Abstract
Mutations in PDE6 genes encoding the effector enzymes in rods and cones underlie severe retinal diseases including retinitis pigmentosa (RP), autosomal dominant congenital stationary night blindness (adCSNB), and achromatopsia (ACHM). Here we examined a spectrum of pathogenic missense mutations in PDE6 using the system based on co-expression of cone PDE6C with its specialized chaperone AIPL1 and the regulatory Pγ subunit as a potent co-chaperone. We uncovered two mechanisms of PDE6C mutations underlying ACHM: (a) folding defects leading to expression of catalytically inactive proteins and (b) markedly diminished ability of Pγ to co-chaperone mutant PDE6C proteins thereby dramatically reducing the levels of functional enzyme. The mechanism of the Rambusch adCSNB associated with the H258N substitution in PDE6B was probed through the analysis of the model mutant PDE6C-H262N. We identified two interrelated deficits of PDE6C-H262N: disruption of the inhibitory interaction of Pγ with mutant PDE6C that markedly reduced the ability of Pγ to augment the enzyme folding. Thus, we conclude that the Rambusch adCSNB is triggered by low levels of the constitutively active PDE6. Finally, we examined PDE6C-L858V, which models PDE6B-L854V, an RP-linked mutation that alters the protein isoprenyl modification. This analysis suggests that the type of prenyl modifications does not impact the folding of PDE6, but it modulates the enzyme affinity for its trafficking partner PDE6D. Hence, the pathogenicity of PDE6B-L854V likely arises from its trafficking deficiency. Taken together, our results demonstrate the effectiveness of the PDE6C expression system to evaluate pathogenicity and elucidate the mechanisms of PDE6 mutations in retinal diseases.
Keywords: retina, photoreceptor, phosphodiesterases, AIPL1, chaperone, achromatopsia
1. Introduction
Cyclic-nucleotide phosphodiesterases of the sixth family (PDE6) are the effectors in the phototransduction cascade in rods and cones [1–3]. The rod PDE6 catalytic core is a heterodimer of PDE6A and PDE6B subunits, whereas cone PDE6C subunits form a catalytic homodimer. Each catalytic PDE6 subunit in a holoenzyme is associated with a small inhibitory Pγ subunit [4]. Light-induced activation of PDE6 involves a G-protein mediated displacement of Pγ leading to a spike in cGMP hydrolysis and generation of an electrical signal by a cGMP-gated channel in the plasma membrane [1, 3]. Mutations in the PDE6A and PDE6B genes account for a sizable fraction of cases of autosomal recessive retinitis pigmentosa (RP), a progressive degeneration of rods leading to blindness [5, 6]. The H258H mutation in PDE6B was identified in patients with autosomal dominant congenital stationary night blindness (adCSNB) [7]. Mutations in PDE6C lead to a loss of cone function and cause autosomal recessive achromatopsia (ACHM) [8–10]. Certain PDE6 mutations, such as nonsense mutations, splice defects, and frame shifts, are predicted to cause loss-of-function, thus leading to elevation of intracellular cGMP levels. Increased cGMP causes photoreceptor cell death primarily via excessive opening of the cGMP- gated channels in photoreceptor plasma membrane and unrestrained influx of Ca2+ [11–13]. Activation of PKG may also contribute to cGMP-induced death of photoreceptors [14, 15]. However, the initial mechanisms by which many missense PDE6 mutations presumably lead to elevation of cGMP are largely unknown. Furthermore, pathogenicity of PDE6 mutations it is not always known, thereby blocking development of potential patient-specific therapies.
Progress in investigating the mechanisms of PDE6 mutations has been slow due to a lack of a heterologous expression system [16–20]. Chimeras between cone PDE6C and the related cGMP-specific PDE5 (PDE5/6) or chimeric PDE5/6C catalytic domains have been employed to overcome this limitation [17, 19, 21]. In particular, several missense mutations linked to ACHM were previously introduced into chimeric PDE5/PDE6C proteins that were expressed in sf9 cells [10]. These mutants indicated either a loss or reduction of the catalytic activity [10]. However, the use of PDE5/PDE6C chimeras to study effects of PDE6 mutations has severe limitations. PDE5/PDE6C chimeras contained the catalytic domain of PDE5 and thus, they are essentially PDE5-like enzymes. Another approach has been ectopic expression of EGFP-PDE6C in the rods of transgenic X. laevis [22–24]. However, comprehensive biochemical studies of PDE6 mutants using this system have been impractical due to the need to produce large numbers of transgenic tadpoles. Recently we developed a simple and robust system for PDE6 expression in HEK293T cells [25]. We found that co-expression of PDE6C with its specialized chaperone AIPL1 in cultured HEK293T cells yields low levels of functional enzyme, which are greatly elevated in the presence of Pγ [25]. Here we applied this system to investigate the mechanisms of PDE6C mutations that were identified in patients with ACHM or model PDE6B mutations causing adCSNB and RP (Fig. 1A). Our analysis identified a novel mechanism whereby mutations in PDE6 attenuate its folding by abrogating the ability Pγ to co-chaperone the mutant proteins. Overall, our study establishes the heterologous expression system of PDE6C as a sensitive and versatile tool to investigate the mechanisms of PDE6 mutations underlying retinal diseases.
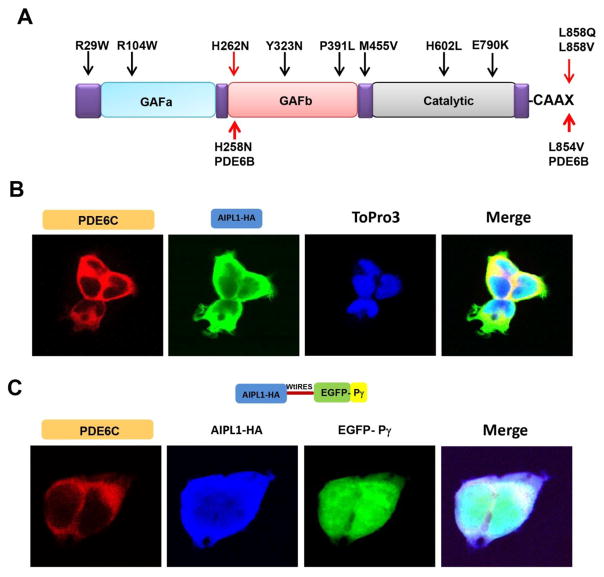
Fig. 1. Heterologous expression system to study disease-linked mutations in PDE6 genes.
A. Diagram of selected mutations in genes encoding the catalytic subunits of PDE6. Mutations in PDE6C linked to ACHM are indicated by black arrows. The H258N and L854V substitutions in PDE6B underlying the Rambusch adCSNB and RP, respectively, and the corresponding model substitutions in PDE6C are indicated by red arrows. (B, C) Confocal immunofluorescence images of HEK293T cells (B) co-transfected with PDE6C (red, anti-PDE6C) and AIPL1 (green, anti-HA) (blue, TO-PRO3 nuclear stain), or (C), co-transfected with PDE6C (red, anti-PDE6C) and the AIPL1-Pγ vector (AIPL1: blue, anti-HA; Pγ: green, EGFP fluorescence).
2. Materials and Methods
2.1. Plasmids/cloning
DNA sequence encoding the full-length mouse AIPL1 was PCR amplified from pET15b vector harboring mouse AIPL1 gene [26] using 5′ primer with BamHI site and 3′ primer that encodes an HA-tag and an XbaI site. The PCR product was then cloned into pcDNA3.1(+) (Invitrogen). Subsequently, IRES-EGFP-Pγ sequence was amplified by PCR from pFIV3.2CAGmcswtIRESeGFP- Pγ and cloned into the XbaI site of the pcDNA3.1-AIPL1-HA plasmid. DNA coding the Flag-tagged human PDE6C was PCR amplified from the PDE6C transgene [27] and cloned into pcDNA3.1(+) using BamHI/Eco RV sites. Mutations were introduced into PDE6C gene using standard QuikChange site-directed mutagenesis protocol. The sequences of all constructs were verified by automated DNA sequencing at the UI DNA Core Facility.
2.2. Cell culture and fractionation of cell lysates
HEK293T cells were cultured and maintained in DMEM containing 10% FBS (Gibco). Cells were cotransfected with either mouse AIPL1 and human PDE6C (2 μg) or with mouse AIPL1, human PDE6C and mouse Pγ (2 μg) plasmids using FuGene6 (Promega) according to manufacturer’s instructions. Transfected cells were collected 48h post- transfection. Cell lysates, prepared in 20 mM Tris-HCl buffer (pH 7.5) containing 120 mM KCl and 1 mM MgCl2 were analyzed by Western blotting for protein expression and assayed for PDE activity. For immunofluorescence, 48h post-transfection, cells were seeded onto poly-D-lysine coated (0.1 mg/mL) 4-well chambered glass slide and allowed to grow for additional 24h before fixation with 4% formaldehyde.
For the PDE6C activity assay, PDE6C, AIPL1, or AIPL1 and Pγ co-transfected HEK293T cell lysates were prepared in hypotonic buffer (5 mM Tris-HCl, pH 7.5, 1 mM MgCl2, buffer A) and centrifuged at 125,000 x g for 30 min at 4°C in Beckman Optima TLX Ultracentrifuge. Supernatants thus obtained were used for all PDE6C activity measurements.
2.3. Pull-down assay
His6-PDE6D was cloned, expressed and purified as described previously [28]. 80μg of purified His6-PDE6D was incubated with 80 μl of Ni-NTA resin for 30 min at 4°C. PDE6D- bound resin was washed three times with buffer A to remove unbound protein. For pull-down assay, 20 μl of PDE6D -bound Ni-NTA resin was incubated with 80 μl each of untreated or trypsin-treated (see 2.6) hypotonic extracts (3–4 μg protein/μl) of HEK293T cell lysates expressing wild type FLAG-PDE6C or L858V mutant at 25°C for 30 min. Resin was washed three times with 250 μl of buffer A to remove unbound proteins and then 30μl of SDS-PAGE sample buffer was added to the resin. The samples were separated on 4–12% gradient Bis-tris NuPage gels (Invitrogen) and analyzed by Western blotting.
2.4. Western Blotting
Proteins separated by 4–12% SDS-PAGE were transferred to a nitrocellulose membrane using iBlot Western blot kit (Invitrogen) and analyzed using mouse monoclonal anti-FLAG antibody (Sigma), (1:2000 dilution), mouse anti-HA (BioLegend) (1:1000 dilution), mouse anti-EGFP (Sigma) (1:2000) and His-probe (H-15, Santa Cruz Biotechnology, 1:2000) primary antibodies. The antibody-antigen complexes were detected using horseradish peroxidase-conjugated goat anti-mouse (1:10000 dilution) secondary antibody and enhanced chemiluminescence (ECL) reagents obtained from GE Healthcare.
2.5. Immunofluorescence
HEK293T cells on chambered glass slide were washed once with PBS and fixed for 15 min in 4% formaldehyde in PBS at 25°C. Cells were then washed once in PBS and permeabilized in 0.1% Triton X-100 in PBS for 3–5 min at 25°C. Permeabilized cells were washed three times for 5 min each in PBS, blocked for 60 minutes in blocking solution (1% BSA in 1x PBS) and then incubated at 4°C overnight with either rabbit anti-PDE6C [27] and mouse anti-HA antibodies (BioLegend), or mouse anti-FLAG (Sigma) diluted (1:2000) in the blocking solution. After washing in PBS three times for 5 min each, where indicated, cells were incubated in the dark for 1 hour in either Alexa Fluor 568-conjugated goat anti-rabbit or Alexa Fluor 488-conjugated goat anti-mouse (Life Technologies) or Alexa Fluor 647-conjugated F(ab′)2-Goat anti-mouse IgG (H+L) secondary antibody (Thermo Scientific) (1:2000) diluted in the blocking solution. Cells were washed three times for 5 min each in PBS, and the nuclei were counter stained with To-Pro-3 (1:1000) (ThermoFisher) for 30 min in the dark at 25°C. Cells were mounted using Vectashield mounting medium (Vector Laboratories, Inc) and imaged using Plan-Neofluar 40×/1.3 oil lens and a LSM 510 confocal microscope (Zeiss).
2.6. PDE6 activity assay
cGMP hydrolysis was measured in cell extracts obtained from HEK293T cells 48 hr post- transfection. Where indicated, samples were treated with 0.1 mg/mL TPCK-Trypsin (Sigma) on ice for 10 min to selectively degrade Pγ, after which trypsin was inhibited with the addition of 10-fold excess of soybean trypsin inhibitor (Sigma) and incubation for 5 min at 25°C. Cell extracts (protein concentration 3–6 mg/ml) were diluted 600 fold into 40 μl (final volume) of 20 mM Tris–HCl (pH 7.5) buffer containing 120 mM NaCl, 2 mM MgSO4, 1 mM 2-mercaptoethanol, 0.1 U bacterial alkaline phosphatase, 10 μM [3H]cGMP (100,000 cpm) (PerkinElmer) for 10–15 min at 37°C. For Pγ-inhibition assays, bovine rod Pγ was expressed and purified as described previously [23]. The reaction was stopped by the addition of AG1-X2 cation exchange resin (0.5 mL of 20% bed volume suspension). Samples were incubated for 6 min at 25°C with occasional mixing, and spun at 10,000g for 3 min. 0.25 ml of the supernatant was removed for counting in a scintillation counter.
3. Results
3.1. Expression and subcellular distribution of PDE6C mutants in HEK293T cells
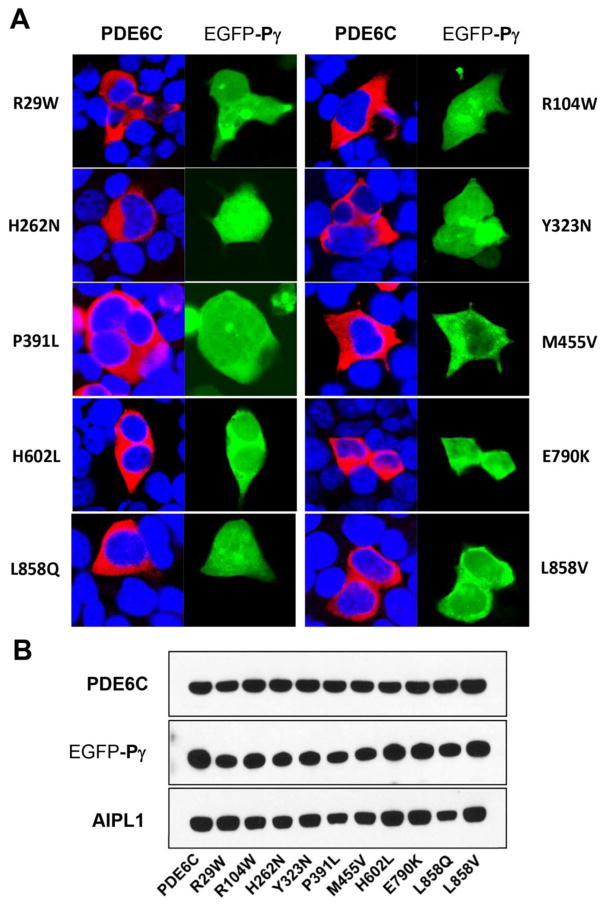
Seven missense mutations in PDE6C have been identified in ACHM patients, resulting in the following substitutions: R29W, R104W, Y323N, P391L, M455V, H602L, and E790K (Fig. 1A) [8–10]. We examined these mutations as well as H262N and L858V (L858Q) that mimic the Rambusch adCSNB [7] and RP [6], respectively, using heterologous expression of functional PDE6C in HEK293T cells [25]. We used two experimental paradigms, (a) where PDE6C is co-expressed with AIPL1 alone, and (b) PDE6C is-co-expressed with both AIPL1 and Pγ. AIPL1 alone provides for low-level expression of active PDE6C in cultured cells, whereas the AIPL1-directed production of the folded enzyme is markedly elevated in the presence of Pγ [25]. For consistent co-expression of PDE6C mutants with AIPL1 and Pγ in the same population of HEK293T cells, we generated a pcDNA3.1(+) expression vector coding C-terminally HA-tagged AIPL1 and EGFP-tagged Pγ separated by wild-type IRES. HEK293T cells were transfected with the Flag-tagged PDE6C, alone or in combination with AIPL1, or both AIPL1 and Pγ. 48 hrs post-transfection, the cells were examined by immunofluorescence microscopy. As previously reported [25], PDE6C expressed in HEK293T in the absence or the presence of AIPL1 is distributed throughout the cell excluding the nucleus (Fig. 1B). The intracellular localization of PDE6C was similar on co-expression with AIPL1 alone or with both AIPL1 and Pγ (Fig. 1B,C). The signal for AIPL1 was observed mainly in cytoplasm but also to a lesser degree in the nucleus (Fig. 1B,C). EGFP-Pγ was distributed diffusely throughout the cells (Fig. 1B,C). Interestingly, the pattens of subcellular distribution of every tested PDE6C mutant were similar to that of the wild-type protein (Fig. 2A). These patterns did not reveal any gross abnormalities such as formation of inclusion bodies or strongly reduced immunofluorescence signals. The immunoblot analysis of cell extracts of transfected HEK293T cells revealed robust expression of both EGFP- Pγ and AIPL1 proteins from the single plasmid (Fig. 2B). PDE6C mutants appeared to be proteolytically stable with a single 100 kDa band detected in immunoblots (Fig. 2B). The levels of expression for all PDE6C mutant proteins were comparable to that of the WT PDE6C.
Fig. 2. Expression and subcellular distribution of mutant PDE6C proteins in HEK293T cells.
A. Confocal immunofluorescence images of HEK293T cells co-transfected with mutant PDE6C (red, anti-Flag) and the AIPL1-Pγ vector (Pγ: green, EGFP fluorescence; blue, TO-PRO3 nuclear stain). B. Western blot analysis of extracts of HEK293T cells co-transfected with mutant PDE6C and the AIPL1-Pγ vector using anti-Flag (PDE6C), anti-EGFP (Pγ), and anti-HA (AIPL1) antibodies. Lanes contain equal amounts of protein.
3.2 Mechanisms of ACHM-linked PDE6C mutations
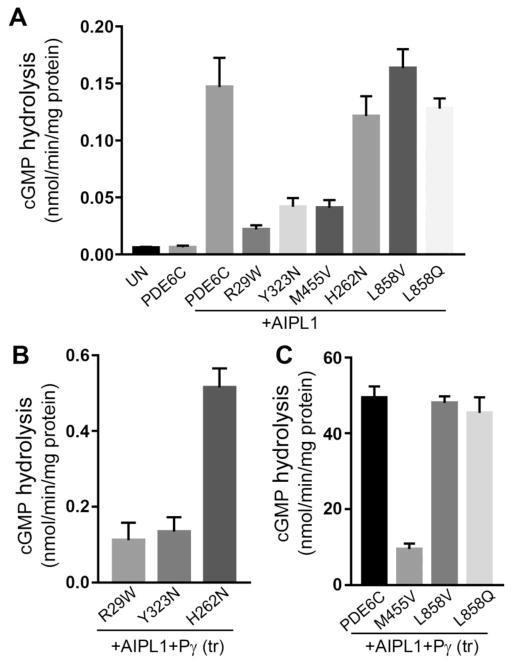
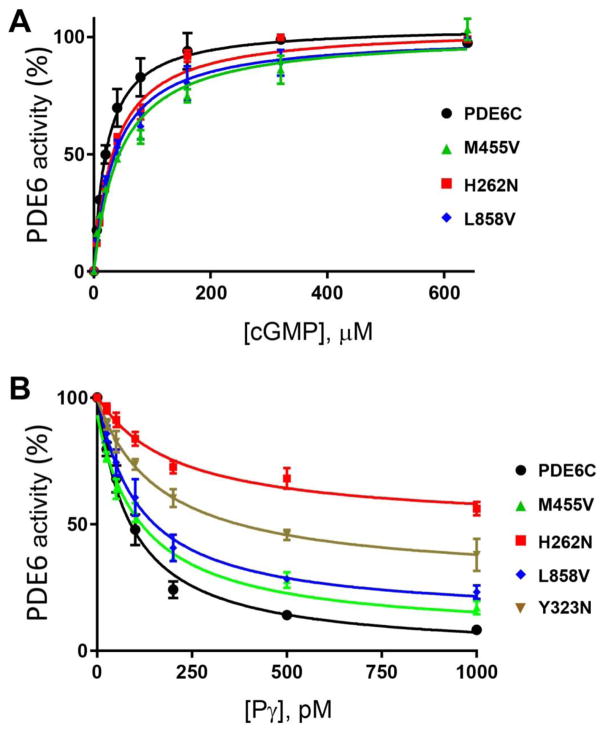
In accordance with previous observations, cGMP-hydrolysis in extracts from HEK293T cells co-transfected with PDE6C and AIPL1 exceeded the background activity in extracts from untransfected cells or cells transfected with PDE6C alone by about 25-fold (Fig. 3A). The PDE6C activity measured in the presence of AIPL1 alone was further elevated by ~ 335 fold when HEK293T cells were co-transfected with PDE6C, AIPL1 and Pγ (Fig. 3C). Note that since Pγ is the inhibitory subunit of PDE6C, the activity assays were performed after removing Pγ from the PDE6C-AIPL1-Pγ cell extracts with limited tryptic proteolysis. cGMP hydrolysis in extracts from HEK293T cells transfected with the ACHM mutants R104W, P391L, H602L, and E790K did not exceed the very low background level regardless of whether these mutants were co-transfected with AIPL1 alone or with AIPL1 and Pγ (not shown). Thus, these four mutations rendered PDE6C catalytically inactive. However, AIPL1 was capable of chaperoning the ACMH-linked R29W, Y323N, and M455V mutant PDE6C proteins. cGMP hydrolysis in extracts from cells transfected with AIPL1 and R29W, Y323N, or M455V was noticeably lower compared to PDE6C-AIPL1 extract, but significantly higher than the background activity in untransfected cells (Fig. 3A). Folding deficiencies of R29W and Y323N became more evident upon co-transfection of these mutants with AIPL1 and Pγ. The yields of functional R29W and Y323N were enhanced in the presence of Pγ by only ~5 and ~3 fold, respectively (Fig. 3B). Consistent with this observation, the inhibition of activity of Y323N by Pγ was impaired (Fig. 4B). In contrast, Pγ potentiated folding of functional M455V by ~230 fold. As a result, the rate of cGMP hydrolysis in the AIPL1-Pγ extract of M455V was only ~5-fold lower than that for WT PDE6C (Fig. 3C). Furthermore, the key properties for M455V such as the KM for cGMP (42±11 μM) and the Ki for the inhibition by Pγ (98±4 nM) were comparable to those of WT PDE6C (Fig. 4B).
Fig. 3. Catalytic activities of mutant PDE6C proteins expressed in HEK293T cells.
cGMP hydrolysis in extracts of HEK293T cells co-transfected with mutant PDE6C and AIPL1 alone (A) or the AIPL1-Pγ vector (B, C) (UN, untransfected control; (tr), limited treatment with trypsin to remove Pγ) (mean ± S.E., n ≥3). Statistical significance of the differences in (A) due to AIPL1 compared to PDE6C alone was initially assessed by t-tests, which showed *p=0.006 for PDE6C+AIPL1, and p values of < 0.02 for all of the tested PDE6C mutants. Statistical significance of the differences in (A) between the samples with AIPL1 present and the samples in (C) was assessed by ANOVA with Tukey’s post-hoc test. The AIPL1-mediated activities (A) of PDE6C, H262N, L858V and L858Q (group I) were not significantly different (p>0.05), as were the activities of R29W, Y232N, and M455V) (group II). However, each group I activity was significantly higher than each group II activity (p<0.05). The activities of PDE6C, L858V and L858Q in (C) were not significantly different from each other (p>0.05), but these were significantly higher than the activity of M455V (****P<0.0001). The differences in activities in (B) compared to those in (A) were analyzed with t-tests for the selected pairs (R29W, p=0.08, n.s.; Y323N, *p=0.04; H262N, ****p<0.0001).
Fig. 4. Characterization of mutant PDE6C proteins.
A. The rates of cGMP hydrolysis were measured in the presence of increasing concentrations of cGMP in hypotonic extracts obtained from HEK293T cells co-transfected with the AIPL1-Pγ vector and the WT or mutant PDE6C. PDE6C activity is expressed as a percentage of maximal activity. PDE6C, KM=24±2 μM; PDE6C-M455V, 42±11 μM; PDE6C-H262N, KM=30±2 μM; PDE6C-L858V, KM=32±3 μM. Results are shown as Mean±SE. B. Pγ-inhibition of PDE6C activity in hypotonic extracts of HEK293T cells co-transfected with AIPL1 and the WT or mutant PDE6C. PDE6C, Ki=88±21 pM; PDE6C-Y323N, Ki=176±28 pM (maximal inhibition 72±6%); PDE6C-M455V, Ki=98±4 pM; PDE6C-H262N, Ki=222±49 pM (maximal inhibition 51±1%); PDE6C-L858V, Ki=114±37 pM. Results are shown as Mean±SE.
3.3. Molecular dysfunctions of PDE6 underlying the Rambusch form of adCSNB
The H262N mutation in PDE6C was utilized as a surrogate for the H258N mutation in PDE6B that causes the Rambusch form of adCSNB (Fig. 1A) [7]. Our rationale for using PDE6C as a template to model this PDE6B mutation was several-fold. The mutated His residue is strictly conserved in cone and rod PDE6. Also, because of heterodimerization of mutant PDE6B with the WT PDE6A in the rod enzyme, the effects of mutation may have not been as apparent as they would have been using homomeric PDE6C. Finally, the use of PDE6C streamlines the expression system by minimizing variability due to transfections with fewer plasmids. The activity of PDE6C-H262N on its co-expression with AIPL1 was robust and comparable to the activity of WT PDE6C expressed under similar conditions (Fig. 3A). Remarkably, Pγ was found to be a very poor co-chaperone in the AIPL1-assisted folding of H262N, as cGMP-hydrolysis in the H262N-AIPL1-Pγ extract was only ~4-fold higher compared to that in the H262N-AIPL1 extract. PDE6C-H262N hydrolyzed cGMP with the KM value of 30±2 μM, which is similar to that for WT PDE6C (24±2 μM) (Fig. 4A). However, the analysis of PDE6C-H262N inhibition by Pγ revealed two significant defects. First, the Ki for the inhibition by Pγ was increased by ~2.5-fold compared to WT PDE6C (220±49pM vs 88±21 pM, *p=0.026). More importantly, the Pγ-inhibition of PDE6C-H262N was incomplete (maximal inhibition of 51±1%, Fig. 4B). Thus, the diminished ability of Pγ to serve as AIPL1 co-chaperone in the folding of PDE6C-H262N was apparently due to disruption of Pγ interaction with the mutant enzyme.
3.4. The identity of prenyl modification of PDE6 does not influence its AIPL1/Pγ-mediated folding, but alters its affinity for PDE6D
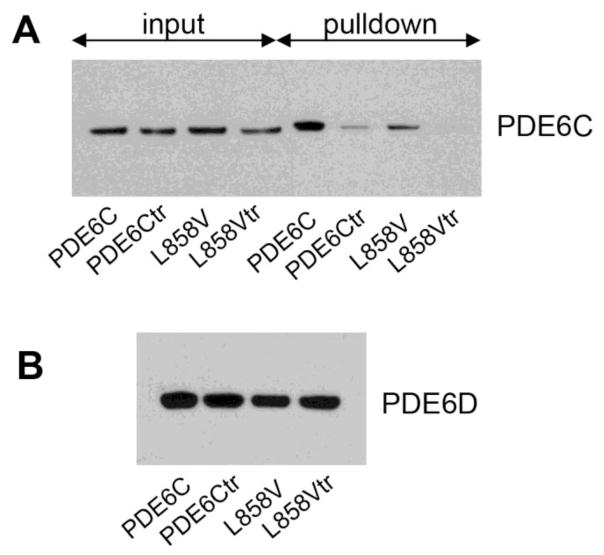
The L854V mutation in PDE6B is particularly interesting, because unlike most of RP mutations that affect the regulatory GAF domains or the catalytic domain, this mutation alters the isoprenylation site at the C-terminus (Fig. 1A) [6]. PDE6B-Leu854 is the X residue in the C-terminal CAAX-box that specifies the geranylgeranyl modification of the Cys residue in the box, whereas the C-terminal Val directs farnesylation [29, 30]. To probe the mechanism of PDE6B L854V mutation, we generated an analogous mutation in PDE6C, L858V. This mutation is predicted to be a functional equivalent of the rod PDE6 mutant, because both mutations lead to the PDE6 catalytic dimers in which each subunit is farnesylated [30]. Analyses of the L858V mutant demonstrated that the levels of correctly folded enzyme in the presence of AIPL1 alone or in combination of AIPL1 and Pγ were comparable to the respective levels of the WT PDE6C (Fig. 3A,C). Moreover, the KM value (cGMP) of 32±3 μM and the Ki (Pγ) value of 114±37 pM were similar to those of WT PDE6C. To strengthen our findings, we produced another PDE6C variant L858Q with the C-terminal Q residue as in the farnesylated PDE6A subunit [30]. Similarly to PDE6C-L858V, functional expression of PDE6C-L858Q was similar to that of WT PDE6C (Fig. 3A,C). We next examined the possibility that the L858V mutation alters the interaction of PDE6C with PDE6D, a prenyl-binding protein essential for trafficking of PDE6 in photoreceptor cells. Pulldowns of WT PDE6C and PDE6C-L858V were performed with His-tagged PDE6D from untreated or trypsin-treated extracts obtained from HEK293T cells transfected with the PDE6C and AIPL1-Pγ constructs (Fig. 5). The trypsin-treated extracts served as the controls for the prenyl-dependent binding of PDE6D because such treatment is known to rapidly remove the C-terminal lipid modifications from PDE6 catalytic subunits [31]. The Western blot analysis with anti-FLAG antibody indicated that the L858V mutation significantly reduced the amounts of PDE6C protein co-precipitated with PDE6D in the prenyl-dependent manner (Fig. 5). Thus, the affinity of PDE6D for farnesylated PDE6C-L858V is apparently lower than for the geranylgeranylated WT PDE6C enzyme.
Fig. 5. The L858V mutation reduces binding of PDE6C to the prenyl-binding protein PDE6D.
A. Inputs of the untreated or trypsin-treated (tr) PDE6C and L858V extracts (25% of total input) and the proteins co-precipitated with the His6-PDE6D resin were analyzed by Western blotting using anti-Flag antibody. B. The pulldowns in (A) analyzed for His6-PDE6D by Western blotting using anti-His antibody.
4. Discussion
In this study, we analyzed for the first time the consequences of the disease-causing mutations in PDE6 in the context of the enzyme heterologously expressed in HEK293T cells. This analysis became possible due to a recently established expression system that is based on co-expression of PDE6C with its chaperone AIPL1 and co-chaperone Pγ [25]. Our analyses revealed that the ACHM-linked mutations, R104W, P391L, H602L, and E790K, abolish PDE6C catalytic activity. The H602L substitution directly alters the metal-coordination site that is indispensable for cGMP hydrolysis [32]. Substitutions R104W, P391L, and E790K apparently prevented folding of the catalytically competent enzyme. The finding that E790K mutant lacks catalytic activity highlights the efficacy of the WT PDE6C template as opposed to the PDE5/PDE6 chimera to screen PDE6 mutations for pathogenicity. Previously, the E790K mutant of the PDE5/PDE6 chimera had been shown to retain a significant fraction of the catalytic activity [10]. Thus, identical mutations within the PDE6 and PDE5-like catalytic domains had a different impact on the catalytic activity. The second key advantage of the PDE6C expression system is the ability to probe the effects of mutations on the co-chaperone function of Pγ [25]. In the presence of AIPL1, ACHM mutants R29W, Y323N, and M455V hydrolyzed cGMP with the rates that were significantly higher than the background hydrolysis in untransfected cells. However, co-transfection with AIPL1 and Pγ demonstrated that the major deficit of the R29W and Y323N proteins originates from the markedly reduced ability of Pγ to co-chaperone these mutants. As consequence, the yields of functional R29W and Y323N in the presence of AIPL1 and Pγ are two-three orders of magnitude lower compared to the yields of WT PDE6C. We hypothesize that Pγ fails to effectively co-chaperone R29W and Y323N because these mutations disrupt the PDE6C-Pγ interface. In support of our hypothesis, the inhibitory interaction of Pγ with PDE6C-Y323N was impaired. Furthermore, Y323 is located in proximity to K328, a counterpart of the PDE6A K326 and PDE6B K324 that were found to crosslink to Pγ [33] (Fig. 6). Thus, our analysis suggests a novel mechanism of pathogenic PDE6 mutations whereby defective co-chaperone function of Pγ is caused by disrupted interaction of the nascent PDE6 catalytic subunits with Pγ. In contrast to R29W and Y323N, the AIPL1-mediated folding of M455V was potentiated by Pγ comparably to the effect of Pγ on WT PDEC, resulting in the overall robust functional expression of this mutant. Therefore, our results predict that M455V is either a benign variant of PDE6C or the phenotype of this mutation is milder compared to other ACHM mutations.
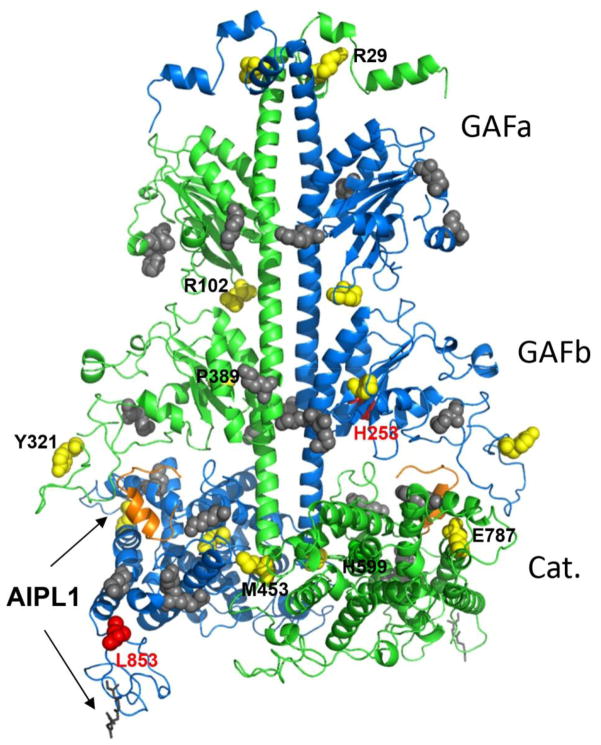
Fig. 6. Map of disease-causing PDE6 mutations and the Pγ-interaction sites.
Residues corresponding to pathogenic substitutions in human PDE6 catalytic subunits are mapped on the cluster 1 model of bovine rod PDE6AB derived from a high-density crosslinking study, which also identified residues cross-linked with Pγ (shown as grey spheres) [33]. The backbones of PDE6A and PDE6B are shown in green and blue, respectively. PDE6A and PDE6B residues corresponding to the ACMH-linked residues in human PDE6C are shown as yellow spheres, but labeled only for the PDE6A subunit for clarity. Bovine PDE6B residues corresponding to those mutated in Rambusch adCSNB and RP are shown as red spheres. The farnesyl and geranylgeranyl lipid modifications of the C-terminal CAAX box are shown as grey sticks. The C-terminal inhibitory fragment of Pγ superimposed from the structure of the PDE5/6 catalytic domain (PDB ID 3JWR) [35] is shown as an orange cartoon. Arrows indicate probable AIPL1-PDE6 interface based on known interactions of AIPL1 with the prenyl moieties and Pγ [26, 39, 46].
Further, our study demonstrates the utility of the cone PDE6C expression system to study mutations in rod PDE6 implicated in RP and/or adCSNB. Certain findings with the ACHM-linked PDE6C mutants should be directly applicable to mutations in rod PDE6. Substitutions R102H and R102S in PDE6A cause RP [6]. The lack of discernable catalytic activity of the counterpart mutant R104W of PDE6C predicts a similar deficiency for the RP mutants. More important, however, is the ability to model rod PDE6 mutations using PDE6C. Our analysis with PDE6C-H262N (H258N in PDE6B) not only provides novel clues to the mechanism of the Rambusch form of adCSNB, but it also has intriguing implications for the mechanism of PDE6 folding. Functional expression of H262N in the presence of AIPL1 alone was comparable to that of WT PDE6C, but the ability of Pγ to co-chaperone H262N was dramatically reduced, resulting in ~100-fold decrease in the yield of functional enzyme in the presence of AIPL1 and Pγ. Such a drastic impairment of the Pγ co-chaperone function was not likely caused by the lower affinity of Pγ for H262N: the Ki value was increased by only ~2.5-fold compared to that for WT PDE6C. Rather, the impairment reflects the inability of Pγ to fully inhibit the catalytic activity of PDE6C-H262N. Partial maximal inhibition of H262N implies that the catalytic pocket of the mutant is not fully occluded by the C-terminus of Pγ [34, 35] (Fig. 6). Based on current models of PDE6 [33, 35, 36], binding of AIPL1 to the prenyl moieties of nascent PDE6 during the enzyme assembly brings AIPL1 close to the catalytic site where it apparently interacts with Pγ to cooperatively chaperone PDE6 (Fig. 6). Thus, the disruption of the interaction between the catalytic domain of PDE6C-H262N and the C-terminus of Pγ is entirely consistent with the abrogation of the Pγ function as a co-chaperone of PDE6. PDE6C His262 (PDE6B His258) is located in the GAFb domain of PDE6 which is thought to bind to the central region of Pγ [33, 37] (Fig. 6). It remains to be determined how the structural change caused by mutation of this His is transmitted to the interface between the Pγ C-terminus and the PDE6 catalytic domain (Fig. 6). An important ramification of our findings is that the level of functional mutant rod PDE6 in Rambusch adCSNB is predicted to be significantly downregulated. Because of its constitutive activity, the mutant PDE6 that is present may still be sufficient to desensitize rods. Yet, downregulation of the protein may explain a puzzling phenotype often observed in Rambusch patients, i. e. preservation ERG a-waves [38]. If rod ERG a-waves are indeed preserved and the b-waves are disproportionally affected, alternative mechanisms of the mutant PDE6 must be considered, such as inappropriate PDE6 activity in the inner compartments of rod photoreceptors.
Finally, we examined the expression and properties of the PDE6C L858V and L858Q mutants in which the lipid modification is switched from geranylgeranyl to farnesyl. Since the mechanism of AIPL1 chaperone activity involves the binding of its FKBP domain to the prenyl moieties of PDE6 [25, 26, 39], the prenylation switch may have impacted the protein folding. Contrary to this assumption, the AIPL1/Pγ-assisted folding of L858V and L858Q in HEK293T cells was equivalent to that of the WT PDE6C. Thus, the type of isoprenylation of the PDE6 catalytic subunits does not influence the folding and assembly of the enzyme. Consequently, unless the L854V substitution in PDE6B is a benign variation, this mutant PDE6 fails to traffic to the outer segment of rod photoreceptors. Trafficking of PDE6 involves its interaction with the prenyl-binding protein PDE6D [40–43]. PDE6D binds geranylgeranyl moiety deeper in its hydrophobic pocket compared to the farnesyl moiety, and additional hydrophobic contacts are likely to increase the binding affinity [44]. Indeed, we demonstrated that the L858V mutation reduced PDE6C binding to PDE6D. High affinity of PDE6D for the geranylgeranylated PDE6 catalytic subunit might be essential for the ARL3-dependent cargo sorting into the photoreceptor cilium [45].
5. Conclusion
Our study reveals two general mechanisms of missense PDE6 mutations underlying retinal diseases: (a) inability of AIPL1 to fold mutant PDE6 proteins leading to complete catalytic inactivity and (b) failure of Pγ to serve as co-chaperone with AIPL1 in folding of mutant PDE6, which markedly reduces the levels of functional enzyme. A third potential mechanism is indicated by the analysis of PDE6C-L858V that involves abnormal trafficking of PDE6 mutants.
Highlights.
Robust system to probe pathogenicity and mechanisms of PDE6 mutations is established
A Novel mechanism of PDE6 mutations involves diminished co-chaperone ability of Pg.
Rambuch night blindness is triggered by low levels of the constitutively active PDE6
The folding of PDE6 is not dependent on the type of its prenylation.
Acknowledgments
This work was supported by the National Institutes of Health grant EY-10843 to N.O.A.
Abbreviations
- PDE6
photoreceptor phosphodiesterase-6
- AIPL1
aryl hydrocarbon receptor-interacting protein-like 1
- LCA
Leber congenital amaurosis
- FKBP
FK506-binding proteins
- TPR
tetratricopeptide repeat
- GAF
domains named for their presence in cGMP-regulated PDEs, adenylyl cyclases, and the E. coli protein Fh1A
- PDE6D
prenyl-binding protein PDE6δ also known as PrBP/δ
Footnotes
Author contribution
K.G. and N.O.A. designed research; K.G. and K.B. performed research; K.G and N.O.A. analyzed data; N.O.A. wrote the paper.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Arshavsky VY, Lamb TD, Pugh EN., Jr G proteins and phototransduction. Annu Rev Physiol. 2002;64:153–87. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 2.Cote RH. Characteristics of photoreceptor PDE (PDE6): similarities and differences to PDE5. Int J Impot Res. 2004;16(Suppl 1):S28–33. doi: 10.1038/sj.ijir.3901212. [DOI] [PubMed] [Google Scholar]
- 3.Fu Y, Yau KW. Phototransduction in mouse rods and cones. Pflugers Arch. 2007;454(5):805–19. doi: 10.1007/s00424-006-0194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deterre P, Bigay J, Forquet F, Robert M, Chabre M. cGMP phosphodiesterase of retinal rods is regulated by two inhibitory subunits. Proc Natl Acad Sci USA. 1988;85(8):2424–8. doi: 10.1073/pnas.85.8.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLaughlin ME, Ehrhart TL, Berson EL, Dryja TP. Mutation spectrum of the gene encoding the beta subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. Proc Natl Acad Sci USA. 1995;92(8):3249–53. doi: 10.1073/pnas.92.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dryja TP, Rucinski DE, Chen SH, Berson EL. Frequency of mutations in the gene encoding the alpha subunit of rod cGMP-phosphodiesterase in autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1999;40(8):1859–65. [PubMed] [Google Scholar]
- 7.Gal A, Orth U, Baehr W, Schwinger E, Rosenberg T. Heterozygous missense mutation in the rod cGMP phosphodiesterase beta-subunit gene in autosomal dominant stationary night blindness. Nat Genet. 1994;7(1):64–8. doi: 10.1038/ng0594-64. [DOI] [PubMed] [Google Scholar]
- 8.Chang B, Grau T, Dangel S, Hurd R, Jurklies B, Sener EC, Andreasson S, Dollfus H, Baumann B, Bolz S, Artemyev N, Kohl S, Heckenlively J, Wissinger B. A homologous genetic basis of the murine cpfl1 mutant and human achromatopsia linked to mutations in the PDE6C gene. Proc Natl Acad Sci USA. 2009;106(46):19581–6. doi: 10.1073/pnas.0907720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiadens AA, den Hollander AI, Roosing S, Nabuurs SB, Zekveld-Vroon RC, Collin RW, De Baere E, Koenekoop RK, van Schooneveld MJ, Strom TM, van Lith-Verhoeven JJ, Lotery AJ, van Moll-Ramirez N, Leroy BP, van den Born LI, Hoyng CB, Cremers FP, Klaver CC. Homozygosity mapping reveals PDE6C mutations in patients with early-onset cone photoreceptor disorders. Am J Hum Genet. 2009;85(2):240–7. doi: 10.1016/j.ajhg.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grau T, Artemyev NO, Rosenberg T, Dollfus H, Haugen OH, Cumhur Sener E, Jurklies B, Andreasson S, Kernstock C, Larsen M, Zrenner E, Wissinger B, Kohl S. Decreased catalytic activity and altered activation properties of PDE6C mutants associated with autosomal recessive achromatopsia. Hum Mol Genet. 2011;20(4):719–30. doi: 10.1093/hmg/ddq517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farber DB, Lolley RN. Cyclic guanosine monophosphate: elevation in degenerating photoreceptor cells of the C3H mouse retina. Science. 1974;186(4162):449–51. doi: 10.1126/science.186.4162.449. [DOI] [PubMed] [Google Scholar]
- 12.Doonan F, Donovan M, Cotter TG. Activation of multiple pathways during photoreceptor apoptosis in the rd mouse. Invest Ophthalmol Vis Sci. 2005;46(10):3530–8. doi: 10.1167/iovs.05-0248. [DOI] [PubMed] [Google Scholar]
- 13.Paquet-Durand F, Beck S, Michalakis S, Goldmann T, Huber G, Muhlfriedel R, Trifunovic D, Fischer MD, Fahl E, Duetsch G, Becirovic E, Wolfrum U, van Veen T, Biel M, Tanimoto N, Seeliger MW. A key role for cyclic nucleotide gated (CNG) channels in cGMP-related retinitis pigmentosa. Hum Mol Genet. 2011;20(5):941–7. doi: 10.1093/hmg/ddq539. [DOI] [PubMed] [Google Scholar]
- 14.Paquet-Durand F, Hauck SM, van Veen T, Ueffing M, Ekstrom P. PKG activity causes photoreceptor cell death in two retinitis pigmentosa models. J Neurochem. 2009;108(3):796–810. doi: 10.1111/j.1471-4159.2008.05822.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang T, Tsang SH, Chen J. Two pathways of rod photoreceptor cell death induced by elevated cGMP. HumMol Genet. 2017 doi: 10.1093/hmg/ddx121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin N, Baehr W. Expression and mutagenesis of mouse rod photoreceptor cGMP phosphodiesterase. J Biol Chem. 1994;269(5):3265–71. [PubMed] [Google Scholar]
- 17.Granovsky AE, Natochin M, McEntaffer RL, Haik TL, Francis SH, Corbin JD, Artemyev NO. Probing domain functions of chimeric PDE6alpha’/PDE5 cGMP-phosphodiesterase. The J Biol Chem. 1998;273(38):24485–90. doi: 10.1074/jbc.273.38.24485. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, Bulgakov OV, Wen XH, Woodruff ML, Pawlyk B, Yang J, Fain GL, Sandberg MA, Makino CL, Li T. AIPL1, the protein that is defective in Leber congenital amaurosis, is essential for the biosynthesis of retinal rod cGMP phosphodiesterase. Proc Natl Acad Sci USA. 2004;101(38):13903–8. doi: 10.1073/pnas.0405160101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muradov H, Boyd KK, Artemyev NO. Analysis of PDE6 function using chimeric PDE5/6 catalytic domains. Vision Res. 2006;46(6–7):860–8. doi: 10.1016/j.visres.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Cahill KB, Quade JH, Carleton KL, Cote RH. Identification of amino acid residues responsible for the selectivity of tadalafil binding to two closely related phosphodiesterases, PDE5 and PDE6. J Biol Chem. 2012;287(49):41406–16. doi: 10.1074/jbc.M112.389189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granovsky AE, Artemyev NO. Partial reconstitution of photoreceptor cGMP phosphodiesterase characteristics in cGMP phosphodiesterase-5. J Biol Chem. 2001;276(24):21698–703. doi: 10.1074/jbc.M100626200. [DOI] [PubMed] [Google Scholar]
- 22.Muradov H, Boyd KK, Haeri M, Kerov V, Knox BE, Artemyev NO. Characterization of human cone phosphodiesterase-6 ectopically expressed in Xenopus laevis rods. J Biol Chem. 2009;284(47):32662–9. doi: 10.1074/jbc.M109.049916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muradov H, Boyd KK, Artemyev NO. Rod phosphodiesterase-6 PDE6A and PDE6B subunits are enzymatically equivalent. J Biol Chem. 2010;285(51):39828–34. doi: 10.1074/jbc.M110.170068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheguru P, Zhang Z, Artemyev NO. The GAFa domain of phosphodiesterase-6 contains a rod outer segment localization signal. J Neurochem. 2014;129(2):256–63. doi: 10.1111/jnc.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gopalakrishna KN, Boyd K, Yadav RP, Artemyev NO. Aryl Hydrocarbon Receptor-interacting Protein-like 1 Is an Obligate Chaperone of Phosphodiesterase 6 and Is Assisted by the gamma-Subunit of Its Client. J Biol Chem. 2016;291(31):16282–91. doi: 10.1074/jbc.M116.737593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majumder A, Gopalakrishna KN, Cheguru P, Gakhar L, Artemyev NO. Interaction of aryl hydrocarbon receptor-interacting protein-like 1 with the farnesyl moiety. J Biol Chem. 2013;288(29):21320–8. doi: 10.1074/jbc.M113.476242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majumder A, Pahlberg J, Muradov H, Boyd KK, Sampath AP, Artemyev NO. Exchange of Cone for Rod Phosphodiesterase 6 Catalytic Subunits in Rod Photoreceptors Mimics in Part Features of Light Adaptation. J Neurosci. 2015;35(24):9225–35. doi: 10.1523/JNEUROSCI.3563-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gopalakrishna KN, Doddapuneni K, Boyd KK, Masuho I, Martemyanov KA, Artemyev NO. Interaction of transducin with uncoordinated 119 protein (UNC119): implications for the model of transducin trafficking in rod photoreceptors. J Biol Chem. 2011;286(33):28954–62. doi: 10.1074/jbc.M111.268821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M, Casey PJ. Protein prenylation: unique fats make their mark on biology. Nat Rev Mol Cell Biol. 2016;17(2):110–22. doi: 10.1038/nrm.2015.11. [DOI] [PubMed] [Google Scholar]
- 30.Anant JS, Ong OC, Xie HY, Clarke S, O’Brien PJ, Fung BK. In vivo differential prenylation of retinal cyclic GMP phosphodiesterase catalytic subunits. J Biol Chem. 1992;267(2):687–90. [PubMed] [Google Scholar]
- 31.Catty P, Pfister C, Bruckert F, Deterre P. The cGMP phosphodiesterase-transducin complex of retinal rods. Membrane binding and subunits interactions. J Biol Chem. 1992;267(27):19489–93. [PubMed] [Google Scholar]
- 32.Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 33.Zeng-Elmore X, Gao XZ, Pellarin R, Schneidman-Duhovny D, Zhang XJ, Kozacka KA, Tang Y, Sali A, Chalkley RJ, Cote RH, Chu F. Molecular architecture of photoreceptor phosphodiesterase elucidated by chemical cross-linking and integrative modeling. JMol Biol. 2014;426(22):3713–28. doi: 10.1016/j.jmb.2014.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Granovsky AE, Natochin M, Artemyev NO. The gamma subunit of rod cGMP-phosphodiesterase blocks the enzyme catalytic site. J Biol Chem. 1997;272(18):11686–9. doi: 10.1074/jbc.272.18.11686. [DOI] [PubMed] [Google Scholar]
- 35.Barren B, Gakhar L, Muradov H, Boyd KK, Ramaswamy S, Artemyev NO. Structural basis of phosphodiesterase 6 inhibition by the C-terminal region of the gamma-subunit. EMBO J. 2009;28(22):3613–22. doi: 10.1038/emboj.2009.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, He F, Constantine R, Baker ML, Baehr W, Schmid MF, Wensel TG, Agosto MA. Domain Organization and Conformational Plasticity of the G Protein Effector, PDE6. J Biol Chem. 2015;290(20):12833–43. doi: 10.1074/jbc.M115.647636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo LW, Muradov H, Hajipour AR, Sievert MK, Artemyev NO, Ruoho AE. The inhibitory gamma subunit of the rod cGMP phosphodiesterase binds the catalytic subunits in an extended linear structure. J Biol Chem. 2006;281(22):15412–22. doi: 10.1074/jbc.M600595200. [DOI] [PubMed] [Google Scholar]
- 38.Tsang SH, Woodruff ML, Jun L, Mahajan V, Yamashita CK, Pedersen R, Lin CS, Goff SP, Rosenberg T, Larsen M, Farber DB, Nusinowitz S. Transgenic mice carrying the H258N mutation in the gene encoding the beta-subunit of phosphodiesterase-6 (PDE6B) provide a model for human congenital stationary night blindness. Hum Mutat. 2007;28(3):243–54. doi: 10.1002/humu.20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramamurthy V, Roberts M, van den Akker F, Niemi G, Reh TA, Hurley JB. AIPL1, a protein implicated in Leber’s congenital amaurosis, interacts with and aids in processing of farnesylated proteins. Proc Natl Acad Sci USA. 2003;100(22):12630–5. doi: 10.1073/pnas.2134194100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillespie PG, Prusti RK, Apel ED, Beavo JA. A soluble form of bovine rod photoreceptor phosphodiesterase has a novel 15-kDa subunit. J Biol Chem. 1989;264(21):12187–93. [PubMed] [Google Scholar]
- 41.Norton AW, Hosier S, Terew JM, Li N, Dhingra A, Vardi N, Baehr W, Cote RH. Evaluation of the 17-kDa prenyl-binding protein as a regulatory protein for phototransduction in retinal photoreceptors. J Biol Chem. 2005;280(2):1248–56. doi: 10.1074/jbc.M410475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, Li S, Doan T, Rieke F, Detwiler PB, Frederick JM, Baehr W. Deletion of PrBP/delta impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments. Proc Natl Acad Sci USA. 2007;104(21):8857–62. doi: 10.1073/pnas.0701681104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baehr W. Membrane protein transport in photoreceptors: the function of PDEdelta: the Proctor lecture. Invest Ophthalmol Vis Sci. 2014;55(12):8653–66. doi: 10.1167/iovs.14-16066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fansa EK, O’Reilly NJ, Ismail S, Wittinghofer A. The N- and C-terminal ends of RPGR can bind to PDE6delta. EMBO Rep. 2015;16(12):1583–5. doi: 10.15252/embr.201541404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fansa EK, Kosling SK, Zent E, Wittinghofer A, Ismail S. PDE6delta-mediated sorting of INPP5E into the cilium is determined by cargo-carrier affinity. Nat Commun. 2016;7:11366. doi: 10.1038/ncomms11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yadav RP, Majumder A, Gakhar L, Artemyev NO. Extended conformation of the proline-rich domain of human aryl hydrocarbon receptor-interacting protein-like 1: implications for retina disease. J Neurochem. 2015;135(1):165–75. doi: 10.1111/jnc.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]