Abstract
T cells mediating autoimmune diseases, such as rheumatoid arthritis (RA), are difficult to characterize because they are likely to be deleted or inactivated in the thymus if the self-antigens they recognize are ubiquitously expressed. One way to obtain and analyze these autoimmune T cells is to alter T cell receptor (TCR) signaling in developing T cells to change their sensitivity to thymic negative selection, thereby allowing their thymic production. From mice thus engineered to generate T cells mediating autoimmune arthritis, we have isolated arthritogenic TCRs and characterized the self-antigens they recognized. One of them was the ubiquitously expressed 60S ribosomal protein L23a (RPL23A), with which T cells and autoantibodies from RA patients reacted. This strategy may improve our understanding of the underlying drivers of autoimmunity.
T cells mediate a variety of autoimmune diseases (1, 2), likely through the recognition of self-antigens. However, identification of the self-antigens targeted by T cells in systemic autoimmune diseases like RA has been technically difficult (3–5). This is because pathogenic T cells expressing high affinity TCRs for ubiquitous self-antigens may be largely deleted (i.e., negatively selected) in the thymus and scarcely detectable in the periphery or, if detected, in an inactivated state (6). This can be circumvented by altering TCR signaling, which changes the sensitivity of developing T cells to thymic selection and results in new dominant self-reactive TCR specificities that are causative of systemic autoimmune diseases (7–11). For example, a hypomorphic point mutation of ζ-associated protein 70 (ZAP-70), a TCR-proximal signaling molecule, causes T cell-mediated spontaneous autoimmune arthritis in mice, which resembles RA (8).
To identify ubiquitously expressed self-antigens commonly targeted in mouse and human systemic autoimmune disease, we first examined whether the arthritogenic CD4+ T helper (TH) cells in BALB/c SKG mice, which develop autoimmune arthritis due to the ZAP-70 mutation, utilized a specific dominant TCR. We compared the arthritogenic capacity of SKG CD4+ T cells expressing different TCR Vβ subfamilies (fig. S1). Transfer of SKG CD4+ T cells expressing Vβ6, 8.1/8.2 or 10 into BALB/c Rag2−/− mice induced arthritis with similar severities. In addition, CDR3 gene segments of Vβ6+ CD4+ T cells in arthritic joints were diverse with few common sequences among individual arthritic SKG mice (table S1–2 and fig. S2). Thus assuming that arthritogenic SKG CD4+ T cells were highly polyclonal and utilized various Vα and Vβ TCR chains, we attempted to isolate a single arthritogenic CD4+ T cell from a particular CD4+ T cell subpopulation, for example, those expressing Vα2 and Vβ6, which constituted ~1% of joint-infiltrating CD4+ T cells. To differentiate arthritogenic CD4+ T cells from forkhead box P3 (Foxp3)-expressing CD4+ regulatory T (Treg) cells (1), we used SKG mice with knock-in of enhanced green fluorescent protein (EGFP)-Foxp3 fusion protein, designated eFOX SKG mice, which also spontaneously developed arthritis (fig. S3). We cloned a single TCR pair from individual GFP− Vα2+ Vβ6+ CD4+ T cells present in arthritic joints of eFOX SKG mice, transfected Rag2−/− SKG bone marrow (BM) cells with the TCR gene, and transferred the BM cells into Rag2−/− mice to construct retrogenic mice expressing the TCR pair in developing T cells (12–15). Among nine retrogenic strains each expressing a distinct TCR, the ones expressing 7–39 or 6–39 TCR spontaneously developed arthritis at the incidence of 80.0% and 27.3%, respectively (Fig. 1A–C and fig. S4A–C). The two arthritogenic TCRs and a control non-arthritogenic 1–23 TCR used the same Vα and Vβ gene segments but different Jα and Jβ genes and CDR3 sequences (Fig. 1A). Arthritic joints in retrogenic 7–39 (R7–39) mice showed mononuclear cell infiltration, pannus formation and cartilage destruction (Fig. 1D). Some (66.7%) of R7–39, but not R6–39, mice also developed chronic dermatitis, which exhibited hyperkeratosis and parakeratosis, histopathological features of human psoriasis (16) (Fig. 1E–F and fig. S5). Other organs were histologically intact (fig. S6).
Fig. 1. Arthritis-inducing activity of 2 TCRs individually expressed in retrogenic mice.
(A) Amino acid sequences and frequencies of arthritogenic 7–39 TCR, 6–39 TCR and non-arthritogenic 1–23 TCR. These 3 TCRs were obtained from 3 different mice. CDR, complementarity determining region. (B) Joint swelling in R7–39 retrogenic mice. (C) Incidence and scores of spontaneous arthritis in R7–39 (n=11), R1–23 (n=14), and W7–39 mice (n=8). Error bars indicates the means ± s.d. (D) Hematoxylin and eosin (HE) staining of arthritic joints (bar, 1 mm). (E) Ears and hind paws of R7–39 and R1–23 mice. (F) HE staining of ears from R7–39 and R1–23 mice (bar, 500 μm). Results in B and D–F represent three independent experiments.
In R7–39 mice, 7–39 TCR-transduced cells preferentially differentiated into monoclonal CD4+ T cells with an activated and memory phenotype (fig. S7), and were able to transfer both arthritis and dermatitis into other Rag2−/− mice. Both arthritic R7–39 and non-arthritic R1–23 mice failed to develop Foxp3+ Treg cells (fig. S8). In contrast to 7–39 TCR gene-transfected Rag2−/− BM cells with the SKG ZAP-70 mutation, 7–39 TCR gene-transfected ZAP-70-intact Rag2−/− BALB/c BM cells did not cause arthritis in retrogenic mice (designated W7–39 mice). In W7–39 mice, the majority of 7–39 TCR-expressing CD4+ T cells were negatively selected in the thymus, and those which had escaped thymic negative selection, exhibited a naïve non-activated phenotype, indicating their dormant or anergic state (Fig. 1C and fig. S9).
Taken together, these results demonstrate that CD4+ T cells with a specific TCR are required to mediate autoimmune arthritis and also dermatitis, and that more than one TCR specificity is individually able to confer T cell arthritogenicity.
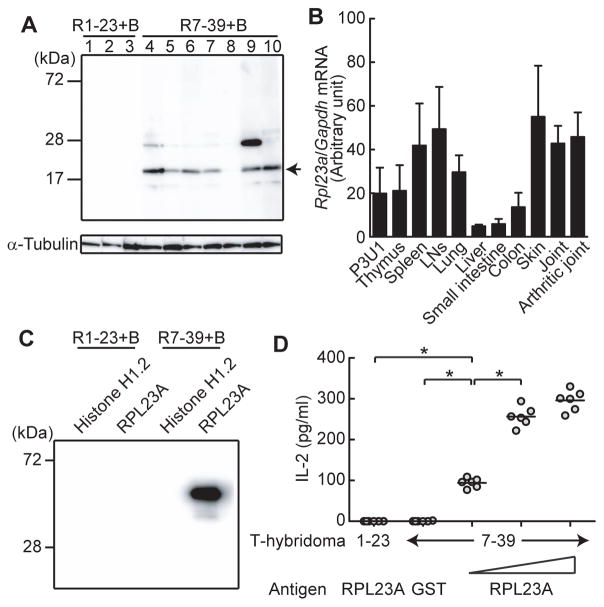
We next constructed T cell hybridomas expressing the 7–39 or 6–39 TCR and attempted to determine the self-antigens recognized by these TCRs. The 7–39 hybridomas cells produced interleukin (IL)-2 when stimulated by cell extracts not only from SKG fibroblast-like synoviocytes (FLSs) but also from P3U1 cells, a BALB/c plasma cell-derived cell line (fig. S10). In contrast, syngeneic antigen presenting cells (APCs) were sufficient to induce IL-2 production by 6–39 hybridoma cells, indicating that the 6–39 TCR recognized a self-antigen constitutively displayed by APCs (fig. S4D). To further characterize the self-antigen recognized by 7–39 TCR, we reconstituted Rag2−/− mice with a mixture of 7–39 TCR-transfected Rag2−/− SKG BM cells and TCRβ−/− BALB/c BM cells on the assumption that the autoantibodies produced by B cells might specifically react with the self-antigen recognized by 7–39 TCR because T-cell help came solely from 7–39 TH cells. The sera from these ‘B cell-reconstituted’ mice specifically reacted with a protein of 18 kD from the cell extract of P3U1 cells (Fig. 2A). Mass spectrometric analysis identified this protein as RPL23A, a component of the 60S subunit of ribosome (17, 18) (fig. S11). Various organs expressed RPL23A mRNA at high levels (Fig. 2B). The amino acid sequence of RPL23A is 100% conserved between mice and humans (18). The sera from the B cell-reconstituted R7–39 mice indeed recognized recombinant RPL23A, but not histone H1.2 protein, another candidate protein indicated by the mass spectrometric analysis (Fig. 2C). In addition, recombinant RPL23A protein specifically stimulated the 7–39 hybridoma cells in a dose dependent, class II major histocompatibility complex (MHC) I-Ad dependent manner (Fig. 2D and fig. S12). Among 20mer RPL23A peptides with consecutive overlapping of 5 amino acid residues, RPL23A71–90 peptide stimulated 7–39 TCR most potently (table S3 and fig. S13A).
Fig. 2. Identification of the self-antigen recognized by arthritogenic 7–39 TCR.
(A) Immunoblot analysis by sera from B cell-reconstituted R7–39 mice (n=7) and B cell-reconstituted R1–23 mice (n=3). Arrow indicates the commonly recognized protein. (B) Quantitative real time polymerase chain reaction (qPCR) analysis for RPL23A gene expression in various tissues from SKG mice (n=3). Error bars indicate the means±s.d. (C) Recombinant RPL23A protein revealed by immunoblotting with sera from indicated mice. (D) IL-2 production by 7–39 or 1–23 T-cell hybridomas stimulated with indicated recombinant proteins (n=6). Horizontal bars indicate the means. Error bars indicate the means±s.d. *P<0.05 (Kruskal-Wallis test followed by Steel-Dwass test). Results represent three (D) or two (A–C) independent experiments.
B cell-reconstituted R7–39 mice and arthritic SKG mice developed antibodies reacting with cyclic citrullinated peptides (CCP) as observed in RA patients (19) (fig. S14A); yet, there was no significant difference in anti-RPL23A antibody titer whether it was assessed with citrullinated or non-citrullinated RPL23A protein (fig. S14B–C). In addition, the RPL23A71–90 peptide recognized by 7–39 TCR contained no arginine residue to be converted to citrulline (table S3).
Taken together, these results indicate that the ubiquitously expressed protein RPL23A can be a target antigen of both arthritis and dermatitis. Furthermore, more than one systemic antigen can be targeted for arthritis induction because the 6–39 TCR did not react to peptides derived from RPL23A (fig. S13B).
Upon transfer, CD4+ T cells, but not sera, from B cell-reconstituted R7–39 mice induced arthritis in Rag2−/− mice (fig. S15). Indeed, CD4+ T cells from arthritic joints or the regional lymph nodes of R7–39 mice produced inflammatory cytokines including IL-17A and interferon (IFN)-γ, and granulocyte macrophage-colony stimulating factor (GM-CSF) upon activation with phorbol 12-myristate 13-acetate (PMA) and ionomycin, RPL23A protein or RPL23A71–90 peptide (fig. S16A–D, Fig. 3A–D and fig. S17). In addition, RPL23A stimulated non-arthritic SKG, but not BALB/c, CD4+ T cells to produce IL-17A in vitro (Fig. 3E). It also augmented the production of IL-17 by CD4+ T cells from SKG mice treated with mannan, which can trigger autoimmune arthritis in SKG mice by promoting TH17 differentiation of arthritogenic CD4+ T cells (20, 21). An arthritic joint of SKG mice indeed harbored CD4+ T cells possessing the Vβ CDR3 of 7–39 TCR (table S2).
Fig. 3. RPL23A-reactive TH cells in R7–39 mice.
(A) Cytokine production by CD4+ T cells from regional lymph nodes (RLNs) of R7–39 or R1–23 mice after in vitro stimulation with recombinant RPL23A or control glutathione S-transferase (GST) protein. Stim, Stimulation. (B) Percentages of cytokine-producing CD4+ T cells in A (n=3). (C) Cytokine amounts in culture supernatants in A (n=6). (D) IL-17A production by RPL23A-stimulated lymphocytes from R7–39 mice in the presence or absence of anti-MHC class I or class II (anti-I-A/E) blocking antibody (n=6). (E) IL-17A production by lymphocytes stimulated with recombinant RPL23A or control GST proteins (n=8). Lymphocytes were taken from SKG or BALB/c mice with or without mannan treatment. Data are from 3 (A and B), 6 (C and D) or 8 (E) mice. In B, results are shown as the means±s.d. For C–E, horizontal bars indicate the means. *P<0.05 (Kruskal-Wallis test followed by Steel-Dwass test). NS, not significant. Results represent two independent experiments.
We next evaluated the contribution of Treg cells to controlling arthritogenic CD4+ T cells. Treg cells from either ZAP-70-intact BALB/c or ZAP-70-mutant SKG mice failed to suppress arthritis development in Rag2−/− mice when co-transferred with phenotypically activated or memory R7–39 TCR+ CD4+ T cells (fig. S7 and S18), albeit Treg cells were capable of suppressing naïve arthritogenic T cells effectively (9).
These results collectively indicate that RPL23A is able to stimulate CD4+ T cells in R7–39 mice via RPL23A-derived peptide/MHC class II complexes, driving them to differentiate into arthritogenic effector TH cells (20), which are capable of mediating arthritis even in the presence of Treg cells.
Lastly, we examined possible immune responses to RPL23A in RA patients. RPL23A mRNA was ubiquitously expressed in healthy human tissues (Fig. 4A). In synovial tissues of RA and also apparently normal synovial tissues of osteoarthritis (OA), RPL23A was detected in the cytoplasm of synovial cells, including CD55+ FLSs (Fig. 4B). A significantly higher proportion of RA patients (16.8%, n=374) were positive for serum anti-RPL23A IgG-type autoantibodies compared with healthy controls (1.3%, n=74)) (Fig. 4C). Two out of 23 psoriatic arthritis (PsA) patients (8.7%) but none of OA (n=11), systemic lupus erythematosus (SLE) (n=30), or polymyositis/dermatomyositis (PM/DM) (n=10) patients were positive for the autoantibody. In addition, in synovial fluid of a subset of RA patients, we detected CD4+ T cells producing IFN-γ upon stimulation with RPL23A (Fig. 4D–E). These findings in humans, together with the key role of anti-RPL23A T-cell responses for autoimmune arthritis and psoriasis-like dermatitis in mice, suggest that the responses may play a pathogenic role at least in a subset of patients with RA or PsA.
Fig. 4. Anti-RPL23A humoral and cellular immune responses in RA patients.
(A) qPCR analysis of RPL23A gene expression in various tissues from healthy human subject (n=3). Results are shown as the means±s.d. and represent two independent experiments. (B) Immunohistochemical staining of synovial tissues from RA or OA patients for RPL23A or CD55 expression (bar, 200 μm in left 4 pictures and 50 μm in right 2 pictures). Serial sections were stained by anti-RPL23A, anti-CD55 or control antibody. Arrows indicate cells that are both RPL23A and CD55 positive. Representative results from 3 patients are shown. (C) Serum levels of anti-RPL23A autoantibodies assessed by enzyme-linked immunosorbent assay (ELISA) in RA, PsA, OA, SLE, PM/DM, or healthy individuals. Horizontal bars indicate the medians. ***P<0.001, (Kruskal-Wallis test followed by Dunn’s multiple comparison test). (D) Cytokine production from CD4+ T cells stimulated with recombinant RPL23A or GST protein. (E) Ratios of IFN-γ+ cells in RPL23A-stimulated or GST-stimulated CD4+ T cells in RA patients (n=24) or healthy individuals (n=9). *P<0.05, (Chi-square test). Dotted bars show the threshold.
We have thus shown that, by attenuating TCR signal intensity in developing T cells, hence reducing their sensitivity to thymic negative selection by natural self-ligands, T cells reactive with ubiquitously expressed self-antigens can be generated as dominant pathogenic clones causing systemic autoimmune disease. Since similar attenuation of TCR signal at various degrees in conjunction with Treg cell depletion is able to produce a variety of other autoimmune diseases in mice (9, 22), this strategy of generating pathogenic T cells and characterizing the self-antigens they recognize would facilitate our understanding of the mechanisms of other autoimmune diseases of currently unknown etiology. In addition, given that genetic polymorphism in a signaling molecule in T cells is a major determinant of genetic susceptibility to various human autoimmune diseases including RA (23), such a genetic variation might, at least in part, alter thymic selection, hence forming a TCR repertoire for causing autoimmune disease. Our approach is also instrumental in deciphering how T-cell autoimmunity to a ubiquitous self-antigen should trigger localized tissue damage in RA and other human autoimmune diseases, and devising effective ways of their systemic or local intervention.
Supplementary Material
Acknowledgments
We thank D. O. Adeegbe, Y. Kitagawa, and K. Chen for critical reading of the manuscript; E. Yamamoto, M. Matsuura, R. Ishii, Y. Tada, Y. Funabiki, technical support team (Graduate School of Medicine, Kyoto University) and K. Saito of DNA-chip Development Center for Infectious Diseases (RIMD, Osaka University) for technical assistance; T. Matsushita for histology; T. Kitamura for gifts of the packaging cell line Plat-E; the members of department of Rheumatology and Clinical Immunology in Kyoto University for providing us with patients’ sera; and the members of our laboratories for comments. The data presented in this paper are tabulated in the main paper and in the supplementary materials. This study was supported by Grants-in-Aid for Specially Promoted Research 20002007 (S.S. and Y.I.), for Young Scientists (B) 24790996 (Y.I.) from Japan Society for the Promotion of Science, and Core Research for Evolutional Science and Technology from the Japan Science and Technology Agency (S.S.). M.H., T.F., M.F., H.I., and T.M. are affiliated with a department that is supported financially by five pharmaceutical companies (Mitsubishi Tanabe Pharma Co., Bristol-Myers K.K., Chugai Pharmaceutical Co., Ltd., AbbVie GK., Eisai Co., Ltd.). The sponsors were not involved in the study design; in the collection, analysis, interpretation of data; in the writing of this manuscript; or in the decision to submit the article for publication.
Footnotes
Materials and Methods
References
- 1.Sakaguchi S, Powrie F, Ransohoff RM. Re-establishing immunological self-tolerance in autoimmune disease. Nature medicine. 2012 Jan;18:54. doi: 10.1038/nm.2622. [DOI] [PubMed] [Google Scholar]
- 2.Marrack P, Kappler J, Kotzin BL. Autoimmune disease: why and where it occurs. Nature medicine. 2001 Aug;7:899. doi: 10.1038/90935. [DOI] [PubMed] [Google Scholar]
- 3.Feldmann M, Brennan FM, Maini RN. Rheumatoid arthritis. Cell. 1996 May 3;85:307. doi: 10.1016/s0092-8674(00)81109-5. [DOI] [PubMed] [Google Scholar]
- 4.Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003 May 15;423:356. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 5.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011 Dec 8;365:2205. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 6.Chang CX, et al. Sources of diversity in T cell epitope discovery. Front Biosci. 2011;16:3014. doi: 10.2741/3895. [DOI] [PubMed] [Google Scholar]
- 7.Hsu LY, Tan YX, Xiao Z, Malissen M, Weiss A. A hypomorphic allele of ZAP-70 reveals a distinct thymic threshold for autoimmune disease versus autoimmune reactivity. The Journal of experimental medicine. 2009 Oct 26;206:2527. doi: 10.1084/jem.20082902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakaguchi N, et al. Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nature. 2003 Nov 27;426:454. doi: 10.1038/nature02119. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka S, et al. Graded attenuation of TCR signaling elicits distinct autoimmune diseases by altering thymic T cell selection and regulatory T cell function. J Immunol. 2010 Aug 15;185:2295. doi: 10.4049/jimmunol.1000848. [DOI] [PubMed] [Google Scholar]
- 10.Sommers CL, et al. A LAT mutation that inhibits T cell development yet induces lymphoproliferation. Science. 2002 Jun 14;296:2040. doi: 10.1126/science.1069066. [DOI] [PubMed] [Google Scholar]
- 11.Siggs OM, et al. Opposing functions of the T cell receptor kinase ZAP-70 in immunity and tolerance differentially titrate in response to nucleotide substitutions. Immunity. 2007 Dec;27:912. doi: 10.1016/j.immuni.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holst J, et al. Generation of T-cell receptor retrogenic mice. Nature protocols. 2006;1:406. doi: 10.1038/nprot.2006.61. [DOI] [PubMed] [Google Scholar]
- 13.Lennon GP, et al. T cell islet accumulation in type 1 diabetes is a tightly regulated, cell-autonomous event. Immunity. 2009 Oct 16;31:643. doi: 10.1016/j.immuni.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bettini ML, Bettini M, Nakayama M, Guy CS, Vignali DA. Generation of T cell receptor-retrogenic mice: improved retroviral-mediated stem cell gene transfer. Nature protocols. 2013 Oct;8:1837. doi: 10.1038/nprot.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Materials and methods are available as supplementary materials on Science Online.
- 16.Nestle FO, Kaplan DH, Barker J. Psoriasis. The New England journal of medicine. 2009 Jul 30;361:496. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki K, Wool IG. The primary structure of rat ribosomal protein L23a. The application of homology search to the identification of genes for mammalian and yeast ribosomal proteins and a correlation of rat and yeast ribosomal proteins. J Biol Chem. 1993 Feb 5;268:2755. [PubMed] [Google Scholar]
- 18.Fan W, Christensen M, Eichler E, Zhang X, Lennon G. Cloning, sequencing, gene organization, and localization of the human ribosomal protein RPL23A gene. Genomics. 1997 Dec 1;46:234. doi: 10.1006/geno.1997.5038. [DOI] [PubMed] [Google Scholar]
- 19.van Venrooij WJ, van Beers JJ, Pruijn GJ. Anti-CCP antibodies: the past, the present and the future. Nature reviews. Rheumatology. 2011 Jul;7:391. doi: 10.1038/nrrheum.2011.76. [DOI] [PubMed] [Google Scholar]
- 20.Hirota K, et al. T cell self-reactivity forms a cytokine milieu for spontaneous development of IL-17+ Th cells that cause autoimmune arthritis. The Journal of experimental medicine. 2007 Jan 22;204:41. doi: 10.1084/jem.20062259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashimoto M, et al. Complement drives Th17 cell differentiation and triggers autoimmune arthritis. The Journal of experimental medicine. 2010 Jun 7;207:1135. doi: 10.1084/jem.20092301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ono M, Shimizu J, Miyachi Y, Sakaguchi S. Control of autoimmune myocarditis and multiorgan inflammation by glucocorticoid-induced TNF receptor family-related protein(high), Foxp3-expressing CD25+ and CD25− regulatory T cells. Journal of immunology. 2006 Apr 15;176:4748. doi: 10.4049/jimmunol.176.8.4748. [DOI] [PubMed] [Google Scholar]
- 23.Bottini N, Vang T, Cucca F, Mustelin T. Role of PTPN22 in type 1 diabetes and other autoimmune diseases. Seminars in immunology. 2006 Aug;18:207. doi: 10.1016/j.smim.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Hirota K, et al. Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. The Journal of experimental medicine. 2007 Nov 26;204:2803. doi: 10.1084/jem.20071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006 May 11;441:235. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 26.Bolotin DA, et al. MiTCR: software for T-cell receptor sequencing data analysis. Nature methods. 2013 Sep;10:813. doi: 10.1038/nmeth.2555. [DOI] [PubMed] [Google Scholar]
- 27.Yoshitomi H, et al. A role for fungal (1)-glucans and their receptor Dectin-1 in the induction of autoimmune arthritis in genetically susceptible mice. The Journal of experimental medicine. 2005 Mar 21;201:949. doi: 10.1084/jem.20041758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White J, Pullen A, Choi K, Marrack P, Kappler JW. Antigen recognition properties of mutant V beta 3+ T cell receptors are consistent with an immunoglobulin-like structure for the receptor. The Journal of experimental medicine. 1993 Jan 1;177:119. doi: 10.1084/jem.177.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aletaha D, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis and rheumatism. 2010 Sep;62:2569. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 30.Taylor W, et al. Classification criteria for psoriatic arthritis: development of new criteria from a large international study. Arthritis and rheumatism. 2006 Aug;54:2665. doi: 10.1002/art.21972. [DOI] [PubMed] [Google Scholar]
- 31.Altman R, et al. Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthritis and rheumatism. 1986 Aug;29:1039. doi: 10.1002/art.1780290816. [DOI] [PubMed] [Google Scholar]
- 32.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis and rheumatism. 1997 Sep;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 33.Tanimoto K, et al. Classification criteria for polymyositis and dermatomyositis. The Journal of rheumatology. 1995 Apr;22:668. [PubMed] [Google Scholar]
- 34.Feitsma AL, et al. Identification of citrullinated vimentin peptides as T cell epitopes in HLA-DR4-positive patients with rheumatoid arthritis. Arthritis and rheumatism. 2010 Jan;62:117. doi: 10.1002/art.25059. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama M, et al. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 2005 May 12;435:220. doi: 10.1038/nature03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stadinski BD, et al. Chromogranin A is an autoantigen in type 1 diabetes. Nature immunology. 2010 Mar;11:225. doi: 10.1038/ni.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.