Abstract
Preparations of purified full-length fusion (F) protein of human respiratory syncytial virus (HRSV) expressed in recombinant vaccinia-F infected cells, or of an anchorless mutant (FTM−) lacking the C-terminal 50 amino acids secreted from vaccinia-FTM−-infected cells contain a minor polypeptide that is an intermediate product of proteolytic processing of the F protein precursor F0. N-terminal sequencing of the intermediate demonstrated that it is generated by cleavage at a furin-motif, residues 106–109 of the F sequence. By contrast, the F1 N terminus derives from cleavage at residue 137 of F0 which is also C-terminal to a furin recognition site at residues 131–136. Site-directed mutagenesis indicates that processing of F0 protein involves independent cleavage at both sites. Both cleavages are required for the F protein to be active in membrane fusion as judged by syncytia formation, and they allow changes in F structure from cone- to lollipop-shaped spikes and the formation of rosettes by anchorless F.
Human respiratory syncytial virus (HRSV) is the main cause of severe lower respiratory tract infections in infants and young children (1), and it is also a pathogen of considerable importance in the elderly (2). HRSV is an enveloped, nonsegmented negative-strand RNA virus, classified within the Pneumovirus genus of the Paramyxoviridae family. The virion has two main surface glycoproteins in the viral membrane: the attachment (G) protein, which mediates virus binding to the cell receptor (3), and the fusion (F) protein, which is responsible for fusion of the viral and cell membranes (4). A third small hydrophobic (SH) surface glycoprotein of unknown function is expressed abundantly at the surface of infected cells, but is incorporated only in small amounts in the virus particle (5).
Fusion of HRSV and cell membranes is thought to occur at the cell surface and is a necessary step for transfer of the viral ribonucleoprotein into the cell cytoplasm. The F protein, which mediates this process, also promotes fusion of infected cell membranes with those of adjacent cells, leading to syncytia formation. Whereas the G protein of HRSV shares neither sequence nor structural features with the attachment protein of related viruses (6), the F protein shares structural elements with its counterpart in other paramyxoviruses, and all of the F proteins have a low but significant level of sequence relatedness (7).
The F protein is synthesized as an inactive precursor of 574 amino acids (see Fig. 1A for a diagram of its primary structure) that is cleaved by furin-like proteases during transport to the cell surface to yield two disulfide-linked polypeptides, F2 from the N terminus and F1 from the C terminus (8). There are three hydrophobic sequences in the F polypeptide. The first is the signal peptide, located at the N terminus of the F2 chain. The second is the fusion peptide at the N terminus of the F1 chain, and the third is the transmembrane region, located near the C terminus of F1. Adjacent to the fusion peptide and transmembrane regions are two heptad repeat sequences, HRA and HRB, that are predicted to form coiled-coil structures (9). HRA and HRB peptides form trimeric complexes in solution (10, 11), and x-ray crystallography of these complexes reveals an internal core of three HRA α-helices bounded by three antiparallel HRB α-helices packed in the grooves of the HRA coiled-coil trimer (11). This structure is very similar to that described for analogous peptides from the paramyxovirus SV5 fusion protein (12), confirming the structural similarities of the HRSV F with its counterparts in related viruses.
Figure 1.
Identification of two cleavage sites in anchorless F protein. (A) Diagram of the F protein primary structure denoting cysteine residues (●), potential glycosylation sites (▴), hybrophobic regions (■), heptad repeat sequences (▨) and cleavage sites by furin-like proteases (↓). Inserted is a partial amino acid sequence of the F protein where the furin-sequence motifs (boldface) and the cleavage sites I and II are indicated. (B) Western blot of purified FTM− developed with anti-F255–275 and anti-F104–117 sera. The location of bands is shown at right (see text for explanation). (C) Coomassie blue-stained poly(vinylidene difluoride) membrane of purified FTM− after SDS/PAGE. N-terminal sequences of FΔ1–109 and F1 bands, determined by Edman degradation (see Materials and Methods), are shown at right. The band with the asterisk was identified as a contaminant of bovine apolipoprotein I by N-terminal sequencing. The F0 and F2 bands yielded no sequence. (D) The mass spectrum of tryptic peptides of the F2 band denotes peaks with masses matching the expected values of peptides from F2 (arrows). (Insert) Magnified spectrum of peptides N88-R106 and R88-A107 denoting by-products with oxidized methionines (+16 m/z) and metastable degradation products (Δ56.3 m/z) of these peptides. The location of the F2 peptides in the F primary structure is shown in A, below the protein diagram.
Recently, we described electron microscopy of the purified full-length F glycoprotein of HRSV and a truncated anchorless mutant lacking the C-terminal 50 amino acids (13), both expressed from recombinant vaccinia viruses. Full-length F, in the absence of detergent, formed rosettes that contained two distinct types of protein rods, one cone-shaped and the other lollipop-shaped. In contrast, the anchorless F mutant that was excreted by cells remained largely as individual cone-shaped rods, although a low proportion of rosettes containing lollipop-shaped rods was also observed. We proposed that these two types of rods might represent different structures that the F molecule may adopt before and after activation for its role in membrane fusion. Additional polypeptides to F1 and F2 that reacted with antibodies specific for the F protein were reported to be present in preparations of both full-length and anchorless F, and were particularly prominent in anchorless F (13). In this paper we have characterized these polypeptides as intermediates in F proteolytic processing. Analysis of their sequences indicate that the F precursor is cleaved at two sites, both preceded by motifs recognized by furin-like proteases. Cleavage at both sites is required for acquisition of membrane fusion potential by the F protein, and electron microscopy shows that, when cleavage is completed at both sites, there is a transition from cone-shaped to lollipop-shaped rods.
Materials and Methods
Cells and Viruses.
All cells were grown in DMEM supplemented with 10% FCS. The Long strain of HRSV and recombinant vaccinia viruses were grown in HEp-2 cells as described (13). HRSV was purified from the culture supernatant of infected cells by precipitation with 6% polyethyleneglycol, centrifugation in a 20–60% sucrose gradient, and pelleting of the virus band through a 30% sucrose cushion (14).
Plasmids and Recombinant Vaccinia Viruses.
pTM1-derived plasmids carrying inserts of the F gene (Long strain) with either the wild-type sequence (pTM1/F) or the sequence with the change I525Stop (ATC to TAA) that encodes the FTM− protein (pTM1/FTM−) have been reported (15). These plasmids have the F gene insert cloned under a T7 promoter. The F inserts were subcloned in the pRB21 plasmid (a gift of R. Blasco, Madrid, Spain) to generate vaccinia recombinants (VAC-F and VAC-FTM−) of the vRB12 strain (15).
The following mutations were introduced in the F gene insert of plasmids pTM1/F, pTM1/FTM−, pRB21/F and pRB21/FTM−: R108N (AGA to AAC), R109N (AGA to AAC), R108N/R109N, K131Q (AAG to CAG), and Δ131–134 (deletion of nucleotides 404–415). The nucleotide changes for each mutant are shown between parentheses. Complementary oligonucleotides with the appropriate changes were synthesized and used in the Quik-Change site-directed mutagenesis kit (Stratagene) to generate the different mutants. pRB21-derived plasmids were used to generate vaccinia recombinants by the method of Blasco and Moss (16).
Antibodies and Peptides.
Monoclonal antibodies raised against the F protein of HRSV (Long strain) were described by García-Barreno et al. (17). All these antibodies recognize epitopes in the F1 chain. A polyclonal rabbit serum raised against a peptide of the F1 chain, spanning residues 255–275, bound to keyhole limpet hemocyanin (KLH) (anti-F255–275) has been described (18). A peptide spanning residues 104–117 of the F sequence with an extra cysteine added at the C terminus was synthesized in an Applied Biosystems 430A synthesizer, by using fluorenylmethoxycarbonyl chemistry. The peptide was deprotected and cleaved off the resin with trifluoroacetic acid, purified by HPLC (19), coupled to KLH through the extra cysteine, and used to raise a rabbit serum (anti-F104–117) as described (18).
Purification, Western Blotting, Sucrose Gradient Centrifugation, and Electron Microscopy of F Proteins.
The full-length F protein was purified from extracts of HEp-2 cells infected with either Long virus or the vaccinia recombinant VAC-F, harvested 48 h after infection. The anchorless F protein was purified from the supernatant of HEp-2 cells infected with the vaccinia recombinant VAC-FTM− and harvested 48 h after infection. Preparation of extracts and purification of the F protein by immunoaffinity chromatography have been described (13). Proteins were separated by SDS/PAGE, transferred to Immobilon membranes (Millipore), and blotted with the antibodies indicated in the figure legends.
Samples of F proteins (≈100 μg) were loaded onto preformed 10–25% sucrose gradients in PBS, and were centrifuged at 39,000 rpm for 15 h at 4°C in a Beckman SW40 rotor. Fractions (1 ml) were collected from the top of the tube, and 20-μl aliquots were analyzed by Western blotting. Pools of fractions were concentrated and dialyzed against PBS in a Centricon-30 (Amicon).
F-protein samples in PBS were absorbed onto carbon films and stained with 1% sodium silicotungstate (pH 7.0). A JEOL 1200 electron microscope, operated at 100 kV, was used to view the samples. Micrographs were taken under minimum-dose, accurate defocus conditions to preserve details to ≈1.5 nm (20).
N-Terminal Sequencing and MS.
Automated Edman degradation was performed on poly(vinylidene difluoride) (PVDF)-immobilized proteins after SDS/PAGE by using an Applied Biosystem Procise 494 sequenator.
Protein bands from Coomassie blue-stained gels were excised manually and processed automatically for trypsin digestion by using an Investigator ProGest Protein Digestion Station (Genomic Solutions, Cambridgeshire, U.K.), following the protocol of Schevchenko et al. (21). Briefly, after reduction and alkylation, protein bands were digested with sequencing-grade modified porcine trypsin (final concentration, 16 ng/ml; Promega) for 12 h at 37°C. Peptides were eluted with 25 mM ammonium bicarbonate/10% formic acid (vol/vol), and acetonitrile in a final volume of 100 μl. A 0.5-μl aliquot was mixed with α-cyano-4-hydroxy-trans-cinnamic acid matrix and processed for MS. Spectra were acquired by using a Bruker Reflex III matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometer (Bruker-Franzen Analytik, Bremen, Germany) equipped with delayed extraction and operated in the reflector mode. The equipment was first externally calibrated by employing protonated mass signals from a peptide mixture covering the 1000- to 4000-m/z range, and thereafter every spectrum was internally calibrated by using selected signals arising from trypsin autoproteolysis. The measured tryptic peptide masses were transferred through MS BioTools program to Mascot Peptide Mass Fingerprinting search engine (Matrix Science, London) to search automatically the National Center for Biotechnology Information nonredundant database. No restrictions were placed on the species of origin of the protein and the allowed protein molecular mass was 1–200 kDa.
Syncytia Formation Assay.
BSR-T7/5 cells (22), a BHK-derived cell line that expresses constitutively the T7 RNA polymerase (a gift of K.-K. Conzelmann, Munich, Germany), were transfected with 1.5 μg of the plasmids indicated in the figure legends by the calcium phosphate precipitation method (MBS, mammalian transfection kit, Stratagene). Forty hours later, the cells were fixed with methanol/acetone and processed for indirect immunofluorescence by using anti-F monoclonal antibodies, as described (23).
Results
Identification of Two Processing Sites in the F Polypeptide.
We have described before two vaccinia recombinant viruses that express either the full-length F glycoprotein of HRSV (VAC-F) or a truncated membrane anchorless mutant (VAC-FTM−) lacking the C-terminal 50 amino acids (15). Full-length F is incorporated in the cell membrane; the FTM− mutant is secreted into the cell culture medium. Both proteins were purified by immunoaffinity chromatography from either extracts of HEp-2 cells infected with VAC-F or culture supernatants of HEp-2 cells infected with VAC-FTM− (13).
When the purified FTM− protein was analyzed by SDS/PAGE and Western bloting by using an antiserum raised against a synthetic peptide of the F1 chain (residues 255–275 of the F polypeptide) three protein bands were observed (Fig. 1B). The smallest, and most abundant, band corresponds to the previously characterized F1 polypeptide (24), which lacks the C-terminal 50 amino acid of intact F1. The other two bands were not characterized in our previous work (13).
An antiserum was raised against a synthetic peptide containing residues 104–117 of the F sequence plus a cysteine added at the C terminus end for coupling to carrier proteins. When preparations of FTM− were blotted with the anti-F104–117 serum, the two upper bands, but not F1, were detected (Fig. 1B).
To characterize the different polypeptides present in the preparations of purified FTM−, we determined their N-terminal sequences (Fig. 1C). The largest band gave no sequence, suggesting a blocked N terminus. Analysis of tryptic peptides derived from this band by MALDI-TOF MS confirmed it contained sequences of F2 and F1, in agreement with its reactivity with anti-F104–117 and anti-F255–275 sera (Fig. 1B). Consequently, this band was identified as the uncleaved F0 precursor of the anchorless protein. The first eight Edman degradation cycles of the intermediate band indicated a sequence that matched residues 110–117 of the F polypeptide (Fig. 1C). Because this band also contained sequences of the F1 chain—inferred from its reactivity with the anti-F255–275 serum (Fig. 1B) and from tryptic peptide mass analysis—it was identified as a polypeptide starting at residue 110 of the F sequence and ending at the C terminus of F1TM−. This band was named FΔ1–109. Finally, the sequence of the six N-terminal amino acids of the F1 band matched the first residues of the fusion peptide (Fig. 1C), confirming that it is the mature F1 chain generated by cleavage of the F precursor after amino acid 136. These results, therefore, demonstrated that the F-protein precursor could be cleaved at two sites (Fig. 1A): site I, after residue 109, and site II, after residue 136. Both sites are preceded by sequence motifs recognized by furinlike proteases.
N-terminal sequencing of a diffuse band of FTM− near the bottom of the gel was also attempted (Fig. 1C) but no amino acid sequence was detected. Peptide mass fingerprinting of the band showed that it corresponded to the F2 chain (Fig. 1D). Peptides derived from the F sequence between cleavage sites I and II, described above, were not detected in the mass spectrum of the F2 band. However, tryptic peptides with masses corresponding to those preceding amino acid 109 were clearly identified (Fig. 1D Inset). Among these peptides there was one, N88-A107, which is unlikely to be generated by the in vitro trypsin digestion of the F2 band, implying that it was generated during the in vivo processing of the anchorless F protein.
To determine whether the FΔ1–109 polypeptide was present in preparations of the F molecule, in addition to FTM−, full-length F protein purified from extracts of HEp-2 cells infected with either VAC-F or HRSV (Long strain) or from purified HRSV virions were analyzed by Western blotting with anti-peptide sera (Fig. 2). The anti-F255–275 serum recognized an F1 band in all preparations containing full-length F protein that migrated slightly less than the F1 band of FTM−, as expected by their size differences (Fig. 2A). The anti-F104–117 serum reacted with a band, corresponding to FΔ1–109, in the preparations that contained full-length F protein (Fig. 2B). Relative to F1, the amounts of F0 and FΔ1–109 depended on the origin of each protein sample. It has been reported that processing of F protein is less efficient in cells infected with VAC-F than with HRSV (25). Accordingly, F0 and FΔ1–109 were less abundant in preparations of virus or full-length F protein obtained from HRSV infected HEp-2 cells than in preparations of F protein from VAC-F- infected cultures. The amounts of F0 and FΔ1–109 were highest in the preparation of anchorless F protein. This abundance may reflect the different intracellular pathway that this form of F protein follows in its transport to the extracellular medium.
Figure 2.
Immunoblots of full-length and anchorless F protein preparations. (A) Purified HRSV virus or F protein purified from extracts of HEp-2 cells infected with either HRSV or VAC-F or FTM− protein purified from the supernatant of HEp-2 cells infected with VAC-FTM− were blotted with the anti-F255–275 serum. The amount of each sample was standardized to yield F1 bands of similar intensity. (B) The same protein samples from A were blotted with the anti-F2104–117 serum. The amount of each sample loaded in the gel relative to the blot of part A was as follows: purified virus, 10:1; F(HRSV), 2:1; F(VAC-F), 1:1; F(VAC-FTM−), 1:1.
The F Protein Precursor Is Cleaved Independently at Sites I and II.
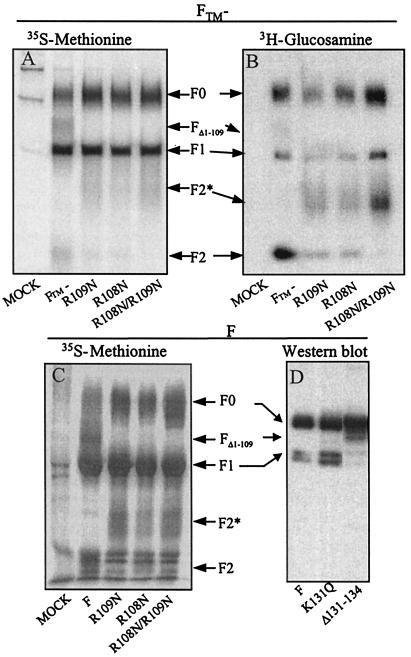
To investigate whether or not cleavage of F0 at site I was required for cleavage at site II, mutants of anchorless F protein and full-length F protein were generated at both sites. HEp-2 cells were infected with vaccinia recombinants expressing either the anchorless F protein or the anchorless F mutants R108N, R109N, or R108N/R109N. After labeling with either [35S]methionine (Fig. 3A) or [3H]glucosamine (Fig. 3B), proteins were immunoprecipitated from the culture supernatants with a monoclonal antibody specific for the F1 chain. Whereas the F0, FΔ1–109, and F1 polypeptides were observed in the immunoprecipitates of anchorless F, the FΔ1–109 band was absent in the three mutants, indicating that cleavage at site I was inhibited in these cases. In the same gels, a diffuse band corresponding to the F2 chain of anchorless F protein was less intense in the single mutants and totally absent in the immunoprecipitate of the double mutant. Very diffuse bands that migrated less than F2 (F2*) were observed in the immunoprecipitates of the three mutant proteins (more clearly seen in the samples labeled with 3H-glucosamine). Absence of FΔ1–109 and F2 and presence of F2* chains was also observed in immunoprecipitates of cell extracts infected with vaccinia recombinants expressing full-length F gene with the mutations R108N, R109N, or R108N/R109N (Fig. 3C). These results, therefore, demonstrated that mutations in the furin-target sequence preceding site I-inhibited cleavage after residue 109 of the F polypeptide. However, inhibition of that cleavage did not prevent processing at site II, after residue 136, to generate the F1 chain.
Figure 3.
Analysis of F-protein mutants with changes at sites I or II. (A) HEp-2 cells were infected with vaccinia recombinants (multiplicity of infection, 5 plaque-forming units per cell) expressing the FTM− protein with the wild-type sequence or with the mutations R109N, R108N, or R108N/R109N. Cells were labeled from 4 to 24 h after infection with [35S]methionine (100 μCi/ml). Cell supernatants were harvested at the end of the labeling period and aliquots were immunoprecipitated with a monoclonal antibody (47F) specific for the F1 chain (17). Proteins were resolved by SDS/PAGE and visualized by fluorography. (B) Immunoprecipitate of cell culture supernatants from cells infected as indicated in A, but labeled with [3H]glucosamine (100 μCi/ml) from 4 to 24 h after infection. (C) HEp-2 cells were infected with vaccinia recombinants expressing either wild-type F or the same mutants from A but in the full-length protein. Cells were labeled from 4 to 24 h after infections with [35S]methionine, and extracts were immunoprecipitated as in A. (D) HEp-2 cells were infected with the vaccinia virus MVA-T7 (26) and transfected with pTM1 plasmids carrying either the full-length F gene or mutants K131Q or Δ131–134, by using Lipofectin (23). Extracts were made 48 h after infection and analyzed by SDS/PAGE and Western blotting with monoclonal antibody 47F.
Mutations K131Q and Δ131–134, which alter the sequence preceding site II, were introduced in the full-length F gene of pTM1-derived plasmids. HEp-2 cells were infected with the recombinant vaccinia MVA-T7 (26), which expresses the T7 RNA polymerase, and transfected with pTM1 plasmids carrying the different F gene constructs. Extracts of the cells were subjected to SDS/PAGE and blotted with a monoclonal antibody specific for the F1 chain. Similar amounts of the F1 polypeptide were detected in the extracts containing the wild-type F or the mutant K131Q (Fig. 3D), but the amount of F1 was clearly reduced in the case of mutant Δ131–134. An increase in the amount of FΔ1–109 was observed for mutant Δ131–134, indicating that inhibition of cleavage at site II did not prevent cleavage at site I. Thus, F0 is cleaved at both sites after the furin motifs and cleavage at one site is not required for cleavage at the other site.
Structural Changes Associated with Proteolytic Processing of the F Protein.
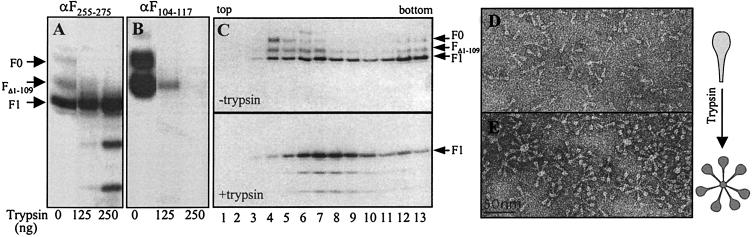
Purified anchorless F protein was incubated with increasing amounts of trypsin (Figs. 4 A and B) and then analyzed by Western blotting with anti-peptide sera. Whereas F0 and FΔ1–109 were progressively digested with increasing doses of trypsin, the F1 band remained essentially unchanged, although some faster migrating bands reacting with anti-F255–275 antibodies were observed with the highest amounts of trypsin (Fig. 4A).
Figure 4.
Changes associated with trypsin digestion of purified F protein. Aliquots of purified FTM− (1 μg) were incubated with the indicated amounts of trypsin for 60 min at 37°C. Subsequently, a large excess of sample buffer was added and the proteins were resolved by SDS/PAGE and blotted with the anti-F255–275 (A) or anti-F104–117 (B) sera. (C) Samples of FTM− (≈100 μg) were digested with trypsin in PBS (equivalent to the conditions of the 250-ng lane of A), or mock-digested. Then, one-fifth volume of a trypsin inhibitor (complete miniEDTA, Roche Diagnostics) was added and the samples were loaded on top of a 10–25% sucrose gradient and centrifuged as indicated in Materials and Methods. Fractions (1-ml) were analyzed by Western blotting with the anti-F255–275 serum. Pools of fractions from the gradients shown at the top (fractions 4–7) or at the bottom (fractions 6–9) of C were observed under the electron microscope by negative staining (D and E, respectively). A diagram of the changes observed in FTM− molecules is shown at right.
In agreement with our previous results (13), the anchorless F protein, mock-treated with trypsin, sedimented in a 10–25% sucrose gradient as a broad band (Fig. 4C), with a peak near the top of the gradient (fractions 4–7); some aggregated forms sedimented toward the bottom of the gradient. F0 and FΔ1–109 were more prominent in the fractions near the top of the gradient. Densitometric quantitation of the proteins in the gradient fractions yielded the following figures: the material in fraction 4 was 53% F1 and 47% F0 plus FΔ1–109; fraction 9, 91% was F1 and 9% F0 plus FΔ1–109. Samples of anchorless F protein, incubated with trypsin under conditions in which F0 and FΔ1–109 were fully digested, sedimented toward the middle of the gradient (Fig. 4C). This change in the sedimentation profile of anchorless F protein after trypsin treatment was accompanied by changes in the aggregation state of the molecules in electron microscopy. Although the mock-treated sample contained a majority of individual, unassociated molecules with <25% in rosettes in the top fraction (13) and about 47% in pooled fractions 4–7 (Fig. 4D), most of the molecules (86%) in pooled fractions 6–9 of the gradient of the trypsin-treated sample were in rosettes (Fig. 4E). The rosettes of the trypsin-treated sample also contained more spikes than those of the untreated anchorless F protein. These results indicate that fully processed anchorless F protein forms rosettes in aqueous media. In agreement with our previous observations, the anchorless F molecules in rosettes are lollipop-shaped compared with unassociated molecules, which are cone-shaped.
Cleavage of the F Protein at Sites I and II Is Needed for Membrane Fusion.
The membrane fusion capacity of full-length F proteins, and mutants derived from it, was evaluated in a syncytia formation assay. BRS-T7/5 cells (22) transfected with F-containing plasmids were fixed 40 h after transfection and stained with a monoclonal antibody specific for the F1 chain. Large syncytia, containing several nuclei in the same cytoplasm, were clearly visible in cultures expressing either the wild type or the K131Q mutant (Fig. 5). In contrast, BSR-T7/5 cells transfected with plasmids carrying the F genes of the double mutant R108N/R109N, which is not processed at site I, or the mutant Δ131–134, which is not processed at site II, showed individual cells stained with the anti-F1 antibody but no syncytia. These results indicate that inhibition of F-protein cleavage at either site removes the capacity of the F protein to promote membrane fusion.
Figure 5.
Syncytia formation in BSR-T7/5 cells transfected with pTM1 plasmids carrying wild type or mutant full-length F genes. Cells were transfected with the indicated plasmids and processed for immunofluorescence 40 h later with an antibody specific for the F1 chain, as indicated in Materials and Methods. (Magnification, ×200.)
Discussion
The results presented here demonstrate that cleavage of the biosynthetic precursor F0 of HRSV after two furin-recognition sequence motifs is required for membrane fusion activity. The fate of the F segment located between the two sites is not known. The F segment was not found in immunoprecipitates of either full-length or anchorless F proteins following processing, suggesting that it is released from the processed molecule. An analogous segment is present in the F protein of the closely related bovine respiratory syncytial virus; however, the F proteins of other pneumoviruses and paramyxoviruses lack the sequence between the two cleavage sites (9).
The mechanism of activation of membrane fusion by paramyxoviruses is unknown. Enveloped viruses, such as influenza virus (27), which are endocytosed after binding of the attachment protein to specific receptors present on the surface of target cells, are activated at endosomal pH. In contrast, paramyxoviruses fuse their membranes to those of target cells at the cell surface. It has been postulated that the interaction of paramyxovirus attachment protein with the cell receptor triggers a conformational change in the F protein that promotes membrane fusion (28). It has also been reported that the G and SH glycoproteins of HRSV enhance the fusion activity of F proteins when coexpressed in transfected cells (29). However, a deletion mutant of HRSV lacking both the G and SH genes is able to replicate efficiently in certain cell lines and to produce syncytia (30). In addition, the expression of the F glycoprotein alone is sufficient to induce formation of syncytia in transfected cells (Fig. 5). Thus, the question arises, is cleavage at the two furin-motif sites of HRSV F the trigger for membrane fusion? If this were so, by analogy with viruses that can be inactivated by triggering at low pH in the absence of a target membrane, inactivation of F might occur before its expression at the cell surface. Perhaps the process of activation occurs slowly after cleavage, ensuring that, for a significant time, some noninactivated molecules survive to maintain virus infectivity. Perhaps activation after processing might involve the slow release of the intervening peptide and perhaps the rate of activation might be enhanced by, for example, recognition of target cells by F2 or some other region of F.
The anchorless F mutant described here remains mainly as single trimeric molecules when a significant proportion of the molecules are not cleaved at either site I to generate FΔ1–109 or sites I and II to generate F1 + F2. On trypsin digestion of FTM−, under conditions in which FΔ1–109 and F0 are digested, the protein forms rosettes and changes its shape from cone-shaped to lollipop-shaped. There are reports from other paramyxovirus that cleavage of F0 results in exposure of a new hydrophobic region (31) and in accessibility to site specific antibodies (32). Rosettes of anchorless F proteins of HRSV are assumed to form as a result of interactions between exposed fusion peptides, which are the major conserved hydrophobic regions remaining in the FTM− molecules. If this is so, processing of F0 at both sites might be needed for fusion peptide exposure. Because cleavage at both sites is also required for the F protein to acquire its potential to induce syncytia, the change in shape of F, its ability to form rosettes, and the capacity to fuse membranes may be directly related.
In the case of some influenza and parainfluenza viruses, cleavability of the fusion protein has been clearly shown to correlate with pathogenicity (for a review, see ref. 33). For avian influenza hemagglutinins, for example, insertions at the site of cleavage increase cleavability and virulence for chickens (34). By analogy, the sequences located between sites I and II may confer unique pathogenic properties to human and bovine respiratory syncytial viruses. The feasibility of constructing mutant viruses (35) should enable the relevance of sites I and II for pathogenicity and attenuation to be assessed.
Acknowledgments
We are grateful to Rafael Blasco (Madrid) for the pRB21 plasmid and for the vRB12 vaccinia virus, and to Klaus-K. Conzelmann (Munich) for the BSR-T7/5 cells. This work was supported in part by Grants PM99–0014 from Ministerio de Ciencia y Tecnología and QLK2-CT-1999–00443 from the European Union (to J.A.M.), by the Medical Research Council (to J.J.S. and L.C.) and by the Howard Hughes Medical Institute. D.C.W. is an Investigator of the Howard Hughes Medical Institute. L.G.-R. was recipient of a predoctoral fellowship from Ministerio de Educación y Cultura (Spain) and M.B.R.-A. was recipient of a postdoctoral fellowship from the Comunidad de Madrid (Spain).
Abbreviations
- HRSV
human respiratory syncytial virus
- F
fusion
- SH
small hydrophobic
- VAC
vaccinia
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Collins P L, Chanock R M, Murphy B R. In: Fields Virology. Fields BN, Knipe DM, Howley PM, editors. Philadelphia: Lippincott; 1996. pp. 1313–1351. [Google Scholar]
- 2.Han L L, Alexander J P, Anderson L J. J Infect Dis. 1999;179:25–30. doi: 10.1086/314567. [DOI] [PubMed] [Google Scholar]
- 3.Levine S, Klaiber-Franco R, Paradiso P R. J Gen Virol. 1987;68:2521–2524. doi: 10.1099/0022-1317-68-9-2521. [DOI] [PubMed] [Google Scholar]
- 4.Walsh E E, Hruska J. J Virol. 1983;47:171–177. doi: 10.1128/jvi.47.1.171-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olmsted R A, Collins P L. J Virol. 1989;63:2019–2029. doi: 10.1128/jvi.63.5.2019-2029.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wertz G W, Collins P L, Huang Y, Gruber C, Levine S, Ball L A. Proc Natl Acad Sci USA. 1985;82:4075–4079. doi: 10.1073/pnas.82.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins P L, Huang Y T, Wertz G W. Proc Natl Acad Sci USA. 1984;81:7683–7687. doi: 10.1073/pnas.81.24.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins P L, Mottet G. J Gen Virol. 1991;72:3095–3101. doi: 10.1099/0022-1317-72-12-3095. [DOI] [PubMed] [Google Scholar]
- 9.Chambers P, Pringle C R, Easton A J. J Gen Virol. 1992;73:1717–1724. doi: 10.1099/0022-1317-73-7-1717. [DOI] [PubMed] [Google Scholar]
- 10.Matthews J M, Young T F, Tucker S P, Mackay J P. J Virol. 2000;74:5911–5920. doi: 10.1128/jvi.74.13.5911-5920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao X, Singh M, Malashkevich N, Kim P S. Proc Natl Acad Sci USA. 2000;97:14172–14177. doi: 10.1073/pnas.260499197. . (First Published December 5, 2000; 10.1073/pnas.260499197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker K A, Dutch R E, Lamb R A, Jardetzky T S. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 13.Calder L J, González-Reyes L, García-Barreno B, Wharton S A, Skehel J J, Wiley D C, Melero J A. Virology. 2000;271:122–131. doi: 10.1006/viro.2000.0279. [DOI] [PubMed] [Google Scholar]
- 14.García-Barreno B, Jorcano J L, Aukenbauer T, López-Galindez C, Melero J A. Virus Res. 1988;9:307–322. doi: 10.1016/0168-1702(88)90090-1. [DOI] [PubMed] [Google Scholar]
- 15.Bembridge G P, López J A, Bustos R, Melero J A, Cook R, Mason H, Taylor G. J Virol. 1999;73:10086–10094. doi: 10.1128/jvi.73.12.10086-10094.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blasco R, Moss B. Gene. 1995;158:157–162. doi: 10.1016/0378-1119(95)00149-z. [DOI] [PubMed] [Google Scholar]
- 17.García-Barreno B, Palomo C, Peñas C, Delgado T, Perez-Breña P, Melero J A. J Virol. 1989;63:925–932. doi: 10.1128/jvi.63.2.925-932.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.López J A, Andreu D, Carreño C, Whyte P, Taylor G, Melero J A. J Gen Virol. 1993;74:2567–2577. doi: 10.1099/0022-1317-74-12-2567. [DOI] [PubMed] [Google Scholar]
- 19.Carreño C, Roig X, Cairo J, Camarero J, Mateu M G, Domingo E, Giralt E, Andreu D. Int J Pept Protein Res. 1992;39:41–47. doi: 10.1111/j.1399-3011.1992.tb01554.x. [DOI] [PubMed] [Google Scholar]
- 20.Wrigley N G, Brown E B, Skehel J J. In: Electron Microscopy of Proteins, Viral Structure. Harris J R, Horne R W, editors. Vol. 5. London: Academic; 1986. pp. 103–163. [Google Scholar]
- 21.Schevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 22.Buchholz U J, Finke S, Conzelmann K-K. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Barreno B, Delgado T, Melero J A. J Virol. 1996;70:801–808. doi: 10.1128/jvi.70.2.801-808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.López J A, Peñas C, García-Barreno B, Melero J A, Portela A. J Virol. 1990;64:927–930. doi: 10.1128/jvi.64.2.927-930.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wertz G W, Stott E J, Young KK-Y, Anderson K, Ball L A. J Virol. 1987;61:293–301. doi: 10.1128/jvi.61.2.293-301.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyatt L S, Moss B, Rozenblatt S. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]
- 27.Skehel J J, Wiley D C. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 28.Lamb R A. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 29.Heminway B R, Yu Y, Tanaka Y, Perrine K G, Gustafson E, Bernstein J M, Galinski M S. Virology. 1994;200:801–805. doi: 10.1006/viro.1994.1245. [DOI] [PubMed] [Google Scholar]
- 30.Karon R A, Buonagurio D A, Georgiu A F, Whitehead S S, Adamus J E, Clements-Mann M L, Harris D O, Randolph V B, Udem S A, Murphy B R, Sidhu M S. Proc Natl Acad Sci USA. 1997;94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu M, Scheid A, Choppin P W. J Biol Chem. 1991;256:3557–3563. [PubMed] [Google Scholar]
- 32.Dutch R E, Hagglund R N, Nagel M A, Paterson R G, Lamb R A. Virology. 2001;281:138–150. doi: 10.1006/viro.2000.0817. [DOI] [PubMed] [Google Scholar]
- 33.Steinhauer D A. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 34.Khatchikian D, Orlich M, Rott R. Nature (London) 1989;340:156–157. doi: 10.1038/340156a0. [DOI] [PubMed] [Google Scholar]
- 35.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]