leaf sheath of each tiller at about 3 to 4 cm above the water line and tied with rubber bands. Water was sprayed every morning so as to maintain a moistened environment. Disease assessment was done 7, 14 and 21 days after infection; a scale of 0–9 as described in Standard Evaluation System for Rice (IRRI 2002) was used for the purpose.
Seed germination assay
The germination capacity of T3 transgenic seeds was compared to their non-transgenic counterpart using the controlled germination test (CGT) as described previously63. Seeds were soaked in sterile distilled water for 8 h at 30 °C, and transferred to fresh water (CGT) for an additional 12 h. After incubation, the seeds were rinsed two to three times in sterile distilled water and were germinated on Whatman filter paper soaked with distilled water and kept at 30 °C in a growth chamber.
Agronomic evaluation of transgenic plants
Different agronomic parameters such as plant height (cm), panicle length (cm), the number of effective tillers, number of panicles per plant (PPP) and dry weight of 1000 grains were recorded for transgenic as well as non-transgenic wild type plants. Ten randomly chosen plants from each transgenic line grown under greenhouse conditions were considered for evaluation.
Statistical analysis
All statistical analyses were conducted using the GraphPad Prism 6 software. The data were represented as the mean value derived from three independent experiments wherein each experiment was done in triplicate. The results are represented as means ± standard error (SE). Additionally, the differences between mean were analyzed by the Bonferroni Post-tests.
Electronic supplementary material
Acknowledgements
The work was funded by Department of Biotechnology (DBT), Govt. of India in the form of DBT Programme Support [Sanction no. BT/PR12656/COE/34/22/2015] and Indian Council of Agricultural Research (ICAR), Govt. of India [Sanction No. CS11/7/2014-1A-IV dated 26.11.2014]. We are thankful to Prof. Xinian Dong of Duke University, USA for donating the pKEXNPR1 vector. We also thank Dr. G.S. Laha (Indian Institute of Rice Research, Hyderabad) for providing the R. solani isolate.
Author Contributions
S.K., K.M. planned and performed the experiments. S.K., K.M., K.D., S.N.S. analyzed the data. S.K. and K.M. prepared the manuscript. K.D. and S.K.D. edited the manuscript and made substantial contributions to conceptually design the research work and have revised the manuscript. All authors discussed the results and commented on the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08180-x
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ou, S. H. Rice diseases (2nd ed.). Kew, England: Common wealth Mycological Institute (1985).
- 2.Datta K, Baisakh N, Thet KM, Tu J, Datta SK. Pyramiding transgenes for multiple resistance in rice against bacterial blight, yellow stem borer and sheath blight. Theor. Appl. Genet. 2002;106:1–8. doi: 10.1007/s00122-002-1014-1. [DOI] [PubMed] [Google Scholar]
- 3.Molla KA, et al. Rice oxalate oxidase gene driven by green tissue‐specific promoter increases tolerance to sheath blight pathogen (Rhizoctonia solani) in transgenic rice. Mol. Plant Pathol. 2013;14:910–922. doi: 10.1111/mpp.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banniza, S. & Holderness, M. Rice sheath blight pathogen biology and diversity. In S. Sreenivasa prasad (Ed.), Major fungal diseases of rice (pp. 201–211). Dordrecht: Springer (2001).
- 5.Damodaram Naidu V, Srijvasa Rao B, Murty PSS. Influence of sheath blight infection on the levels of chlorophyll and CO2 uptake in rice. Indian Phytopathol. 1981;34:30–33. [Google Scholar]
- 6.Savary, S. & Mew, T. W. Analyzing crop losses due to Rhizoctonia solani: rice sheath blight, a case study. In: Sneh, B., Jabaji-Hare, S., Neate, S., Dijst, G., eds. Rhizoctonia Species: Taxonomy, Ecology, Pathology and Disease Control. Dordrecht, Netherlands: Kluwer Academic Publishers, 237 ± 45 (1996).
- 7.Pinson SRM, Capdevielle FM, Oard JH. Confirming QTLs and finding additional loci conditioning sheath blight resistance in rice using recombinant inbred lines. Crop Sci. 2005;45:503–510. doi: 10.2135/cropsci2005.0503. [DOI] [Google Scholar]
- 8.Kuc J. Induced immunity to plant disease. Bio Sci. 1982;32:854–860. [Google Scholar]
- 9.Durrent WE, Dong X. Systemic aquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 10.Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mou Z, Fan W, Dong X. Inducers of Plant Systemic Acquired Resistance Regulate NPR1 Function through Redox Changes. Cell. 2003;113:935–944. doi: 10.1016/S0092-8674(03)00429-X. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Fan W, Kinkema M, Li X, Dong X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. 1999;96:6523–6528. doi: 10.1073/pnas.96.11.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan W, Dong X. In Vivo Interaction between NPR1 and Transcription Factor TGA2 Leads to Salicylic Acid–Mediated Gene Activation in Arabidopsis. Plant Cell. 2002;14:1377–1389. doi: 10.1105/tpc.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin WC, et al. Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 2004;13:567–581. doi: 10.1007/s11248-004-2375-9. [DOI] [PubMed] [Google Scholar]
- 15.Meur G, et al. Constitutive expression of Arabidopsis NPR1 confers enhanced resistance to the early instars of Spodoptera litura in transgenic tobacco. Physiol. Plant. 2008;133:765–775. doi: 10.1111/j.1399-3054.2008.01098.x. [DOI] [PubMed] [Google Scholar]
- 16.Chern M, Canlas PE, Fitzgerald HA, Ronald PC. Rice NRR, a negative regulator of disease resistance, interacts with Arabidopsis NPR1 and rice. NH1. Plant J. 2005;43:623–635. doi: 10.1111/j.1365-313X.2005.02485.x. [DOI] [PubMed] [Google Scholar]
- 17.Molla KA, et al. Tissue-specific expression of Arabidopsis NPR1 gene in rice for sheath blight resistance without compromising phenotypic cost. Plant Sci. 2016;250:105–114. doi: 10.1016/j.plantsci.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Xu G, et al. ORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature. 2017;545:491–494. doi: 10.1038/nature22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartnicki-Garcia S. Cell Wall Chemistry, Morphogenesis, and Taxonomy of Fungi. Annu. Rev. Microbiol. 1968;22:87–108. doi: 10.1146/annurev.mi.22.100168.000511. [DOI] [PubMed] [Google Scholar]
- 20.Muthukrishnan S, Liang GH, Trick HN, Gill BS. Pathogenesis related proteins and their genes in cereals. Plant Cell, Tissue Organ Cult. 2001;64:93–114. doi: 10.1023/A:1010763506802. [DOI] [Google Scholar]
- 21.Datta K, et al. Enhanced resistance to sheath blight by constitutive expression of infection-related rice chitinase in transgenic elite indica rice cultivars. Plant Sci. 2001;160:405–414. doi: 10.1016/S0168-9452(00)00413-1. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto T, et al. Transgenic grapevine plants expressing a rice chitinase with enhanced resistance to fungal pathogens. Plant Cell Rep. 2000;19:639–646. doi: 10.1007/s002999900174. [DOI] [PubMed] [Google Scholar]
- 23.Kishimoto K, et al. Detailed analysis of rice chitinase gene expression in transgenic cucumber plants showing different levels of disease resistance to gray mold (Botrytis cinerea) Plant Sci. 2002;162:655–662. doi: 10.1016/S0168-9452(01)00602-1. [DOI] [Google Scholar]
- 24.Fukuoka S, et al. Gene pyramiding enhances durable blast disease resistance in rice. Scientific reports. 2015;5:7773. doi: 10.1038/srep07773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karmakar S, et al. Green tissue-specific co-expression of chitinase and oxalate oxidase 4 genes in rice for enhanced resistance against sheath blight. Planta. 2016;243:115–130. doi: 10.1007/s00425-015-2398-x. [DOI] [PubMed] [Google Scholar]
- 26.Singh S, et al. Pyramiding three bacterial blight resistance genes (xa5, xa13 and xa21) using marker-assisted selection into indica rice cultivar PR106. Theor. Appl. Genet. 2001;102:1011–1015. doi: 10.1007/s001220000495. [DOI] [Google Scholar]
- 27.Yasuda N, Mitsunaga T, Hayashi K, Koizumi S, Fujita Y. Effects of pyramiding quantitative resistance genes pi21, pi34, and pi35 on rice leaf blast disease. Plant Dis. 2015;99:904–909. doi: 10.1094/PDIS-02-14-0214-RE. [DOI] [PubMed] [Google Scholar]
- 28.Hamid R, et al. Chitinases: An update. J. Pharmacy Bioallied Sci. 2013;5:21–29. doi: 10.4103/0975-7406.106559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao H, Li X, Dong X. Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. 1998;95:6531–6536. doi: 10.1073/pnas.95.11.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datta K, Koukolikova-Nicola Z, Baisakh N, Oliva N, Datta SK. Agrobacterium-mediated engineering for sheath blight resistance of indica rice cultivars from different ecosystems. Theor. Appl. Genet. 2000;100:832–839. doi: 10.1007/s001220051359. [DOI] [Google Scholar]
- 31.Xu Y, et al. Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell. 1994;6:1077–1085. doi: 10.1105/tpc.6.8.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quilis J, Peñas G, Messeguer J, Brugidou C, Segundo BS. The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol. Plant-Microbe Interact. 2008;21:1215–1231. doi: 10.1094/MPMI-21-9-1215. [DOI] [PubMed] [Google Scholar]
- 33.Kasprzewska A. Plant chitinases-regulation and function. Cell. Mo. Bio. Lett. 2003;8:809–824. [PubMed] [Google Scholar]
- 34.Mei C, Qi M, Sheng G, Yang Y. Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol Plant-Microbe Interact. 2006;19:1127–1137. doi: 10.1094/MPMI-19-1127. [DOI] [PubMed] [Google Scholar]
- 35.Antico CJ, Colon C, Banks T, Ramonell KM. Insights into the role of jasmonic acid-mediated defenses against necrotrophic and biotrophic fungal pathogens. Front. Biol. 2012;7:48–56. doi: 10.1007/s11515-011-1171-1. [DOI] [Google Scholar]
- 36.Kishi-Kaboshi. M, et al. A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant. J. 2010;63:599–612. doi: 10.1111/j.1365-313X.2010.04264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arora A, Sairam R, Srivastava G. Oxidative stress and antioxidative system in plants. Current Sci-Bangalore- 2002;82:1227–1238. [Google Scholar]
- 38.Wu, W., Wan, X., Shah, F., Fahad, S. & Huang, J. The role of antioxidant enzymes in adaptive responses to sheath blight infestation under different fertilization rates and hill densities. Sci. World J. (2014). [DOI] [PMC free article] [PubMed]
- 39.Paranidharan V, Palaniswami A, Vidhyasekaran P, Velazhahan R. Induction of enzymatic scavengers of active oxygen species in rice in response to infection by Rhizoctonia solani. Acta Physiol. Plant. 2003;25:91–96. doi: 10.1007/s11738-003-0041-0. [DOI] [Google Scholar]
- 40.Passardi F, Penel C, Dunand C. Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci. 2004;9:534–540. doi: 10.1016/j.tplants.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 41.Vidhyasekaran P, et al. Host-specific toxin production by Rhizoctonia solani, the rice sheath blight pathogen. Phytopathology. 1997;87:1258–1263. doi: 10.1094/PHYTO.1997.87.12.1258. [DOI] [PubMed] [Google Scholar]
- 42.Brooks SA. Sensitivity to a phytotoxin from Rhizoctonia solani correlates with sheath blight susceptibility in rice. Phytopathology. 2007;97:1207–1212. doi: 10.1094/PHYTO-97-10-1207. [DOI] [PubMed] [Google Scholar]
- 43.Heil M, Baldwin IT. Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci. 2002;7:61–67. doi: 10.1016/S1360-1385(01)02186-0. [DOI] [PubMed] [Google Scholar]
- 44.Fitzgerald HA, Chern M-S, Navarre R, Ronald PC. Overexpression of (At) NPR1 in rice leads to a BTH-and environment-induced lesion-mimic/cell death phenotype. Mol Plant-Microbe Interact. 2004;17:140–151. doi: 10.1094/MPMI.2004.17.2.140. [DOI] [PubMed] [Google Scholar]
- 45.Mao B, Liu X, Hu D, Li D. Co-expression of RCH10 and AGLU1 confers rice resistance to fungal sheath blight Rhizoctonia solani and blast Magnorpathe oryzae and reveals impact on seed germination. World J Microbiol Biotechnol. 2014;30:1229–1238. doi: 10.1007/s11274-013-1546-3. [DOI] [PubMed] [Google Scholar]
- 46.Ganguly M, Molla KA, Karmakar S, Datta K, Datta SK. Development of pod borer-resistant transgenic chickpea using a pod-specific and a constitutive promoter-driven fused cry1Ab/Ac gene. Theor Appl Genet. 2014;127:2555–2565. doi: 10.1007/s00122-014-2397-5. [DOI] [PubMed] [Google Scholar]
- 47.Murashige T, Skoog F. A. revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 48.Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol bio reporter. 1983;1:19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- 49.Sambrook, J. & Russell, D.W. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press (2001).
- 50.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 51.Wirth SJ, Wolf GA. Dye-labelled substrates for the assay and detection of chitinase and lysozyme activity. J Microbiol Methods. 1990;12:197–205. doi: 10.1016/0167-7012(90)90031-Z. [DOI] [Google Scholar]
- 52.Hsu S, Lockwood J. Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. App Microbiol. 1975;29:422–426. doi: 10.1128/am.29.3.422-426.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 54.Kim KJ, Yang YJ, Kim JG. Purification and characterization of chitinase from Streptomyces sp. M-20. J Biochem Mol Biol. 2003;36:185–189. doi: 10.5483/bmbrep.2003.36.2.185. [DOI] [PubMed] [Google Scholar]
- 55.Shi J, et al. Spermine pretreatment confers dehydration tolerance of citrus in vitro plants via modulation of antioxidative capacity and stomatal response. Tree Physiol. 2010;30:914–922. doi: 10.1093/treephys/tpq030. [DOI] [PubMed] [Google Scholar]
- 56.Gupta, A. S., Webb, R. P., Holaday, A. S. & Allen, R. D. Overexpression of superoxide dismutase protects plants from oxidative stress. Plant Physiol. 103, 1067–1073. [DOI] [PMC free article] [PubMed]
- 57.Chen GX, Asada K. Ascorbate peroxidase in tea leaves: occurrence of two isozymes and the differences in their enzymatic and molecular properties. Plant Cell Physiol. 1989;30:987–998. doi: 10.1093/oxfordjournals.pcp.a077713. [DOI] [Google Scholar]
- 58.Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:121–126. doi: 10.1016/S0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 59.Kochba J, Lavee S, Spiegel-Roy P. Differences in peroxidase activity and isoenzymes in embryogenic and non-embryogenic ‘Shamouti’ orange ovular callus lines. Plant Cell Physiol. 1977;18:463–467. doi: 10.1093/oxfordjournals.pcp.a075455. [DOI] [Google Scholar]
- 60.Heath RL, Packer L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys. 1968;125:189–198. doi: 10.1016/0003-9861(68)90654-1. [DOI] [PubMed] [Google Scholar]
- 61.DeLong JM, et al. Using a modified ferrous oxidation-xylenol orange (FOX) assay for detection of lipid hydroperoxides in plant tissue. J Agr Food Chem. 2002;50:248–254. doi: 10.1021/jf0106695. [DOI] [PubMed] [Google Scholar]
- 62.Datta K, et al. Over-expression of the cloned rice thaumatin-like protein (PR-5) gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia solani causing sheath blight disease. Theor Appl Genet. 1999;98:1138–1145. doi: 10.1007/s001220051178. [DOI] [Google Scholar]
- 63.Campion B, et al. Isolation and characterization of an lpa (low phytic acid) mutant in common bean (Phaseolus vulgaris L.) Theor Appl Genet. 2009;118:1211–1221. doi: 10.1007/s00122-009-0975-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


