Abstract
Background
Cadmium (Cd) and selenium (Se) antagonistically influence redox balance and apoptotic signaling, with Cd potentially promoting and Se inhibiting oxidative stress and apoptosis. Alterations to placental redox and apoptotic functions by maternal exposure to Cd and Se during pregnancy may explain some of the Cd and Se associations with fetal development.
Objectives
Investigate associations between Cd and Se concentrations in maternal toenails with placental expression patterns of tumor necrosis factor (TNF) and steroidogenic genes involved in redox reactions and test associations with fetal growth.
Methods
In a sub-sample from the Rhode Island Child Health Study (n = 173), we investigated the relationships between: (1) maternal toenail Cd and Se concentrations and fetal growth using logistic regression, (2) Cd and Se interactions with factor scores from placental TNF and steroidogenic expression patterns (RNAseq) using linear models, and (3) TNF and steroidogenic expression factors with fetal growth via analysis of covariance.
Results
Se was associated with decreased odds of intrauterine growth restriction (IUGR) (OR = 0.27, p-value = 0.045). Cd was associated with increased odds of IUGR (OR = 1.95, p-value = 0.13) and small for gestational age (SGA) births (OR = 1.46, p-value = 0.11), though not statistically significant. Cd and Se concentrations were antagonistically associated with placental TNF and steroidogenic expression patterns, which also differed by birth size.
Conclusions
Se may act as an antagonist to Cd and as a modifiable protective factor in fetal growth restriction, and these data suggest these effects may be due to associated variations in the regulation of genes involved in placental redox balance and/or apoptotic signaling.
1. Introduction
Trace exposures to compounds or elements ubiquitous in the environment have received substantial recent attention for their potential detrimental influences on reproductive health and birth outcomes (Rahman et al., 2016). Cadmium (Cd), a common contaminant that is toxic at relatively low-levels, is well recognized as a human carcinogen and for its toxic effects on the kidneys, liver, and bones (Nair et al., 2013), and has been implicated as a developmental toxicant. Human exposure to Cd primarily occurs through the diet, though among smokers, cigarettes are the primary source of Cd exposure (Järup and Åkesson, 2009; Satarug and Moore, 2004). Selenium (Se), on the other hand, is an essential micronutrient that putatively elicits numerous health benefits including anti-carcinogenic and anti-inflammatory activities, positive effects on the immune and cardiovascular systems, an ability to mitigate the toxic effects of heavy metals, and critical anti-oxidative roles during pregnancy (Pieczyńska and Grajeta, 2015). However, Se also has a narrow window of sufficiency and may elicit toxic effects dependent on dose and interactions with other environmental exposures (Jablonska and Vinceti, 2015). Like Cd, the primary source of human Se exposure is via the diet, through the consumption of plants, or of animals that fed on Se-rich plants (Pieczyńska and Grajeta, 2015; Mehdi et al., 2013).
At the cellular level, Cd can induce delayed apoptosis, apoptosis, or necrosis (Templeton and Liu, 2010; López et al., 2003), whereas Se can inhibit these signals (Zhou et al., 2009). Furthermore, Cd and Se appear to influence the generation and suppression of reactive oxygen species (ROS) in a diametrically opposed a manner (Zhou et al., 2009; El-Sharaky et al., 2007). Though Cd is not a redox-active metal, once it has been absorbed into a cell, it interferes with mitochondrial function, depletes and inhibits antioxidants, and displaces redox-active metals from metal-binding proteins, all of which can result in substantial shifts in cellular redox balance towards ROS generation and increased oxidative stress (Valko et al., 2016). Conversely, Se is a critical co-factor in redox homeostasis, interacting with many selenoproteins that neutralize reactive species and are involved in various anti-oxidative activities, hormone synthesis and reproduction (Mehdi et al., 2013). Se has also been shown to protect against Cd-induced oxidative stress and apoptosis (Zhou et al., 2009; El-Sharaky et al., 2007; Jihen and Imed, 2009; Karabulut-Bulan et al., 2016).
Animal models and in vitro studies have shown that Cd induces preeclampsia-like symptoms, restricts fetal growth (Wang et al., 2014, 2016a), may interfere with placental steroidogenesis (Kawai et al., 2002; Stasenko et al., 2010), inhibits trophoblast proliferation while promoting apoptosis (Wang et al., 2012; Erboga and Kanter, 2016), increases placental oxidative stress (Wang et al., 2012) and interferes with maternal-fetal nutrient transfer across the placenta (Wang et al., 2016a; Mikolić et al., 2015), demonstrating the placenta is a likely target tissue for Cd-associated reproductive toxicity. Indeed, excess ROS generation in the placenta has also been associated with altered placental function and negative pregnancy outcomes (Min et al., 2009; Scifres and Nelson, 2009; Vanderlelie et al., 2005). Furthermore, Cd may influence the distribution of Se across maternal, fetal and placental tissues, and vice versa (Al-Saleh et al., 2015; Kantola et al., 2004).
Epidemiologic studies have observed increasing concentrations of placental, maternal or fetal Cd to be associated with various measures of restricted fetal growth or pregnancy complications (Al-Saleh et al., 2015, 2014; Kippler et al., 2013, 2012; Johnston et al., 2014; Laine et al., 2015; Llanos and Ronco, 2009; Wang et al., 2016b; Menai et al., 2012). Alternatively, many studies have found that maternal, fetal or placental Se concentrations positively correlate with fetal growth and successful pregnancies (Bogden et al., 2006; Klapec et al., 2008; Negi et al., 2012; Sun et al., 2014; Rayman et al., 2011, 2015, 2003). The adverse Cd-associated pregnancy and birth outcomes may be mitigated by concurrently higher levels of Se. While Cd-Se antagonism has been demonstrated at the cellular level with redox and apoptotic activity, only a handful of epidemiologic studies have investigated these responses, and these have produced mixed results. Laine et al. (2015) found that the odds of Cd-associated preeclampsia (PE) were higher among mothers with lower placental concentrations of Se (Laine et al., 2015), while Al-Saleh et al. (2015) observed no antagonism between Se and Cd on birth anthropomorphic measurements (Al-Saleh et al., 2015).
There is biological evidence that Cd and Se can influence steroid biosynthesis, redox balance, apoptotic signaling, fetal growth, and successful pregnancies. Yet there is much left to be unraveled about the mechanisms through which trace exposures to Cd and Se during pregnancy influence fetal growth and pregnancy outcomes in humans. Studies have shown that the tumor necrosis factor super family (TNF-SF) encompasses many potent pro-apoptotic cytokines and their receptors (Zelová and Hošek, 2013), in which some of them have been shown to be perturbed by Cd exposure (Kayama et al., 1995; Kim et al., 2002; Kumar et al., 2016). Cd has also been observed to perturb various steps in progesterone, testosterone, cortisol/cortisone, and estradiol/estrone synthesis (Wang et al., 2014; Kawai et al., 2002; Nagata et al., 2005; Pillai and Gupta, 2005; Pandya et al., 2012; Ronco et al., 2010), and the by-products of these activities present major sources of intracellular ROS, particularly those involved in electron transport at the mitochondrial membrane (Prasad et al., 2014). Based on these observations, we sought to test the hypothesis that maternal Cd and Se exposures during pregnancy might be associated with variations in the placental expression patterns of key genes involved in apoptotic signaling or ROS generation via steroidogenesis, and could be related to fetal growth. Cd and Se were measured in maternal toenails collected postpartum and which represent long-term exposure. In this study, we included all TNF-SF genes that were sufficiently detectable in our placental samples, which comprised of 19 TNF-SF receptors (TNFRSF1A, TNFRSF1B, TNFRSF3, TNFRSF4, TNFRSF5, TNFRSF6, TNFRSF10A, TNFRSF10B, TNFRSF10C, TNFRSF10D, TNFRSF11A, TNFRSF12A, TNFRSF14, TNFRSF16, TNFRSF19, TNFRSF19L, TNFRSF21, TNFRSF25, and TNFRSF27) and 2 TNF-SF ligands (TNFSF10 and TNFSF15). Thorough descriptions of TNF-SF receptor-ligand interactions and their functions are reviewed (Zelová and Hošek, 2013). The TNF-SF genes included in our study, as well as their major structural similarities are highlighted in the supplemental materials (Supplemental Fig. 1A). Additionally, we investigated cytochrome P450 (CYP11A1 and CYP19A1) and hydroxysteroid dehydrogenase (HSD3B1, HSD3B7, HSD11B1, HSD11B2, HSD17B1, and HSD17B2) genes involved in steroid biosynthesis. Moon et al (2014) provide a thorough description of this pathway in association with preeclampsia (Moon et al., 2014); we highlight the genes included in our study in the supplemental materials (Supplemental Fig. 1B).
2. Methods
2.1. Rhode Island Child Health Study
The Rhode Island Child Health Study (RICHS) is a hospital-based birth cohort of healthy mothers with singleton, viable, non-pathologic pregnancies born at ≥ 37 weeks gestation from the Women and Infants’ Hospital in Providence, RI, USA (n = 840). Exclusion criteria comprised of maternal age less than 18 years, life threatening conditions, or infants with congenital/chromosomal abnormalities. This study aimed to investigate factors associated with very large and very small birth sizes. In accordance with the eligibility criteria, mothers of infants born large for gestational age (LGA) (≥ 90th BW percentile) and SGA (≤ 10th BW percentile) during normal working hours and up to two infants born adequate for gestational age (AGA) (between the 10th and 90th BW percentiles) of the same sex, gestational age (± 3 days), and maternal age (± 2 years) were approached; 63% of those approached agreed to participate. All protocols were approved by the institutional review boards at the Women and Infants’ Hospital and Emory University and all participants provided written informed consent. Toenails for trace elements measurements (n = 242), and placental samples for gene expression profiling (n = 200), were collected after delivery. This study included the cross-section of mother-infant pairs for which both toenail trace elements and placental gene expression measurements were available (n = 173).
2.2. Medical records and questionnaire-based data
An interviewer administered questionnaire was used to collect self-reported sociodemographic, lifestyle, and medical history data, and a structured medical records review was employed to collect anthropometric and clinical data. Data on gestational weeks, infant sex, birth weight (grams) and intrauterine growth restriction (IUGR) status, which was determined in utero via ultrasound, were obtained via medical records abstraction. Size at birth was categorized as SGA, AGA, and LGA, calculated as sex-specific birthweight percentiles adjusted for gestational age via the revised Fenton growth chart (Fenton and Kim, 2013).
2.3. Toenail Cd and Se concentrations
Toenail clippings were collected from mothers approximately 2.8 months postpartum via pre-labelled collection envelopes as described in a prior study (Everson et al., 2016). Protocols for toenail cleaning, preparation, and analysis via inductively coupled plasma mass spectrometry (ICP-MS) have been described in detail elsewhere (Punshon et al., 2016). Cd and Se concentrations (micrograms (µg) per gram (g) of toenail) were measured at the Dartmouth Trace Element Analysis Core from 242 maternal toenail samples. Toenails were digested with 9:1 nitric acid: hydrochloric acid (HNO3:HCl). The digestate was then analyzed by ICP-MS (Agilent 7700x, Santa Clara, CA) following EPA 6020A (Method, 1998); QC involved initial and continuing calibration verification and blanks, digestion blanks, and fortified blanks. Hair powder certified reference materials were used to check the accuracy of the method (NIES # 13 and GBW07601, certified at 0.23 and 0.11 µg/g (Cd) and 1.79 and 0.6 µg/g (Se)). Recoveries for NIES # 13 (n = 17) were 102% ± 12% (Cd) and 91% ± 6% (Se) while recoveries for GBW07601 (n = 9) were 114% ± 8% (Cd) and 102% ± 10% (Se). Because each study participant provided a different mass (g) of the toenail (ranging between 0.0011 g and 0.1223 g), the assay had sample-specific detection limits (SSDL) for each metal. Medians for the SSDLs were 0.0059 (IQR = 0.015) µg/g for Cd and 0.017 (IQR = 0.030) µg/g for Se; reported concentrations were above the SSDLs for > 99% of samples for Se, and 49% for Cd.
2.4. Placental gene expression
Within two hours of birth, full-thickness sections of placenta were biopsied from the fetal side, free of maternal decidua, at four quadrants around the cord insertion. These samples were immediately placed in RNAlater™ (Applied Biosystems, Inc., AM7020). Following ≥72 h at 4ºC, samples were blotted dry and snap-frozen in liquid nitrogen. The samples were homogenized and stored at −80 °C until RNA was extracted for analyses via RNA-seq. Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA), ribosomal RNA was removed and the RNA sequencing library was prepared using a RiboZero Kit (Huang et al., 2011) and the rRNA depleted samples were converted to cDNA using random hexamers (Thermo Scientific, Waltham, MA). Transcriptome-wide RNA sequencing was conducted using the HiSeq. 2500 platform (Illumina, San Diego, CA) (Bentley et al., 2008). Approximately 20 million single-end reads (50 bp in length) were generated per sample. The raw RNA sequencing data (fastq files) were assessed for quality control, including read length and GC content, using the FastQC software. Reads that passed the quality control metrics were mapped to the human reference genome (hg19) in a splice-aware manner using the Spliced Transcripts Alignment to a Reference (STAR) aligner, with common SNPs in the reference genome masked prior to alignment. Genes with counts per million < 1 in greater than 30 samples (the sample size of the smallest phenotypic group in this study) were considered unexpressed and removed. Read counts were GC content adjusted using the EDASeq R package, followed by TMM correction for library size differences across samples using the calcNorm-Factors function in edgeR R package. The data was then transformed into logCPM values accounting for the mean-variance relationship in the data using the voom function of the limma R package. The normalized log2 counts per million (logCPM) reads were filtered to genes with expression levels above 2 in the log2 scale in a minimum of 30 samples. The Blom transformation was used to correct for skewness, unequal variance between genes, and potential outliers (Zwiener et al., 2014). The final data-set included 12,137 genes, from which our 21 TNF-SF and 8 steroidogenesis candidate genes were selected. Batch effects were removed from the data via the ‘combat’ function in the ‘sva’ R.
2.5. Statistical analysis
Toenail Cd and Se concentrations that were below the limit of detection (LOD) were assigned a value equal to half of their SSDL. Cd concentrations were log-normally distributed, so Cd was natural log-transformed prior to all analyses. Se concentrations were approximately normally distributed with a few potential outliers in both tails of the distribution. To reduce the potential bias that these outliers may introduce into our regression models, we winsorized (Wilcox and Keselman, 2012) the Se distribution by three times the median absolute deviation, resulting in seven data points being recoded to the maximum and minimum values of the truncated distribution (Supplemental Fig. 2).
Exploratory factor analysis (EFA), a multivariate method that examines the degree to which unmeasured latent variables are described by a set of measured variables, was used to reduce the dimensionality and summarize the expression patterns of the TNF-SF gene set (21 genes) and steroidogenesis gene set (8 genes). The number of factors was determined by sequentially adding in additional factors (starting with one factor) while calculating goodness-of-fit measures and stopping once the minimum value for the Bayes information criterion (BIC) was achieved. To improve interpretability of factor loadings, we utilized the promax rotation, which is an oblique rotation that allows for correlations between factors. We opted for an oblique solution because we assumed that there may be multiple underlying but potentially correlated patterns within the TNF-SF and redox expression structures. We explored the correlations between genes using Pearson correlations and heatmaps, then used Pearson correlations and scatterplots to characterize correlations between factors.
Associations between binary outcomes (odds of IUGR or SGA) and metals concentrations were fitted with penalized logistic regressions models to reduce potential small-sample bias (Firth, 1993). Robust linear models were used to estimate the associations between metals concentrations and factor scores. Standard errors were calculated from covariance matrices estimated via robust sandwich estimators shown to be effective with small samples and potentially influential observations (Long and Ervin, 2000). Multivariate analyses of covariance (MANCOVA) and analyses of covariance (ANCOVA) were used to assess whether the distributions of factor scores were associated with birth outcomes.
Adjustment covariates and effect modifiers were selected based on literature review. Smoking during pregnancy was included as a covariate due to its effects as a reproductive toxicant and its influence on tissue concentrations of Cd (Punshon et al., 2016) and Se (Mistry et al., 2014). Because Cd may accumulate in body tissues with age, and the placenta of older mothers may have higher concentrations of Cd (Punshon et al., 2016), and maternal age can influence pregnancy success and birth outcomes (Kenny et al., 2013), we included maternal age in all models. Gestational age was included due to its strong influence on birth size and a potential inverse relationship with placental apoptosis (Erboga and Kanter, 2016). We also explored potential effect modification by infant sex (Kippler et al., 2013; Romano et al., 2016).
3. Results
3.1. Cd and Se associations with fetal growth
The study sample (n = 173) exhibited very similar distributions of all covariate and outcome variables when compared to the full RICHS cohort (n = 840) (Supplemental Table 1). The median and IQR for toenail concentrations of Cd (median = 0.0084 µg/g, IQR = 0.012 µg/g) were substantially lower than for Se (median = 0.965 µg/g, IQR = 0.226 µg/g) (Supplemental Table 2). Median maternal Cd concentrations were higher among IUGR-complicated pregnancies (median = 0.015 µg/g) or those that resulted in SGA (median = 0.0091 µg/g), and lowest among pregnancies complicated by neither (median = 0.0083 µg/g). Conversely, median maternal Se concentrations were lower among IUGR-complicated pregnancies (median = 0.869 µg/g) or those that resulted in SGA (median = 0.972 µg/g), whereas pregnancies complicated by neither had the highest Se concentrations (median = 0.974 µg/g). White mothers had a lower proportion of SGA and IUGR births, and female infants were more likely to be SGA or IUGR than male infants. Most other covariates and demographic characteristics were similar across babies born SGA, IUGR, or neither SGA or IUGR (Table 1). Maternal toenail Se and Cd concentrations were not significantly associated with smoking during pregnancy, ever-smoking status, highest educational achievement (t-test or correlation p-values > 0.05), maternal age, or weeks of gestation (correlation p-values > 0.05). However, toenail concentrations of Cd and Se were modestly inversely correlated with each other (r = −0.18, p-value = 0.02), Se was higher among white mothers (t-test p-value = 0.0077), and Cd was higher among mothers that gave birth to female infants (t-test p-value = 0.0077).
Table 1.
Distributions of maternal and child demographic, pregnancy, and birth outcome variables, among those complicated by intra-uterine growth restriction (IUGR), born small for gestational age (SGA) but not with IUGR, and neither IUGR or SGA.
| Non-SGA or IUGR (n = 143) Mean (SD) |
SGA, non- IUGR (n = 22) Mean (SD) |
IUGR (n = 8) Mean (SD) |
Comparison P-value† |
|
|---|---|---|---|---|
| Birth Weight (g) | 3747.67 (506.26) | 2672.05 (274.07) | 2400.63 (211.13) | < 0.0001 |
| Maternal Age (yrs) | 31.43 (4.08) | 32.55 (4.09) | 34.88 (4.76) | 0.0034 |
| Gestational Age (weeks) | 39.14 (0.87) | 39.36 (1.05) | 38.00 (0.93) | 0.070 |
| Maternal Se (µg/g)* | 0.97 (0.20) | 0.97 (0.28) | 0.87 (0.23) | 0.23 |
| Maternal Cd (µg/g)* | 0.0083 (0.013) | 0.0091 (0.015) | 0.0153 (0.0095) | 0.23 |
| Freq. (%) | Freq. (%) | Freq. (%) | ||
| Infant Sex | ||||
| Female | 67 (47) | 13 (59) | 7 (88) | 0.051 |
| Male | 76 (53) | 9 (41) | 1 (12) | |
| Ever Smoker | ||||
| No | 103 (72) | 19 (86) | 6 (75) | 0.38 |
| Yes | 40 (28) | 3 (14) | 2 (25) | |
| Smoking During Pregnancy | ||||
| None | 133 (93) | 21 (95) | 7 (88) | 0.65 |
| Any | 10 (7) | 1 (5) | 1 (12) | |
| Maternal Ethnicity | ||||
| Other | 21 (15) | 9 (41) | 4 (50) | 0.0018 |
| White | 122 (85) | 13 (59) | 4 (50) | |
| Highest Educational Attainment | ||||
| No more than High School | 15 (11) | 2 (9) | 1 (12) | 1.0 |
| Some Post-High School | 126 (89) | 20 (91) | 7 (88) |
Median and interquartile range presented in place of mean and sd.
P-values from Kruskal-Wallis tests for continuous variables and Fisher’s Exact tests for categorical variables.
We then tested whether these variations in Cd and/or Se were associated with IUGR or SGA outcomes while controlling for potential confounders (Table 2). Odds of IUGR were significantly lower among mothers with higher Se concentrations (OR = 0.27, 95% CI = (0.08, 0.98) per IQR increase in toenail Se), adjusted for maternal Cd, infant sex, gestational time, maternal age and maternal smoking during pregnancy. Whereas odds of IUGR were greater with increasing maternal Cd, though this association was not statistically significant (OR = 1.95, 95% CI = (0.82, 4.65) per IQR increase in toenail Cd). Neither element was significantly associated with odds of SGA. We then tested whether associations between Cd and IUGR or SGA were dependent on Se concentrations or infant sex. None of the stratified models yielded statistically significant associations, though the odds ratio for Cd and IUGR was substantially larger within the low Se strata (OR = 3.32, 95% CI = (0.88, 12.48)). Since others have observed that Cd associations with fetal growth and pregnancy complications might vary by sex and Se exposure, we then included Se*log-Cd and sex*log-Cd interaction terms within the adjusted logistic regression models. Though none of the interaction terms yielded p-values < 0.05, we present the stratified results to allow for comparisons with other studies (Supplemental Tables 3).
Table 2.
Associations between IUGR and SGA with Cd (µg/g) and Se (µg/g) toenail concentrations within the study sample (n = 173), via bias-reduced logistic regression models.
| IUGR | SGA | |||
|---|---|---|---|---|
|
|
|
|||
| Toenail Metal Conc. |
Odds Ratio (95% CI) |
p-value | Odds Ratio (95% CI) |
p-value |
| Log-Cda | 1.63 (0.98, 2.69) | 0.056 | 1.53 (0.97, 2.39) | 0.065 |
| Log-Cdb,d | 1.95 (0.82, 4.65) | 0.13 | 1.46 (0.92, 2.30) | 0.11 |
| Sea | 0.41 (0.13, 1.34) | 0.14 | 0.90 (0.49, 1.65) | 0.73 |
| Sec,e | 0.27 (0.08, 0.98) | 0.045 | 0.96 (0.53, 1.73) | 0.90 |
Unadjusted model.
IUGR models adjusted for Se, gestational time, infant sex, maternal age, and maternal smoking during pregnancy.
IUGR models adjusted for log(Cd), gestational time, infant sex, maternal age, and maternal smoking during pregnancy.
SGA models adjusted for Se, infant sex, maternal age, and maternal smoking during pregnancy.
SGA models adjusted for log(Cd), infant sex, maternal age, and maternal smoking during pregnancy.
3.2. Cd and Se associations with gene expression
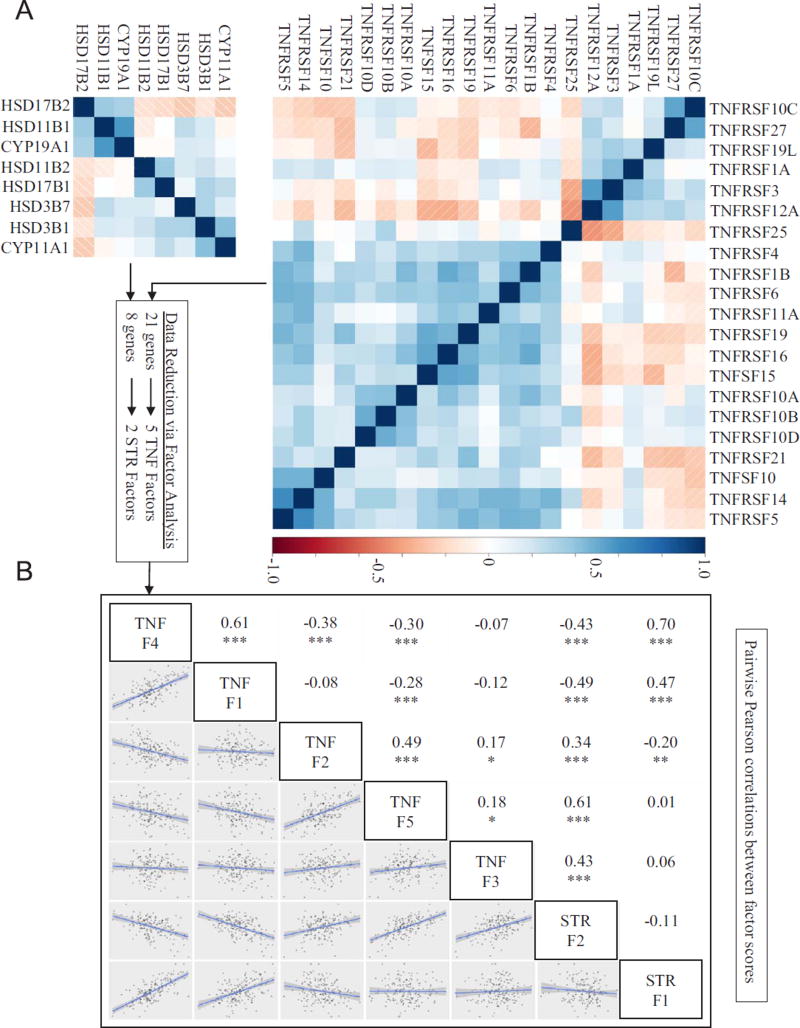
To test whether Cd and/or Se concentrations in maternal toenails were associated with placental redox and TNF-SF signaling, we identified 29 candidate genes, 8 involved in steroidogenesis reactions (STR) and 21 involved in TNF signaling. Many of the expression levels within these gene sets were at least moderately correlated with each other (Fig. 1A), thus we conducted exploratory factor analyses (EFA) to summarize the major axes of variation within each gene set. Using the minimum BIC and the extended BIC (eBIC) to determine the number of factors (Supplemental Table 4), we reduced 21 TNF genes to 5 factors and 8 steroidogenic (STR) genes to 2 factors. Factor loadings and annotations for the TNF (Table 3) and STR (Table 4) gene sets aided in our interpretations of these factors and their potential functional relevance. The CYP11A1 and HSD3B1 genes, which drive the first steps of steroid biosynthesis, were co-expressed and loaded to the same factor (STR Factor 2), whereas genes that drive biosynthesis in opposite directions (HSD11B1 vs HSD11B2, and HSD17B1 vs HSD17B2) tended to be anti-correlated and loaded to different factors. Additionally, all of the TNFSF10 receptors (TNFRSF10A–D) were co-expressed and at least moderately loaded to TNF Factor 1 (loadings > 0.3). The remaining TNF-SF factors were loaded with various combinations of genes involved in both pro- and anti-apoptotic signaling. For instance, TNF Factor 2 was strongly loaded with both a pro-apoptotic gene (TNFRSF27) and a decoy rector (TNFRSF10C) that most likely inhibits apoptosis, though these two receptors interact with different ligands.
Fig. 1.
(A) Pearson correlations for gene expression levels within the TNF and STR gene sets, and (B) relationships between TNF and STR factors. Statistically significant correlations denoted as *, **, or *** for p-values < 0.05, < 0.01, and < 0.001.
Table 3.
TNF factor scores and functional descriptions of candidate genes; factor loadings greater than 0.40 are highlighted in bold to aid in factor interpretation.
| Gene Symbol | F1 | F2 | F3 | F4 | F5 | Relevant Functional Annotations |
|---|---|---|---|---|---|---|
| TNFRSF10A | 0.877 | −0.103 | 0.086 | −0.239 | 0.041 | Receptor (TRAIL-R); binds TRAIL, transduces cell death and apoptosis signals. |
| TNFRSF1B | 0.622 | −0.361 | 0.142 | 0.077 | 0.031 | Receptor (TNFR2); mediator of TNF-α associated metabolic activity; pro- and anti-apoptotic signaling. |
| TNFRSF10B | 0.622 | 0.318 | −0.381 | 0.072 | 0.094 | Receptor (TRAIL-R); binds TRAIL, transduces cell death signals and apoptosis. |
| TNFRSF10D | 0.546 | 0.137 | −0.100 | 0.100 | 0.157 | Receptor (TRAIL-R); may protect cells from TRAIL-induced apoptosis. |
| TNFRSF16 | 0.395 | 0.030 | −0.031 | 0.217 | −0.384 | Receptor (NGFR); mediator of neural cell survival and death pathways. |
| TNFRSF27 | −0.129 | 0.981 | −0.007 | 0.130 | −0.152 | Receptor (EDA2R); binds ectodysplasin A2, mediates apoptosis. |
| TNFRSF10C | 0.331 | 0.570 | 0.279 | −0.321 | −0.091 | Receptor (TRAIL-R); may protect cells from TRAIL-induced apoptosis. |
| TNFRSF21 | 0.249 | −0.305 | −0.075 | 0.051 | −0.139 | Receptor (DR6); can induce apoptosis via death domain; involved in T-helper cell activation. |
| TNFRSF3 | 0.028 | 0.060 | 0.696 | 0.165 | 0.344 | Receptor (LTBR); regulatory role in lipid metabolism, immune response, and programmed cell death. |
| TNFRSF25 | −0.047 | 0.057 | −0.668 | 0.226 | −0.028 | Receptor (DR3); lymphocyte homeostasis and proliferation; pro- and anti-apoptotic signaling. |
| TNFRSF12A | −0.246 | 0.240 | 0.521 | 0.136 | 0.315 | Receptor (TWEAK-R); promotes angiogenesis and the proliferation of endothelial cells. |
| TNFRSF14 | 0.030 | 0.008 | −0.219 | 0.828 | 0.075 | Receptor (HVEM); can promote/inhibit T-cell immune response; mediator of herpes simplex virus (HSV) uptake by cells. |
| TNFRSF5 | 0.001 | 0.015 | 0.102 | 0.799 | 0.044 | Receptor (CD40); broad spectrum of immune and inflammatory functions. |
| TNFRSF11A | −0.195 | 0.148 | 0.206 | 0.729 | −0.185 | Receptor (RANK); can activate NF-kappa B; regulates T cell and dendritic cell interactions. |
| TNFSF10 | −0.160 | −0.049 | −0.109 | 0.604 | 0.228 | Cytokine (TRAIL); can preferentially induce apoptosis in transformed and/or tumor cells. |
| TNFRSF6 | 0.347 | 0.007 | −0.027 | 0.418 | −0.052 | Receptor (FAS); regulator of programmed cell death and T cell proliferation. |
| TNFRSF4 | 0.275 | 0.026 | 0.086 | 0.387 | −0.001 | Receptor (OX40L-R); activator of NF-kappa B and suppressor of apoptosis; involved in T-cell signaling and B cell proliferation and differentiation. |
| TNFRSF1A | 0.204 | −0.203 | 0.222 | 0.258 | 0.692 | Receptor (TNFR1); mediator of apoptosis and regulates inflammation. |
| TNFSF15 | 0.256 | 0.152 | −0.068 | 0.193 | −0.594 | Cytokine (VEGI); abundantly expressed in endothelial cells; induce apoptosis and inhibit proliferation. |
| TNFRSF19L | 0.219 | 0.064 | 0.099 | −0.048 | 0.484 | Receptor (RELT); activator the NF-kappa B pathway and T-cell proliferation. |
| TNFRSF19 | 0.166 | −0.044 | 0.088 | 0.383 | −0.423 | Receptor (TROY); highly expressed during embryonic development; inducer of apoptosis. |
Table 4.
Steroidogenic (STR) factor scores and functional descriptions of candidate genes; factor loadings greater than 0.40 are highlighted in bold to aid in factor interpretation.
| Gene Symbol | F1 | F2 | Relevant Functional Annotations |
|---|---|---|---|
| HSD11B1 | 0.785 | 0.072 | Hydroxysteroid dehydrogenase; microsomal enzyme involved in inter-conversion between cortisol and cortisone. |
| CYP19A1 | 0.733 | 0.102 | Cytochrome P450 enzyme; localized in the endoplasmic reticulum that catalyzes the last steps of estrogen biosynthesis. |
| HSD17B2 | 0.483 | −0.431 | Hydroxysteroid dehydrogenase; involved in catalyzing the conversion of testosterone and androstenedione, as well as estradiol and estrone. |
| HSD3B1 | 0.247 | 0.709 | Hydroxysteroid dehydrogenase; primarily localized to placenta and non-steroidogenic tissues; required for progesterone synthesis. |
| CYP11A1 | −0.078 | 0.574 | Cytochrome P450 enzyme; localized to the inner mitochondrial membrane; integral to the first step in steroidogenesis. |
| HSD17B1 | −0.071 | 0.472 | Hydroxysteroid dehydrogenase; primarily involved in the reduction of estrogens and androgens. |
| HSD3B7 | 0.276 | 0.434 | Hydroxysteroid dehydrogenase; localized at the membrane of the endoplasmic reticulum; involved in bile acid synthesis. |
| HSD11B2 | −0.075 | 0.396 | Hydroxysteroid dehydrogenase; microsomal enzyme; primarily protects cells from the growth-inhibiting and/or pro-apoptotic effects of cortisol. |
Interestingly, many of the STR and TNF factor scores were moderately-to-highly correlated with each other (Fig. 1B). Of particular note, were the strong correlations between STR Factor 1 with TNF Factor 1 (r = 0.47) and TNF Factor 4 (r = 0.70), and the moderate-to-strong correlations between STR Factor 2 with TNF Factor 2 (r = 0.34), TNF Factor 3 (r = 0.43), and TNF Factor 5 (r = 0.61), all with p-value < 0.0001.
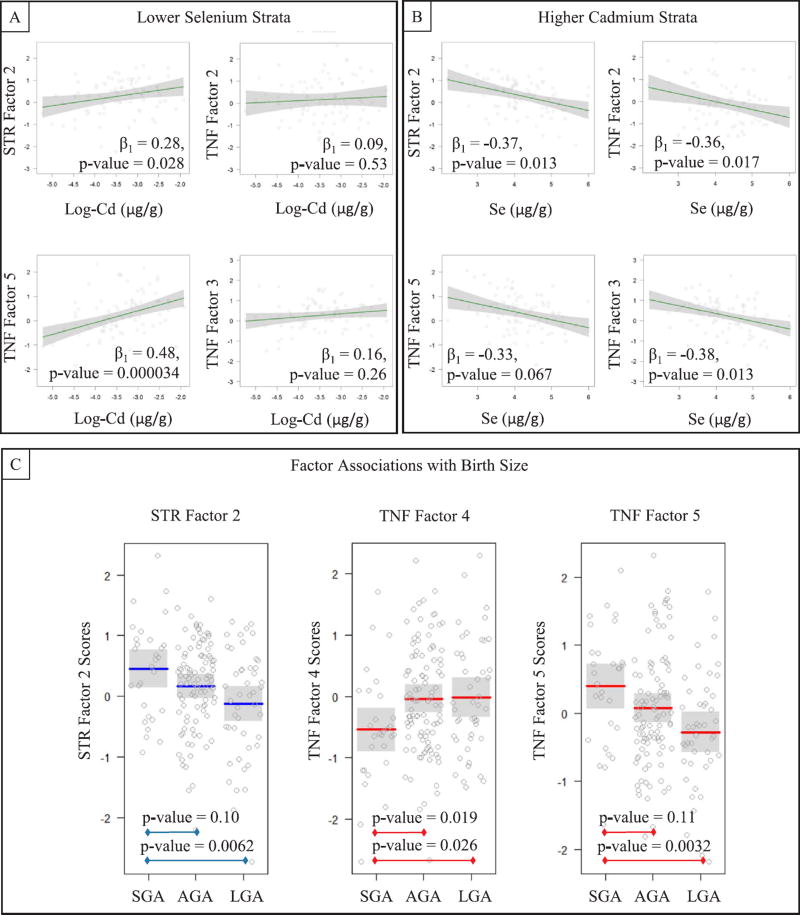
We then tested whether Cd and Se were associated with the TNF and STR factor scores, via adjusted linear regression models while including log(Cd)*Se interaction terms (Table 5). Four of the seven factors yielded interaction p-values < 0.05 (STR Factor 2, TNF-SF Factor 2, TNF-SF Factor 3, and TNF-SF Factor 5). Because we considered that either Se may alter the relationship between Cd and the factor scores, or that Cd may alter the relationship between Se and factor scores, we produced four post-hoc models for each of the four factors with potential Cd-Se interactions: stratified by median Se and then stratified by median Cd (Supplemental Figures 3–6). We observed consistently antagonistic relationships between Cd and Se with STR Factor 2 and TNF Factor 5 in which log(Cd) was positively associated with factor scores within the low-Se strata (Se < 0.97 µg/g) while Se was inversely associated with the factor scores within the higher Cd strata (Cd ≥ 0.0084 µg/g). Se was also inversely associated with TNF Factor 2 and TNF Factor 3 within the high Cd strata, though log(Cd) was not associated with these factors; results are presented in Figs. 2A and B. The factors that were not associated with Se*Cd interaction terms (STR F1, TNF-SF F1 and TNF-SF F4), also were not associated with Cd (p-values = 0.70, 0.17 and 0.64) or Se (p-values = 0.12, 0.93 and 0.42) in adjusted main effects models.
Table 5.
Beta coefficients (p-values) for associations between TNF and steroidogenic factor scores with Cd (µg/g) and Se (µg/g) toenail concentrations and their interaction via linear regressions, adjusted for infant sex, gestational time (weeks), maternal age, and maternal smoking during pregnancy.
| Outcome | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Exposure | STR F1 | STR F2 | TNF F1 | TNF F2 | TNF F3 | TNF F4 | TNF F5 |
| Log-Cd | 0.176 (0.82) | 1.512 (0.0032) | −0.508 (0.43) | 1.175 (0.055) | 1.432 (0.032) | −0.362 (0.63) | 1.751 (0.00026) |
| Se | −0.331 (0.63) | −1.202 (0.0049) | 0.289 (0.58) | −1.154 (0.021) | −1.262 (0.017) | 0.164 (0.79) | −1.306 (0.0013) |
| Interaction Term | −0.052 (0.79) | −0.315 (0.0078) | 0.080 (0.57) | −0.283 (0.041) | −0.328 (0.031) | 0.070 (0.69) | −0.347 (0.0017) |
Fig. 2.
(A) Associations between Se and factor scores (y-axes) dependent on higher Cd concentrations, and (B) associations between Cd and factor scores (y-axes) dependent on lower Se concentrations; (C) average factor scores by birth size groups (AGA, SGA, and LGA); all models adjusted for maternal age, maternal smoking, gestational age, and infant sex.
3.3. Gene expression associations with fetal growth
We then tested whether the set of TNF and STR factors were associated with IUGR and birth size by gestational age (SGA, AGA, and LGA) via multivariate analysis of covariance (MANCOVA), adjusted for maternal age, maternal smoking during pregnancy, and infant sex. The multivariate set of factors did not differ by IUGR (multivariate F-test p-value = 0.44). However, variations in the distribution of TNF and STR factors were significantly associated with birth size by gestational age (multivariate F-test p-value = 0.00016). We then performed individual analyses of covariance (ANCOVA) on each factor separately and found that TNF Factor 5 (p-value = 0.0023), TNF Factor 4 (p-value = 0.0083), and STR Factor 2 (p-value = 0.0021) were associated with birth size. Among those factors with significant differences in factor scores by birth size, larger factor scores were associated with babies born SGA compared to LGA for STR Factor 2 and TNF Factor 5, whereas TNF Factor 4 exhibited the opposite relationship (Fig. 2C).
3.4. Sensitivity analyses
We ran two sensitivity analyses to assess potential biases in our approach. To evaluate possible bias introduced by winsorizing our Se data, which contained apparent influential observations, we performed penalized logistic and robust linear regression models done with and without excluding Se concentrations that were 3*IQR greater or lesser than the median. The results were not affected by these exclusions (data not shown).
Next, to evaluate the influence that Cd concentrations below the limits of detection (51%) may have had on our findings, we repeated our penalized logistic models for IUGR and SGA (Supplemental Table 5), robust linear regression models with log(Cd)*Se interactions (Supplemental Table 6), and robust linear regression models stratified by Cd and Se (Supplemental Table 7), while only including those samples for which both Cd and Se concentrations were above the limits of detection (n = 85). This reduced sample yielded similar associations between IUGR and Se (OR = 0.35, 95%CI = 0.13, 0.91), however the IUGR-association with Cd was attenuated and the confidence interval substantially overlapped the null (OR = 1.21, 95%CI = 0.43, 3.39). Though none of the models with factor scores as the outcomes and log (Cd)*Se interaction terms as the exposures yielded p-values < 0.05 in this sample, three of the four interaction terms did exhibit p-values < 0.15 and TNF Factor 5 which had a p-value = 0.154. Additionally, the directions of effect remained the same for all parameter estimates, and some of the magnitudes of Se-effects actually became stronger for associations with STR Factor 2, TNF Factor 2, and TNF Factor 3 (Table 5; Supplemental Table 6). Likewise, the linear models stratified by Se and Cd produced relationships with the same direction and greater magnitude of association when compared to the original models (Supplemental Table 7) for all factors except for TNF Factor 5.
Last, we explored whether additional adjustments for maternal ethnicity, which was associated with toenail Se concentrations, influenced the observed associations between Cd and Se with IUGR and SGA, or with the interaction terms from our models with factor scores. We found that adjusting for maternal ethnicity did somewhat attenuate the association between Se and IUGR, though mothers with higher Se still tended to have lower odds of IUGR (OR = 0.34, 95% CI: 0.10–1.13). These adjustments also resulted in the interaction p-value for TNF Factor 3 to no longer be significant (p-value = 0.25), though it did not alter the interpretations of interaction terms for the other linear models: STR Factor 2 (p-value = 0.014), TNF Factor 2 (p-value = 0.015), or TNF Factor 5 (p-value = 0.0028).
4. Discussion
In this study, toenail concentrations of the potentially reproductively toxic metal, Cd (median = 0.0084 µg/g, IQR = 0.013 µg/g), were similar but lower than those reported in the Normative Aging Study (Mordukhovich et al., 2012), a case control study of prostate cancer in Italy (Vinceti et al., 2007), and a study of Cd-associations with prostate cancer in Maryland, USA (Platz et al., 2002); median Cd concentrations from these studies ranged between 0.015 µg/g and 0.055 µg/g. This is not unexpected since our study only included healthy women of reproductive age, and had a much higher proportion of non-smokers; 76% of our sample identified as never-smokers, whereas in the above studies prevalence of never-smokers ranged from 29% to 43% (Mordukhovich et al., 2012; Platz et al., 2002). Toenail concentrations of Se in our sample (median = 0.97 µg/g, IQR = 0.23 µg/g) tended to be higher than those reported in previous studies of women in the UK (median = 0.61 µg/g) (Rayman et al., 2015) or the Netherlands (Brandt et al., 1994, 2003) (means = 0.57 µg/g and 0.53 µg/g, respectively), and more similar but still higher than previously observed concentrations in the USA (means ranged between 0.77 µg/g and 0.96 µg/g) (Garland et al., 1993; Hunter et al., 1990; Park et al., 2011; Wallace et al., 2009; Xun et al., 2010; Yoshizawa et al., 1998). The high Se in RICHS could be due to many factors, including the US having substantially higher Se intake compared to Europe which is due, in part, to Se-deficient soils in Europe (Rayman, 2012) Additionally, women, non-smokers, and those of white ethnicity have been observed with higher Se levels (Xun et al., 2010) and our cohort is predominantly composed of non-smoking white mothers, most of whom took prenatal vitamins that likely included Se. Furthermore, the majority of reported toenail-Se levels in the US come from the Nurses’ Health Study (NHS) and the Health Professionals Follow-up Study (HPFS) (Garland et al., 1993; Hunter et al., 1990; Park et al., 2011; Yoshizawa et al., 1998), from which toenails were collected in 1982–83 and 1986–87, respectively. One follow-up study of some NHS participants found significantly higher Se concentrations in 1988 (mean = 0.92 µg/g) when compared to paired samples from the original study in 1982 (mean = 0.83 µg/g) (Garland et al., 1993). These findings likely indicate that Se intake and exposure changes over time, and the values reported in the 1980s may not be representative of the today’s US population.
In our study, associations between Cd and IUGR or SGA were not statistically significant, perhaps due to limited statistical power (relatively small sample size and low Cd concentrations). However, odds ratios ranged between 1.45 and 1.95, associated with a one IQR increase in log(Cd), for all crude and adjusted models of SGA and IUGR. These are comparable to other investigations of Cd on growth restriction and pregnancy outcomes, and similar to a previous study from our lab utilizing a larger sample size (n = 242), which applied different statistical methods and did not adjust for Se exposure, that did observe a significant Cd-SGA association (Everson et al., 2016). Other investigations have observed associations between increasing placental, maternal or fetal Cd concentrations with increased odds of preeclampsia (PE) (Laine et al., 2015) and odds of SGA (Al-Saleh et al., 2015; Johnston et al., 2014; Wang et al., 2016b), and with decreased birth weights (Kippler et al., 2012; Llanos and Ronco, 2009; Menai et al., 2012), birth weight percentiles (Johnston et al., 2014), head circumference (Kippler et al., 2012), Apgar 5-min scores and with placental size (Al-Saleh et al., 2015). Some of these associations were dependent on low maternal Se concentrations (Laine et al., 2015) or varied by infant sex (Kippler et al., 2013, 2012; Romano et al., 2016). We did not observe any sex-dependent associations in our study. However, mothers with lower Se concentrations exhibited a much stronger association between Cd and IUGR, though the 95% CI still included the null.
On the other hand, we observed a clear inverse association between maternal Se concentrations and IUGR. This is on par with other investigations that found cord blood Se to be positively associated with birth weight (Klapec et al., 2008; Sun et al., 2014), and low maternal Se concentrations associated with low birth weight (Bogden et al., 2006), preterm birth (Rayman et al., 2011), and with increased odds and severity of PE (Rayman et al., 2003, 2014). These studies, like ours, highlight a potential role for Se in promoting successful pregnancies and protecting against fetal growth restriction. However, we did not observe an inverse association between maternal Se and SGA, which may be due to residual confounding or the overall high levels of Se measured in our sample, or differences in the definitions of IUGR and SGA in our study. IUGR was clinically diagnosed via ultrasound during pregnancy, whereas SGA was defined after birth for all children based on their birth weight, sex, and gestational age (Fenton and Kim, 2013). Although these infants are considered small, the majority of pregnancies resulting in SGA births are unlikely to be pathological (Peleg et al., 1998). IUGR, on the other hand, is most commonly related to placental insufficiency but may also result from other maternal and/or fetal pathologies (Figueras and Gardosi, 2011). Thus, this discordance of findings, an inverse association between maternal Se and IUGR but no association between Se and SGA, may reflect the potential for Se to protect against pathologically-associated fetal growth restriction (IUGR) but not against more common, non-pathological SGA births.
The factor analyses revealed correlation structures in the placental expression patterns of steroid biosynthesis genes and TNF-SF signaling which reflect their biologic functions. CYP11A1 and HSD3B1 drive the first steps of steroid biosynthesis, from cholesterol to pregnenolone then to progesterone, and appear to be co-expressed in human placental tissue. Whereas the genes that drive synthesis towards cortisone (HSD11B2) versus cortisol (HSD11B1), or towards estrone (HSD17B2) versus estradiol (HSD17B1), appear to be modestly anti-correlated. Additionally, the TNFSF10 receptors appear to be co-expressed in placental tissue, which elicit both pro- (TNFRSF10A and TNFRSF10B) and anti- (TNFRSF10C and TNFRSF10D) apoptotic responses. Likewise, many of the TNF-SF factors were loaded with genes involved in both apoptotic and cell survival signaling. Interestingly, we found that maternal Cd and Se concentrations were differentially, and antagonistically, associated with the placental expression of TNF genes involved apoptotic signaling and steroidogenic activities that may disrupt redox balance. Three of the seven factors representing these gene expression patterns were associated with birth size for gestational age. The two factors most strongly associated with birth size (TNF F5 and STR F2) were also consistently associated with Se and Cd in a diametrically opposed manner.
TNF-SF Factor 5, which was associated with higher Cd, lower Se, and smaller birth size, was primarily loaded with the expression levels of TNFRSF1A (aka. TNFR1), which is one of the two the primary receptors for TNFα. These findings are on par with experimental studies, which have demonstrated that Se and Cd can perturb TNF-signaling, particularly TNFα. Se-deficient macrophages have been shown to overexpress TNFα upon lipopolysaccharide (LPS) stimulation (Vunta et al., 2008), and Se can repress the TNFα-activation of the nuclear factor NF-κB (Zhang et al., 2002; Maehira et al., 2003), an important regulator of cytokine production. Conversely, TNFα signaling plays an important role in Cd-induced hepatotoxicity (Kayama et al., 1995) and Cd-associated cell death has been shown to be potentiated by TNFα and possibly TRAIL, another TNF-SF ligand (Kim et al., 2002). Additionally, the placental and/or maternal expression of TNFα may influence fetal growth. Placenta from idiopathic fetal growth restricted pregnancies have been shown express greater levels of TNFα (Almasry et al., 2012). Furthermore, maternal serum concentrations of soluble TNFR1 (sTNFR1) were elevated in pregnancies complicated by PE, pregnancies also affected by IUGR had the highest levels of sTNFR1 and sTNFR1 was inversely associated with infant weight (Minuz et al., 2015). Our findings provide additional evidence that Cd may be positively associated, and Se inversely associated, with placental TNFR1 expression and that its expression may play a role in fetal growth restriction.
We also provide some evidence that trace element-associated and fetal growth-associated variations in TNF-signaling may not be limited to TNFα and TNFR1. TNF Factor 5 was also moderately loaded with TNFRSF19L (aka. RELT) and inversely loaded with TNFSF15 (aka. VEGI) and TNFRSF19 (aka. TROY). RELT overexpression has been shown to promote apoptosis (Cusick et al., 2010). Whereas, increased placental expression of VEGI may play a role in PE and placental dysfunction, as a potential target of hypoxia-induced micro-RNA (miR-517a/b and miR-517c) expression (Anton et al., 2015). TROY has also been implicated as a promoter of paraptosis, a type programmed cell death (Wang et al., 2004), as well as a promoter of nuclear factor NF-κB mediated transcription via lymphotoxin-α (LTα) binding (Hashimoto et al., 2008) or initiation of the JNK cascade (Mikkola, 2008), both pathways may suggest a role in regulating apoptosis. Activation of TROY has been implicated as being critical to skin appendage development and may have other functions redundant with the Eda-receptor (Hashimoto et al., 2008; Mikkola, 2008). However, the placental-specific functions of RELT, VEGI, and TROY are not well understood.
Furthermore, TNF Factor 2 and TNF Factor 3 were inversely associated with Se concentrations. These factors were primarily loaded with expression levels of TNFRSF27 and TNFRSF3. Interestingly, the TNFRSF27 gene encodes the Eda isoform A2 receptor (EDA2R), which induces apoptotic signaling upon binding to its ligand (Sinha and Chaudhary, 2004; Chang et al., 2007). Likewise, the TNFRSF3 gene encodes the lymphotoxin-β (LTβ) receptor (LTBR), which also an induces cell death upon activation (You et al., 2006) via TNFRSF14, LTβ or a LT-α/β complex. Additionally, though the factor (TNF factor 2) loaded with LTBR did not significantly differ by birth size, placenta of SGA infants did tend to have higher factor scores than those that were AGA or LGA (Supplemental Fig. 7). The expression levels of these receptors have not previously been studied in relation to Se, Cd, or birth outcomes. However, in a study of prostate cancer cells, LTBR, among many other genes, was shown to be differentially expressed in response to some selenium compounds (selenomethionine and methyl-seleninic acid) (Zhao and Brooks, 2007). Placental expression levels of these and other TNF-SF genes deserve consideration from future studies to clarify which TNF-SF pathways may be most responsive to trace-element exposures and how they may influence birth outcomes.
STR Factor 2, which was also associated with increasing Cd, decreasing Se, and smaller birth size, was primarily loaded with the expression levels of HSD3B1 and CYP11A1. This is consistent with other studies that found dysregulation of steroidogenesis genes in relation to Cd exposure. Mouse models have demonstrated Cd exposure may down-regulate the expression of CYP11A1 (Hu et al., 2014). Similarly, the expression of CYP11A1 and HSD3B1 have been shown to be down-regulated in response to Cd in Leydig Cells (Ji et al., 2015) and in cultured trophoblasts (Kawai et al., 2002). Whereas, Wang et al. (2014) observed increased corticosterone synthesis and significantly up-regulated CYP11A1 (and increased HSD3B1 though not statistically significant) in placenta of rats exposed to Cd during pregnancy, and that increased circulating corticosterone was associated with PE-like symptoms and IUGR (Wang et al., 2014). Another study found that selenium deficiency resulted in substantially decreased steroidogenesis in adrenal cells, but did not assess the expression levels of CYP11A1 or HSD3B1 (Chanoine et al., 2001). These studies and ours suggest that Cd and Se may disrupt the expression of steroidogenic genes. Our findings were specific to the up-regulation of CYP11A1 and HSD3B1, both of which localize to the mitochondrial membrane. Further investigations are necessary to clarify whether these trace elements repress or promote the expression of these genes in tissue-specific and/or dose-dependent manners.
Both CYP11A1 and HSD3B1 have been found to be hypo-methylated and more highly expressed in placenta from PE pregnancies compared to control pregnancies (Hogg et al., 2013). Similarly, P450scc, encoded by CYP11A, was more highly expressed in placenta samples complicated by PE compared to normal pregnancies and this upregulation in human trophoblast cell lines reduced cell viability and increased apoptosis under hypoxic conditions (He et al., 2013). While others found that CYP11A1 was up-regulated in serum of women with PE pregnancies (Moon et al., 2014). Our findings provide support that placental CYP11A1 and HSD3B1 may be up-regulated in association with maternal Cd while maternal Se exhibits the opposite relationship, and that increased placental expression of these steroidogenic genes is associated with decreasing size for gestational age.
We also observed strong correlations between some of the TNF and STR factors, particularly among those primarily TNF Factor 5 (loaded with TNFRSF1A) and STR Factor 2 (primarily loaded with HSD3B1). Considering the strong statistical overlap between these factors and the biological overlap between apoptotic signaling and redox balance, it is possible that our findings with TNF and STR genes do not represent independent mechanisms. TNF mediated apoptosis often works through the potentiation of reactive oxygen intermediates (ROI), and anti-oxidative mechanisms can block many, but not all, TNF-SF apoptotic signals (Shakibaei et al., 2005). Unexplained recurrent spontaneous abortions or miscarriages have been associated with lower levels of antioxidants, increased markers of oxidative stress, and higher levels of TNFα (El-Far et al., 2009, 2007). Likewise, trophoblasts from placenta complicated by PE and/or IUGR are more susceptible to hypoxia- and TNFα-induced apoptosis (Crocker et al., 2003; Longtine et al., 2012). We present additional evidence that TNFα- and/or steroidogenic activity that may increase ROS within the placenta likely play important roles in fetal growth, and that these systems may be perturbed by trace element exposures during pregnancy.
Identifying risk factors and protective factors for IUGR and other pregnancy complications, has important public health implications. IUGR-complicated pregnancies and babies born SGA confer an increased risk for perinatal morbidity and mortality, as well as poor cognition, neurodevelopmental disorders, cardiovascular complications and metabolic diseases in adolescence and/or adulthood (Pallotto and Kilbride, 2006; Salam et al., 2014). The potential antagonistic effects between Se and Cd on critical placental signaling pathways and on pregnancy outcomes, presents a notable opportunity since Se exposure is largely a function of diet, and thus is modifiable. Should further studies continue to produce evidence that Se intake can mitigate Cd-associated pregnancy complications, or other oxidative stress-associated effects, recommendations for increasing Se consumption during pregnancy may be developed. Some already argue that the evidence in favor Se supplementation during preconception is compelling enough, at least for those with Se deficiency (Pieczyńska and Grajeta, 2015), but also recommend future Se intervention trials.
The mounting evidence from these studies and ours, suggest that maternal exposures Cd and Se are likely associated with pregnancy outcomes and fetal growth, though these associations may be dependent on some conditions that are still being explored (concurrent exposures and fetal sex). If these associations represent causal effects, they may be potentiated, in part, through disruptions to redox activities and apoptotic signaling in placental tissue. Future studies should consider investigating similar birth outcomes and placental expression patterns, but expand the breadth of the metals exposures to explore other potential interactions or even metal-mixture associations.
Overall, we found that Cd and Se were most strongly associated with IUGR, and less so with SGA, while the Cd- and Se-associated expression patterns were associated with birth size by gestational age but not with IUGR. Though this suggests that the Cd- and Se-associated variations in placental steroidogenic and TNF-SF expression patterns are important in fetal growth, we could not conclude that these variations in expression mediate Cd- and Se-associated fetal growth restriction. This may be due, in part, to some of the limitations of this study, which include possible selection bias, small sample size, and potential for residual confounding. The RICHS cohort only included non-pathological, successful pregnancies with a minimum of 37 weeks gestation. Given that maternal Se deficiency and high Cd exposure have been associated with increased odds of preterm birth (Rayman et al., 2011) and increased odds of PE (Laine et al., 2015; Rayman et al., 2015), it possible that mothers with some of the highest levels of Cd and/or lowest levels of Se would have been more likely to have had pathological pregnancies and thus should have been excluded from this study. If so, exclusion of these mothers could have hindered our ability to detect associations between these metals, pregnancy outcomes, and placental TNF-SF and redox signaling. Likewise, toenail Cd concentrations in our sample were among the lowest compared to previous publications and toenail Se concentrations were among the highest. Because Se may suppress some of the reproductively toxic effects of Cd, it is possible that the metal-exposure profile for our sample impacted our ability observe Cd-related pregnancy and fetal-growth outcomes. This could explain why we observed more robust and consistent associations with maternal Se rather than Cd. Since dietary intake is a common source of both Se and Cd exposure, and nutrient intake can certainly influence fetal development, diet is a potential confounder that we were unable to control for in this study. Additionally, toenails were collected postpartum and should represent long-term cumulative exposures. However, if mothers drastically changed their Cd- and Se-exposures after pregnancy, our measures of these elements may not be as representative of their exposures during pregnancy. We also found that adjusting for maternal ethnicity somewhat attenuated the associations between Se and IUGR, as well as between Se-Cd interaction terms and TNF Factor 2 scores; though most of associations with factor scores were unaffected. Given the already small sample size, we could not reliably stratify by ethnicity and pursue a more detailed analysis of how some of these associations may vary by ethnicity. Also, the Cd-associated variations in placental expression were not as robust as the Se-associated variations in our sensitivity analysis, due to the high proportion of Cd concentrations below the LOD. Because of these limitations, we encourage other cohorts, particularly those with larger sample sizes, greater ranges of Cd exposure levels, detailed dietary intake data, and diverse racial and ethnic backgrounds, to attempt to replicate these findings or explore similar hypotheses. Despite these limitations, which likely inhibited our ability to detect some associations, we observed potential interactions between maternal Cd and Se exposure with the placental expression of TNF and steroidogenic genes in relation to size at birth. These findings may contribute to our evolving understanding of how trace elements may perturb placental function, and influence fetal growth.
5. Conclusions
Maternal exposures to the trace elements Cd and Se during pregnancy are associated with variations in the placental expression of TNF superfamily genes involved in apoptotic signaling, particularly the receptors that bind to TNFα, LTα, LTβ, and Eda, and the expression of the steroidogenic genes HSD3B1 and CYP11A1. Furthermore, the expression patterns of these TNF and steroidogenic genes were also associated with size at birth. These genes and their associated pathways should be investigated further in larger samples, and those with wider ranges of exposure to Cd and Se to further clarify their potential roles in environmentally-associated fetal growth restriction.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health [NIHNIMH R01MH094609, NIH-NIEHS R01ES022223, and NIH-NIEHS P01 ES022832] and by the United States Environmental Protection Agency [US EPA grant RD83544201]. Its contents are solely the responsibility of the grantee and do not necessarily represent the official views of the US EPA. Further, the US EPA does not endorse the purchase of any commercial products or services mentioned in the presentation.
Footnotes
Competing financial interests
The authors have no competing financial interests to declare.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.envres.2017.06.016.
References
- Almasry SM, Eldomiaty MA, Elfayomy AK, Habib FA. Expression pattern of tumor necrosis factor alpha in placentae of idiopathic fetal growth restriction. J. Mol. Histol. 2012;43:253–261. doi: 10.1007/s10735-012-9410-6. http://dx.doi.org/10.1007/s10735-012-9410-6. [DOI] [PubMed] [Google Scholar]
- Al-Saleh I, Shinwari N, Mashhour A, Rabah A. Birth outcome measures and maternal exposure to heavy metals (lead, cadmium and mercury) in Saudi Arabian population. Int. J. Hyg. Environ. Health. 2014;217:205–218. doi: 10.1016/j.ijheh.2013.04.009. http://dx.doi.org/10.1016/j.ijheh.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Al-Saleh I, Al-Rouqi R, Obsum CA, Shinwari N, Mashhour A, Billedo G, et al. Interaction between cadmium (Cd), selenium (Se) and oxidative stress biomarkers in healthy mothers and its impact on birth anthropometric measures. Int. J. Hyg. Environ. Health. 2015;218:66–90. doi: 10.1016/j.ijheh.2014.08.001. http://dx.doi.org/10.1016/j.ijheh.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Anton L, Olarerin-George AO, Hogenesch JB, Elovitz MA. Placental expression of miR-517a/b and miR-517c contributes to trophoblast dysfunction and preeclampsia. PLoS One. 2015;10:e0122707. doi: 10.1371/journal.pone.0122707. http://dx.doi.org/10.1371/journal.pone.0122707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. http://dx.doi.org/10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogden JD, Kemp FW, Chen X, Stagnaro-green A, Stein TP, Scholl TO. Low-normal serum selenium early in human pregnancy predicts lower birth weight. Nutr. Res. 2006;26:497–502. http://dx.doi.org/10.1016/j.nutres.2006.08.008. [Google Scholar]
- Brandt PA, Van Den Goldbohm RA, ’t Veer PV, Bode P, Dorant E, Hermus RJJ, et al. Toenail selenium levels and the risk of breast cancer. Am. J. Epidemiol. 1994;140:20–26. doi: 10.1093/oxfordjournals.aje.a117155. [DOI] [PubMed] [Google Scholar]
- Brandt PA, Van Den Zeegers MPA, Bode P, Goldbohm RA. Toenail selenium levels and the subsequent risk of prostate cancer: a prospective cohort study. Cancer Epidemiol. 2003;12:866–871. [PubMed] [Google Scholar]
- Chang B, Punj V, Shindo M, Chaudhary PM. Adenoviral-mediated gene transfer of ectodysplasin-A2 results in induction of apoptosis and cell-cycle arrest in osteosarcoma cell lines. Cancer Gene Ther. 2007;14:927–933. doi: 10.1038/sj.cgt.7701078. http://dx.doi.org/10.1038/sj.cgt.7701078. [DOI] [PubMed] [Google Scholar]
- Chanoine J-P, Compagnone NA, Wong ACK. Modulation of steroidogenesis by selenium in a novel adrenal cell line developed using targeted tumorigenesis. Biofactors. 2001;14:229–238. doi: 10.1002/biof.5520140129. [DOI] [PubMed] [Google Scholar]
- Crocker IP, Cooper S, Ong SC, Baker PN. Differences in apoptotic susceptibility of cytotrophoblasts and syncytiotrophoblasts in normal pregnancy to those complicated with preeclampsia and intrauterine growth restriction. Am. J. Pathol. 2003;162:637–643. doi: 10.1016/S0002-9440(10)63857-6. http://dx.doi.org/10.1016/S0002-9440(10)63857-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick JK, Mustian A, Goldberg K, Reyland ME. RELT induces cellular death in HEK 293 epithelial cells. Cell. Immunol. 2010;261:1–8. doi: 10.1016/j.cellimm.2009.10.013. http://dx.doi.org/10.1016/j.cellimm.2009.10.013.RELT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Far M, El-Sayed IH, El-Motwally AE-G, Hashem IA, Bakry N. Tumor necrosis factor-alpha and oxidant status are essential participating factors in unexplained recurrent spontaneous abortions. Clin. Chem. Lab. Med. 2007;45:879–883. doi: 10.1515/CCLM.2007.138. http://dx.doi.org/10.1515/CCLM.2007.138. [DOI] [PubMed] [Google Scholar]
- El-Far M, El-Sayed IH, El-Motwally AE, Hashem IA, Bakry N. Serum levels of TNF-αlpha and antioxidant enzymes and placental TNF-αlpha expression in unexplained recurrent spontaneous miscarriage. J. Physiol. Biochem. 2009;65:175–181. doi: 10.1007/BF03179068. [DOI] [PubMed] [Google Scholar]
- El-Sharaky AS, Newairy AA, Badreldeen MM, Eweda SM, Sheweita SA. Protective role of selenium against renal toxicity induced by cadmium in rats. Toxicology. 2007;235:185–193. doi: 10.1016/j.tox.2007.03.014. http://dx.doi.org/10.1016/j.tox.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Erboga M, Kanter M. Effect of cadmium on trophoblast cell proliferation and apoptosis in different gestation periods of rat placenta. Biol. Trace Elem. Res. 2016;169:285–293. doi: 10.1007/s12011-015-0439-8. http://dx.doi.org/10.1007/s12011-015-0439-8. [DOI] [PubMed] [Google Scholar]
- Everson TM, Armstrong DA, Jackson BP, Green BB, Karagas MR, Marsit CJ. Maternal cadmium, placental PCDHAC1, and fetal development. Reprod. Toxicol. 2016;65:263–271. doi: 10.1016/j.reprotox.2016.08.011. http://dx.doi.org/10.1016/j.reprotox.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton TR, Kim JH. A systematic review and meta-analysis to revise the Fenton growth chart for preterm infants. BMC Pediatr. 2013;13 doi: 10.1186/1471-2431-13-59. http://dx.doi.org/10.1186/1471-2431-13-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueras F, Gardosi J. Intrauterine growth restriction: new concepts in antenatal surveillance, diagnosis, and management. Am. J. Obstet. Gynecol. 2011;204:288–300. doi: 10.1016/j.ajog.2010.08.055. http://dx.doi.org/10.1016/j.ajog.2010.08.055. [DOI] [PubMed] [Google Scholar]
- Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80:27–38. http://dx.doi.org/10.1093/biomet/80.1.27. [Google Scholar]
- Garland M, Morris JS, Rosner BA, Stampfer MJ, Spate VL, Baskett CJ, et al. Toenail trace element levels as biomarkers: reproducibility over a 6-year period. Cancer Epidemiol. 1993;2:493–497. [PubMed] [Google Scholar]
- Hashimoto T, Schlessinger D, Cui C-Y. Troy binding to lymphotoxin-alpha activates NF-kappa B mediated transcription. Cell Cycle. 2008;7:106–111. doi: 10.4161/cc.7.1.5135. http://dx.doi.org/10.4161/cc.7.1.5135. [DOI] [PubMed] [Google Scholar]
- He G, Xu W, Chen Y, Liu X, Xi M. Abnormal apoptosis of trophoblastic cells is related to the up-regulation of CYP11A gene in placenta of preeclampsia patients. PLoS One. 2013;8:e59609. doi: 10.1371/journal.pone.0059609. http://dx.doi.org/10.1371/journal.pone.0059609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg K, Blair JD, McFadden DE, Von Dadelszen P, Robinson WP. Early onset pre-eclampsia is associated with altered DNA methylation of cortisol-signalling and steroidogenic genes in the placenta. PLoS One. 2013;8:e62969. doi: 10.1371/journal.pone.0062969. http://dx.doi.org/10.1371/journal.pone.0062969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Lu X, Cen X, Chen X, Li F, Zhong S. RNA-Seq identifies key reproductive gene expression alterations in response to cadmium exposure. Biomed. Res. Int. 2014;2014:529271. doi: 10.1155/2014/529271. http://dx.doi.org/10.1155/2014/529271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Jaritz M, Guenzl P, Vlatkovic I, Sommer A, Tamir IM, et al. An RNA-seq strategy to detect the complete coding and non-coding transcriptome including full-length imprinted macro ncrnas. PLoS One. 2011;6 doi: 10.1371/journal.pone.0027288. http://dx.doi.org/10.1371/journal.pone.0027288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ, Morris JS, Stampfer MJ, Colditz GA, Speizer FE, Willett WC. A prospective study of selenium status and breast cancer risk. JAMA. 1990;264:1128–1131. [PubMed] [Google Scholar]
- Jablonska E, Vinceti M. Selenium and human health: witnessing a copernican revolution? J. Environ. Sci. Heal. Part C. 2015;33:328–368. doi: 10.1080/10590501.2015.1055163. http://dx.doi.org/10.1080/10590501.2015.1055163. [DOI] [PubMed] [Google Scholar]
- Järup L, Åkesson A. Current status of cadmium as an environmental health problem. Toxicol. Appl. Pharmacol. 2009;238:201–208. doi: 10.1016/j.taap.2009.04.020. http://dx.doi.org/10.1016/j.taap.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Ji X, Li Z, Chen H, Li J, Tian H, Li Z, et al. Cytotoxic mechanism related to dihydrolipoamide dehydrogenase in Leydig cells exposed to heavy metals. Toxicology. 2015;334:22–32. doi: 10.1016/j.tox.2015.05.003. http://dx.doi.org/10.1016/j.tox.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Jihen EH, Imed M, Fatima H, Abdelhamid K. Protective effects of selenium (Se) and zinc (Zn) on cadmium (Cd) toxicity in the liver of the rat: effects on the oxidative stress. Ecotoxicol. Environ. Saf. 2009;72:1559–1564. doi: 10.1016/j.ecoenv.2008.12.006. http://dx.doi.org/10.1016/j.ecoenv.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Johnston JE, Valentiner E, Maxson P, Miranda ML, Fry RC. Maternal cadmium levels during pregnancy associated with lower birth weight in infants in a north carolina cohort. PLoS One. 2014;9:e109661. doi: 10.1371/journal.pone.0109661. http://dx.doi.org/10.1371/journal.pone.0109661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantola M, Purkunen R, Kröger P, Tooming A, Juravskaja J, Pasanen M, et al. Selenium in pregnancy: is selenium an active defective ion against environmental chemical stress? Environ. Res. 2004;96:51–61. doi: 10.1016/j.envres.2004.03.003. http://dx.doi.org/10.1016/j.envres.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Karabulut-Bulan O, Bolkent S, Yanardag R, Bilgin-Sokmen B. The role of vitamin C, vitamin E, and selenium on cadmium-induced renal toxicity of rats. Drug Chem. Toxicol. 2016;31:413–426. doi: 10.1080/01480540802383200. http://dx.doi.org/10.1080/01480540802383200. [DOI] [PubMed] [Google Scholar]
- Kawai M, Swan KF, Green AE, Edwards DE, Anderson MB, Henson MC. Placental Endocrine disruption induced by cadmium: effects on P450 cholesterol side-chain cleavage and 3beta-hydroxysteroid dehydrogenase enzymes in cultured human trophoblasts. Biol. Reprod. 2002;67:178–183. doi: 10.1095/biolreprod67.1.178. [DOI] [PubMed] [Google Scholar]
- Kayama F, Yoshida T, Elwell MR, Luster MI. Role of tumor necrosis factor alpha in cadmium-induced hepatotoxicity. Toxicol. Appl. Pharmacol. 1995;131:224–234. doi: 10.1006/taap.1995.1065. [DOI] [PubMed] [Google Scholar]
- Kenny LC, Lavender T, Mcnamee R, O’Neil S, Mills T, Khashan AS. Advanced maternal age and adverse pregnancy outcome: evidence from a large contemporary cohort. PLoS One. 2013;8:e56583. doi: 10.1371/journal.pone.0056583. http://dx.doi.org/10.1371/journal.pone.0056583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, Kim M-S, Kim K-B, Kim K-W, Hong Y-M, Kim I-K, et al. Sensitizing effects of cadmium on TNF-alpha- and TRAIL-mediated apoptosis of NIH3T3 cells with distinct expression patterns of p53. Carcinogenesis. 2002;23:1411–1417. doi: 10.1093/carcin/23.9.1411. [DOI] [PubMed] [Google Scholar]
- Kippler M, Tofail F, Gardner R, Rahman A, Hamadani JD, Bottai M, et al. Maternal cadmium exposure during pregnancy and size at birth: a prospective cohort study. Environ. Health Perspect. 2012;120:284–289. doi: 10.1289/ehp.1103711. http://dx.doi.org/10.1289/ehp.1103711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippler M, Engström K, Mlakar SJ, Bottai M, Ahmed S, Hossain MB, et al. Sex-specific effects of early life cadmium exposure on DNA methylation and implications for birth weight. Epigenetics. 2013;8:494–503. doi: 10.4161/epi.24401. http://dx.doi.org/10.4161/epi.24401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapec T, Ćavar S, Kasač Z, Ručević S, Popinjač A. Selenium in placenta predicts birth weight in normal but not intrauterine growth restriction pregnancy. J. Trace Elem. Med. Biol. 2008;22:54–58. doi: 10.1016/j.jtemb.2007.10.004. http://dx.doi.org/10.1016/j.jtemb.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Kumar KMK, Kumar MN, Patil RH, Nagesh R, Hegde SM, Kavya K, et al. Cadmium induces oxidative stress and apoptosis in lung epithelial cells. Toxicol. Mech. Methods. 2016;26:658–666. doi: 10.1080/15376516.2016.1223240. http://dx.doi.org/10.1080/15376516.2016.1223240. [DOI] [PubMed] [Google Scholar]
- Laine JE, Ray P, Bodnar W, Cable PH, Boggess K, Offenbacher S, et al. Placental Cadmium levels are associated with increased preeclampsia risk. PLoS One. 2015;10:e0139341. doi: 10.1371/journal.pone.0139341. http://dx.doi.org/10.1371/journal.pone.0139341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llanos MN, Ronco AM. Fetal growth restriction is related to placental levels of cadmium, lead and arsenic but not with antioxidant activities. Reprod. Toxicol. 2009;27:88–92. doi: 10.1016/j.reprotox.2008.11.057. http://dx.doi.org/10.1016/j.reprotox.2008.11.057. [DOI] [PubMed] [Google Scholar]
- Long JS, Ervin LH. Using heteroscedasticity consistent standard errors in the linear regression model. Am. Stat. 2000;54:217–224. http://dx.doi.org/10.2307/2685594. [Google Scholar]
- Longtine MS, Chen B, Odibo AO, Zhong Y, Nelson DM. Villous trophoblast apoptosis is elevated and restricted to cytotrophoblasts in pregnancies complicated by preeclampsia, IUGR, or preeclampsia with IUGR. Placenta. 2012;33:352–359. doi: 10.1016/j.placenta.2012.01.017. http://dx.doi.org/10.1016/j.placenta.2012.01.017.Villous. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López E, Figueroa S, Oset-Gasque MJ, González MP. Apoptosis and necrosis: two distinct events induced by cadmium in cortical neurons in culture. Br. J. Pharmacol. 2003;138:901–911. doi: 10.1038/sj.bjp.0705111. http://dx.doi.org/10.1038/sj.bjp.0705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehira F, Miyagi I, Eguchi Y. Selenium regulates transcription factor NF-kB activation during the acute phase reaction. Clin. Chim. Acta. 2003;334:163–171. doi: 10.1016/s0009-8981(03)00223-7. http://dx.doi.org/10.1016/S0009-8981(03)00223-7. [DOI] [PubMed] [Google Scholar]
- Mehdi Y, Hornick J, Istasse L, Dufrasne I. Selenium in the environment, metabolism and involvement in body functions. Molecules. 2013;18:3292–3311. doi: 10.3390/molecules18033292. http://dx.doi.org/10.3390/molecules18033292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menai M, Heude B, Slama R, Forhan A, Sahuquillo J, Charles M-A, et al. Association between maternal blood cadmium during pregnancy and birth weight and the risk of fetal growth restriction: the EDEN mother–child cohort study. Reprod. Toxicol. 2012;34:622–627. doi: 10.1016/j.reprotox.2012.09.002. http://dx.doi.org/10.1016/j.reprotox.2012.09.002. [DOI] [PubMed] [Google Scholar]
- Method, E.P.A. 6020A (SW-846): inductively coupled plasma-mass spectrometry. Revision. 1998;1 [Google Scholar]
- Mikkola ML. TNF superfamily in skin appendage development. Cytokine Growth Factor Rev. 2008;19:219–230. doi: 10.1016/j.cytogfr.2008.04.008. http://dx.doi.org/10.1016/j.cytogfr.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Mikolić A, Piasek M, Grgec SA, Varnai VM, Stasenko S, Oguić SK. Oral cadmium exposure during rat pregnancy: assessment of transplacental micronutrient transport and steroidogenesis at term. J. Appl. Toxicol. 2015;35:508–519. doi: 10.1002/jat.3055. http://dx.doi.org/10.1002/jat.3055. [DOI] [PubMed] [Google Scholar]
- Min J, Park B, Kim YJ, Lee H, Ha E, Park H. Effect of oxidative stress on birth sizes:sizes: consideration of window from mid pregnancy to delivery. Placenta. 2009;30:418–423. doi: 10.1016/j.placenta.2009.02.007. http://dx.doi.org/10.1016/j.placenta.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Minuz P, Fava C, Hao S, Pedraza P, Amen G, Meneguzzi A, et al. Differential regulation of TNF receptors in maternal leukocytes is associated with severe preterm preeclampsia. J. Matern. Noenatal Med. 2015;28:869–875. doi: 10.3109/14767058.2014.937695. http://dx.doi.org/10.3109/14767058.2014.937695. [DOI] [PubMed] [Google Scholar]
- Mistry HD, Kurlak LO, Young SD, Briley AL, Pipkin FB, Baker PN, et al. Maternal selenium, copper and zinc concentrations in pregnancy associated with small-for-gestational-age infants. Matern. Child Nutr. 2014;10:327–334. doi: 10.1111/j.1740-8709.2012.00430.x. http://dx.doi.org/10.1111/j.1740-8709.2012.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J-Y, Moon MH, Kim KT, Jeong DH, Kim YN, Chung BC, et al. Cytochrome P450-mediated metabolic alterations in preeclampsia evaluated by quantitative steroid signatures. J. Steroid Biochem. Mol. Biol. 2014;139:182–191. doi: 10.1016/j.jsbmb.2013.02.014. http://dx.doi.org/10.1016/j.jsbmb.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Mordukhovich I, Wright RO, Hu H, Amarasiriwardena C, Baccarelli A, Litonjua A, et al. Association of toenail arsenic, cadmium, mercury, manganese, and lead with blood pressure in the normative aging study. Environ. Health Perspect. 2012;120:98–104. doi: 10.1289/ehp.1002805. http://dx.doi.org/10.1289/ehp.1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata C, Nagao Y, Shibuya C, Kashiki Y, Shimizu H. Urinary cadmium and serum levels of estrogens and androgens in postmenopausal japanese women. Cancer Epidemiol. Biomark. Prev. 2005;14:705–708. doi: 10.1158/1055-9965.EPI-04-0619. http://dx.doi.org/10.1158/1055-9965.EPI-04-0619. [DOI] [PubMed] [Google Scholar]
- Nair AR, DeGheselle O, Smeets K, Van Kerkhove E, Cuypers A. Cadmiuminduced pathologies: where is the oxidative balance lost (or not)? Int. J. Mol. Sci. 2013;14:6116–6143. doi: 10.3390/ijms14036116. http://dx.doi.org/10.3390/ijms14036116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi R, Pande D, Karki K, Kumar A, Khanna RS, Khanna HD. Trace elements and antioxidant enzymes associated with oxidative stress in the pre-eclamptic/eclamptic mothers during fetal circulation. Clin. Nutr. 2012;31:946–950. doi: 10.1016/j.clnu.2012.04.005. http://dx.doi.org/10.1016/j.clnu.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Pallotto EK, Kilbride HW. Perinatal outcome and later implications of intrauterine growth restriction. Clin. Obstet. Gynecol. 2006;49:257–269. doi: 10.1097/00003081-200606000-00008. [DOI] [PubMed] [Google Scholar]
- Pandya C, Pillai P, Nampoothiri LP, Bhatt N, Gupta S, Gupta S. Effect of lead and cadmium co-exposure on testicular steroid metabolism and antioxidant system of adult male rats. Andrologia. 2012;44:813–822. doi: 10.1111/j.1439-0272.2010.01137.x. http://dx.doi.org/10.1111/j.1439-0272.2010.01137.x. [DOI] [PubMed] [Google Scholar]
- Park K, Rimm E, Siscovick D, Spiegelman D, Morris JS, Mozaffarian D. Demographic and lifestyle factors and selenium levels in men and women in the U.S. Nutr. Res. Pract. 2011;5:357–364. doi: 10.4162/nrp.2011.5.4.357. http://dx.doi.org/10.4162/nrp.2011.5.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg D, Kennedy CM, Hunter SK. Intrauterine growth restriction: Identification and management. Am. Fam. Physician. 1998;58:453–460. [PubMed] [Google Scholar]
- Pieczyńska J, Grajeta H. The role of selenium in human conception and pregnancy. J. Trace Elem. Med. Biol. 2015;29:31–38. doi: 10.1016/j.jtemb.2014.07.003. http://dx.doi.org/10.1016/j.jtemb.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Pillai A, Gupta S. Effect of gestational and lactational exposure to lead and/or cadmium on reproductive performance and hepatic oestradiol metabolising enzymes. Toxicol. Lett. 2005;155:179–186. doi: 10.1016/j.toxlet.2004.09.015. http://dx.doi.org/10.1016/j.toxlet.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Platz EA, Helzlsouer KJ, Hoffman SC, Morris JS, Baskett CK, Comstock GW. Prediagnostic toenail cadmium and zinc and subsequent prostate cancer risk. Prostate. 2002;52:288–296. doi: 10.1002/pros.10115. http://dx.doi.org/10.1002/pros.10115. [DOI] [PubMed] [Google Scholar]
- Prasad R, Kowalczyk JC, Meimaridou E, Storr HL, Metherell LA. Oxidative stress and adrenocortical insufficiency. J. Endocrinol. 2014;221:R63–R73. doi: 10.1530/JOE-13-0346. http://dx.doi.org/10.1530/JOE-13-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punshon T, Li Z, Marsit CJ, Jackson BP, Baker ER, Karagas MR. Placental metal concentrations in relation to maternal and infant toenails in a U.S. cohort. Environ. Sci. Technol. 2016;50:1587–1594. doi: 10.1021/acs.est.5b05316. http://dx.doi.org/10.1021/acs.est.5b05316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Kumarathasan P, Gomes J. Infant and mother related outcomes from exposure to metals with endocrine disrupting properties during pregnancy. Sci. Total Environ. 2016;569–570:1022–1031. doi: 10.1016/j.scitotenv.2016.06.134. http://dx.doi.org/10.1016/j.scitotenv.2016.06.134. [DOI] [PubMed] [Google Scholar]
- Rayman MP. Selenium and human health. Lancet. 2012;379:1256–1268. doi: 10.1016/S0140-6736(11)61452-9. http://dx.doi.org/10.1016/S0140-6736(11)61452-9. [DOI] [PubMed] [Google Scholar]
- Rayman MP, Bode P, Redman CWG. Low selenium status is associated with the occurrence of the pregnancy disease preeclampsia in women from the United Kingdom. Am. J. Obstet. Gynecol. 2003;189:1343–1349. doi: 10.1067/s0002-9378(03)00723-3. http://dx.doi.org/10.1067/S0002-9378(03)00723-3. [DOI] [PubMed] [Google Scholar]
- Rayman MP, Wijnen H, Vader H, Kooistra L, Pop V. Maternal selenium status during early gestation and risk for preterm birth. Can. Med. Assoc. J. 2011;183:549–555. doi: 10.1503/cmaj.101095. http://dx.doi.org/10.1503/cmaj.101095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman MP, Searle E, Kelly L, Johnsen S, Bodman-Smith K, Bath SC, et al. Effect of selenium on markers of risk of pre-eclampsia in UK pregnant women: a randomised, controlled pilot trial. Br. J. Nutr. 2014;112:99–111. doi: 10.1017/S0007114514000531. http://dx.doi.org/10.1017/S0007114514000531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayman MP, Bath SC, Westaway J, Williams P, Mao J, Vanderlelie JJ, et al. Selenium status in UK pregnant women and its relationship with hypertensive conditions of pregnancy. Br. J. Nutr. 2015;113:249–258. doi: 10.1017/S000711451400364X. http://dx.doi.org/10.1017/S000711451400364X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano ME, Enquobahrie DA, Simpson C, Checkoway H, Williams MA. Maternal body burden of cadmium and offspring size at birth. Environ. Res. 2016;147:461–468. doi: 10.1016/j.envres.2016.02.029. http://dx.doi.org/10.1016/j.envres.2016.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronco AM, Llaguno E, Epuñan MJ, Llanos MN. Effect of cadmium on cortisol production and 11 beta-hydroxysteroid dehydrogenase 2 expression by cultured human choriocarcinoma cells (JEG-3) Toxicol. Vitr. 2010;24:1532–1537. doi: 10.1016/j.tiv.2010.07.003. http://dx.doi.org/10.1016/j.tiv.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Salam RA, Das JK, Bhutta ZA. Impact of intrauterine growth restriction on long-term health. Curr. Opin. Clin. Nutr. Metab. Care. 2014;17:249–254. doi: 10.1097/MCO.0000000000000051. http://dx.doi.org/10.1097/MCO.0000000000000051. [DOI] [PubMed] [Google Scholar]
- Satarug S, Moore MR. Adverse health effects of chronic exposure to low-level cadmium in foodstuffs and cigarette smoke. Environ. Health Perspect. 2004;112:1099–1103. doi: 10.1289/ehp.6751. http://dx.doi.org/10.1289/ehp.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scifres CM, Nelson DM. Intrauterine growth restriction, human placental development and trophoblast cell death. J. Physiol. 2009;587:3453–3458. doi: 10.1113/jphysiol.2009.173252. http://dx.doi.org/10.1113/jphysiol.2009.173252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakibaei M, Schulze-Tanzil G, Takada Y, Aggarwal BB. Redox regulation of apoptosis by members of the TNF superfamily. Antioxid. Redox Signal. 2005;7:482–496. doi: 10.1089/ars.2005.7.482. [DOI] [PubMed] [Google Scholar]
- Sinha SK, Chaudhary PM. Induction of apoptosis by X-linked ectodermal dysplasia receptor via a caspase 8-dependent mechanism. J. Biol. Chem. 2004;279:41873–41881. doi: 10.1074/jbc.M407363200. http://dx.doi.org/10.1074/jbc.M407363200. [DOI] [PubMed] [Google Scholar]
- Stasenko S, Bradford EM, Piasek M, Henson MC, Varnai VM, Jurasović J, et al. Metals in human placenta: focus on the effects of cadmium on steroid hormones and leptin. J. Appl. Toxicol. 2010;30:242–253. doi: 10.1002/jat.1490. http://dx.doi.org/10.1002/jat.1490. [DOI] [PubMed] [Google Scholar]
- Sun H, Chen W, Wang D, Jin Y, Chen X, Xu Y. The effects of prenatal exposure to low-level cadmium, lead and selenium on birth outcomes. Chemosphere. 2014;108:33–39. doi: 10.1016/j.chemosphere.2014.02.080. http://dx.doi.org/10.1016/j.chemosphere.2014.02.080. [DOI] [PubMed] [Google Scholar]
- Templeton DM, Liu Y. Multiple roles of cadmium in cell death and survival. Chem. Biol. Interact. 2010;188:267–275. doi: 10.1016/j.cbi.2010.03.040. http://dx.doi.org/10.1016/j.cbi.2010.03.040. [DOI] [PubMed] [Google Scholar]
- Valko M, Jomova K, Rhodes CJ, Kuča K, Musílek K. Redox- and non-redox-metal-induced formation of free radicals and their role in human disease. Arch. Toxicol. 2016;90:1–37. doi: 10.1007/s00204-015-1579-5. http://dx.doi.org/10.1007/s00204-015-1579-5. [DOI] [PubMed] [Google Scholar]
- Vanderlelie J, Venardos K, Clifton VL, Gude NM, Clarke FM, Perkins AV. Increased biological oxidation and reduced anti-oxidant enzyme activity in pre-eclamptic placentae. Placenta. 2005;26:53–58. doi: 10.1016/j.placenta.2004.04.002. http://dx.doi.org/10.1016/j.placenta.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Vinceti M, Venturelli M, Sighinolfi C, Trerotoli P, Bonvicini F, Ferrari A, et al. Case-control study of toenail cadmium and prostate cancer risk in Italy. Sci. Total Environ. 2007;373:77–81. doi: 10.1016/j.scitotenv.2006.11.005. http://dx.doi.org/10.1016/j.scitotenv.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Vunta H, Belda BJ, Arner RJ, Reddy CC, Vanden Heuvel JP, Prabhu KS. Selenium attenuates pro-inflammatory gene expression in macrophages. Mol. Nutr. Food Res. 2008;52:1316–1323. doi: 10.1002/mnfr.200700346. http://dx.doi.org/10.1002/mnfr.200700346. [DOI] [PubMed] [Google Scholar]
- Wallace K, Kelsey KT, Schned A, Morris JS, Andrew AS, Karagas MR. Selenium and risk of bladder cancer: a population-based case-control study. Cancer Prev. Res. 2009;2:70–74. doi: 10.1158/1940-6207.CAPR-08-0046. http://dx.doi.org/10.1158/1940-6207.CAPR-08-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhang Q, Zhang X, Luo S, Ye D, Guo Y, et al. Preeclampsia induced by cadmium in rats is related to abnormal local glucocorticoid synthesis in placenta. Reprod. Biol. Endocrinol. 2014;12:1–9. doi: 10.1186/1477-7827-12-77. http://dx.doi.org/10.1186/1477-7827-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang Y, Bo Q-L, Ji Y-L, Liu L, Hu Y-F, et al. Maternal cadmium exposure reduces placental zinc transport and induces fetal growth restriction in mice. Reprod. Toxicol. 2016a;63:174–182. doi: 10.1016/j.reprotox.2016.06.010. http://dx.doi.org/10.1016/j.reprotox.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Wang H, Liu L, Hu Y-F, Hao J-H, Chen Y-H, Su P-Y, et al. Maternal serum cadmium level during pregnancy and its association with small for gestational age infants: a population-based birth cohort study. Sci. Rep. 2016b;6:1–7. doi: 10.1038/srep22631. http://dx.doi.org/10.1038/srep22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li X, Wang L, Ding P, Zhang Y, Han W, et al. An alternative form of paraptosis-like cell death, triggered by TAJ/TROY and enhanced by PDCD5 overexpression. J. Cell Sci. 2004;15:1525–1532. doi: 10.1242/jcs.00994. http://dx.doi.org/10.1242/jcs.00994. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang H, Xu ZM, Ji Y-L, Chen Y-H, Zhang Z-H, et al. Cadmium-induced teratogenicity: association with ROS-mediated endoplasmic reticulum stress in placenta. Toxicol. Appl. Pharmacol. 2012;259:236–247. doi: 10.1016/j.taap.2012.01.001. http://dx.doi.org/10.1016/j.taap.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Wilcox RR, Keselman HJ. Modern regression methods that can substantially increase power and provide a more accurate understanding of associations. Eur. J. Pers. 2012;26:165–174. doi: 10.1002/per.860. http://dx.doi.org/10.1002/per.860.MODERN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun P, Liu K, Morris JS, Daviglus ML, Stevens J, Jacobs DR, et al. Associations of toenail selenium levels with inflammatory biomarkers of fibrinogen, high-sensitivity C-reactive protein, and interleukin-6: the CARDIA trace element study. Am. J. Epidemiol. 2010;171:793–800. doi: 10.1093/aje/kwq001. http://dx.doi.org/10.1093/aje/kwq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa K, Willett WC, Morris SJ, Stampfer MJ, Spiegelman D, Rimm EB, et al. Study of prediagnostic selenium level in toenails and the risk of advanced prostate cancer. J. Natl. Cancer Inst. 1998;90:1219–1229. doi: 10.1093/jnci/90.16.1219. [DOI] [PubMed] [Google Scholar]
- You R-I, Chen M-C, Wang H-W, Chou Y-C, Lin C-H, Hsieh S-L. Inhibition of lymphotoxin-beta receptor – mediated cell death by survivin-deltaEx3. Cancer Res. 2006;4:3051–3061. doi: 10.1158/0008-5472.CAN-05-2479. http://dx.doi.org/10.1158/0008-5472.CAN-05-2479. [DOI] [PubMed] [Google Scholar]
- Zelová H, Hošek J. TNF-apha signalling and inflammation: interactions between old acquaintances. Inflamm. Res. 2013;62:641–651. doi: 10.1007/s00011-013-0633-0. http://dx.doi.org/10.1007/s00011-013-0633-0. [DOI] [PubMed] [Google Scholar]
- Zhang F, Yu W, Hargrove JL, Greenspan P, Dean RG, Taylor EW, et al. Inhibition of TNF-alpha induced ICAM-1, VCAM-1 and E-selectin expression by selenium. Atherosclerosis. 2002;161:381–386. doi: 10.1016/s0021-9150(01)00672-4. [DOI] [PubMed] [Google Scholar]
- Zhao H, Brooks JD. Selenomethionine induced transcriptional programs in human prostate cancer cells. J. Urol. 2007;177:743–750. doi: 10.1016/j.juro.2006.09.071. http://dx.doi.org/10.1016/j.juro.2006.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y-J, Zhang S-P, Liu C-W, Cai Y-Q. The protection of selenium on ROS mediated-apoptosis by mitochondria dysfunction in cadmium-induced LLC-PK(1) cells. Toxicol. Vitr. 2009;23:288–294. doi: 10.1016/j.tiv.2008.12.009. http://dx.doi.org/10.1016/j.tiv.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Zwiener I, Frisch B, Binder H. Transforming RNA-Seq data to improve the performance of prognostic gene signatures. PLoS One. 2014;9 doi: 10.1371/journal.pone.0085150. http://dx.doi.org/10.1371/journal.pone.0085150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.