Abstract
Objectives
To improve the fermentation production of transglutaminase (TGase) from Streptomyces mobaraensis for applications in the food industry, the atmospheric and room-temperature plasma (ARTP) mutagenesis was applied to breed S. mobaraensis mutants with increased TGase production.
Results
After eight rounds of iterative ARTP mutagenesis, four genetically stable mutants, Sm5-V1, Sm6-V13, Sm2-V10, and Sm7-V12, were identified, which showed increased TGase production by 27, 24, 24, and 19%, respectively. The best mutant Sm5-V1 exhibited a maximum TGase activity of 5.85 U/mL during flask fermentation. Compared to the wild-type strain, the transcription levels of the zymogen TGase genes in the mutants increased significantly as indicated by quantitative real-time PCR, while the gene nucleotide sequences of the mutants did not change at all. It was shown that the overexpression of TGase zymogen gene in the mutants contributes to the increase in TGase production.
Conclusions
ARTP is a potentially efficient tool for microbial mutation breeding to bring some significant changes required for the industrial applications.
Electronic supplementary material
The online version of this article (doi:10.1186/s40643-017-0168-2) contains supplementary material, which is available to authorized users.
Keywords: Atmospheric and room-temperature plasma, Breeding, Streptomyces mobaraensis, Transcription level, Transglutaminase
Background
Transglutaminases (TGase, EC 2.3.2.13), also referred to protein-glutamine γ-glutamyltransferases, are enzymes capable of catalyzing acyl-transfer reactions between the γ-carboxamide group of protein or peptide-bound glutamine and ε-amino group of lysine or other primary amines (Zhu et al. 1995; Martins et al. 2014). The covalent modifications of proteins promoted by TGase facilitate extensive applications in food, medicine, and other industries. TGases are widely distributed in prokaryotes and eukaryotes. TGases from animal tissues are Ca2+-dependent enzymes that lead to the precipitation of proteins from food containing casein, soybean globulin, or myosin (Martins et al. 2014). However, the scarcity of resources as well as complexity of further extraction and purification still limits their applications. Microbial transglutaminases (MTG) that were usually found in Streptomyces (Duran et al. 1998; Marx et al. 2008) and Bacillus (Kobayashi et al. 1998; Soares et al. 2003) species have a wider application due to their advantages as Ca2+-independent activity, thermostability, and broad specificity for acyl donors (Salis et al. 2015). Up to now, MTG for applications in the food processing were mainly produced by fermentation of Streptomyces mobaraensis (Yokoyama et al. 2004). TGase from S. mobaraensis was known to be secreted as a zymogen and then activated by proteolytic processing to the enzymatically active mature form (Zotzel et al. 2003a, b). Its pro-region is essential for efficient protein folding, secretion, and suppression of the enzymatic activity (Yurimoto et al. 2004). Considering the fact that genetically engineered strains are somehow restricted in the food industry, it is more feasible to improve TGase production by mutation breeding.
Microbial mutation breeding by altering the genomes shows great significance for biotechnology researches and applications (Tan et al. 2014; Kumar 2015). Recently, a novel and efficient mutation tool called atmospheric and room-temperature plasma (ARTP) has been successfully employed to cause some significant changes in enzyme activity, biochemical productivity, and metabolism without lethal damages (Guo et al. 2011; Lu et al. 2011; Xu et al. 2012; Wang et al. 2014). Compared to the conventional mutation breeding methods, ARTP mutagenesis shows some distinct advantages, such as low costs, low and controllable plasma temperatures, various active chemical species with a high density, rapid mutation, flexible and secure operations (Laroussi 2005; Zhang et al. 2014, 2015).
In this study, the iterative ARTP mutagenesis was applied to S. mobaraensis for breeding mutants with increased TGase production. Protocols for iterative rapid mutation of S. mobaraensis with helium-driven ARTP system and effective tube screening method of the mutants were established. The transcription levels and the nucleotide sequences of the gene (pro-smtg) encoding TGase zymogen were compared between the mutants and the wild-type strain to explain the reasons for the enhanced TGase production, which may provide a valuable guidance for further researches on mechanisms regarding ARTP effects on the whole cells and the intracellular bio-macromolecules.
Methods
Materials
l-glutamic acid γ-monohydroxamate was purchased from Sigma-Aldrich Co., Ltd. (Shanghai, China). N-α-Carbobenzoxy-l-glutaminyl-glycine (Nα-CBZ-Gln-Gly) was purchased from Civi Chemical Technology Co., Ltd. (Shanghai, China). Reduced glutathione was purchased from Aladdin Reagent Co., Ltd. (Shanghai, China). All-in-One First-Strand cDNA Synthesis SuperMix (One-Step gDNA Removal) and TransStart Tip Green qPCR SuperMix that were used for RT-PCR (Reverse Transcription PCR) and qRT-PCR (quantitative Real-Time PCR) were purchased from TransBionovo Co., Ltd. (Beijing, China). All other reagents were obtained from commercial sources and were of analytical grade.
Strains and media
Escherichia coli DH5α was used as the host strain for the recombinant DNA manipulations. Streptomyces mobaraensis, the wild-type strain (S. mobaraensis ECU7480, stored in our lab) and the mutants generated by ARTP mutagenesis, were grown on a solid medium comprising 20 g/L soluble starch, 3 g/L tryptone, 1 g/L KNO3, 0.5 g/L K2HPO4·3H2O, 0.5 g/L MgSO4·7H2O, 0.5 g/L NaCl, 0.01 g/L FeSO4·7H2O, and 1.5–2.0% (w/v) agar at pH 7.4–7.6 and 30 °C for 5 days. Both tube and flask fermentations were conducted using a seeding medium (20 g/L glycerol, 20 g/L tryptone, 5 g/L yeast extract, 2 g/L MgSO4·7H2O, 2.62 g/L K2HPO4·3H2O, 2 g/L KH2PO4 , pH 7.0) at 30 °C for 24 h to prepare the inoculums and a fermentation medium (20 g/L glycerol, 20 g/L tryptone, 5.5 g/L corn steep powder, 5 g/L yeast extract, 2 g/L MgSO4, 2.62 g/L K2HPO4·3H2O, 10 g/L CaCO3 , pH 7.2) at 30 °C with 8–10% inoculum size for the production of TGase.
Procedures for iterative mutagenesis with ARTP and directed screening
The procedures for iterative mutation of S. mobaraensis genome with ARTP mutation breeding system purchased from Si Qing Yuan Biotechnology Co., Ltd. (Beijing, China) and the following screening are shown in Fig. 1. In this study, pure helium was used as the working gas at a flow rate of 10 slpm (standard liters per minute). The RF power input was 40 W. The distance (D) between the plasma torch nozzle exit and the sample plate was 4 mm and the plasma jet temperature was below 30 °C. To provide different dosages of the active species in the plasma jet region, 10 μL of the spore suspension was pipetted onto the stainless minidisk and then exposed to ARTP jet for different treatment times ranging from 10 to 60 s. Spores without treatment were used as the control.
Fig. 1.
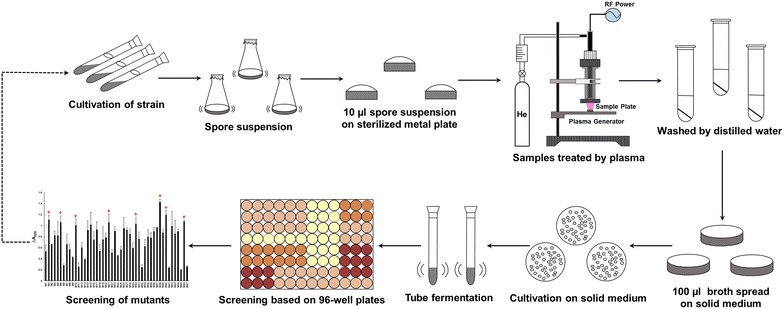
Scheme of ARTP mutagenesis for microbial breeding
After ARTP mutation, the spore suspension was transferred onto solid medium. Single colonies from the solid medium were selected randomly and inoculated into a test tube containing 3 mL fermentation medium for 96 h fermentation. The cell-free supernatant of fermentation broth was withdrawn for TGase activity detection. The top eight mutants with increased TGase production were chosen as the starting strains for the next round of ARTP mutagenesis. After eight rounds of iterative ARTP mutagenesis, flask cultivation was performed to verify the TGase production of selected mutants from shake tube screening. The selected mutants grown on the solid medium were cultivated in 250-mL flasks containing 25 mL seeding medium at 30 °C for 24 h. Then, 2.5 mL of the seeding culture was transferred into 250-mL shake flasks containing 25 mL fermentation medium and cultured at 30 °C. Aliquots of the cell-free supernatant were taken at different time intervals for examination of the TGase activity. Additionally, the genetic stability of the identified mutants was evaluated by eight rounds of subcultures. The TGase production of the mutants was examined by shake flask fermentation in every two subcultures.
Evaluation of ARTP mutagenesis of S. mobaraensis
The lethal rates of the spores under different treatment times were evaluated according to Eq. 1. The mutation rate and the positive mutation rate were calculated based on Eqs. 2 and 3, respectively.
| 1 |
| 2 |
| 3 |
where U is the total colony count of the sample without treatment, T is the total colony count after treatment with ARTP, M is the total colony count of mutants with different TGase production from the wild-type strain, and P is the total colony count of mutants with increased TGase production than that of the wild-type strain. All the colony numbers were obtained by the colony forming unit (CFU) method on a solid medium.
Activity assay of TGase
The activity of TGase was measured according to the colorimetric hydroxamate procedure (Grossowicz et al. 1950) using Nα-CBZ-Gln-Gly as the substrate. The calibration curve was prepared using l-glutamic acid γ-monohydroxamic acid as a standard. One unit (U) of TGase activity was defined as the amount of enzyme that catalyzes the formation of 1 μmol l-glutamic acid γ-monohydroxamate per minute at 37 °C.
Quantitative real-time PCR analysis
Total RNAs of S. mobaraensis ECU7480 and its mutants were extracted after cultivation for different times. The mycelium pellets were collected by centrifugation and then frozen in liquid nitrogen immediately. The extraction of total RNAs was performed using the SV Total RNA Isolation System (Promega, USA) according to the manufacturer’s protocol. The quantity and quality of the isolated RNAs were examined by NanoDrop 2000c UV–Vis spectrophotometer (Thermo Scientific, USA) and agarose gel electrophoresis. Subsequently, the reverse transcription PCR (RT-PCR) was carried out using 1 μg total RNA as the template. The gene transcription level of pro-smtg during fermentation process was investigated by quantitative real-time PCR (qRT-PCR) with primers listed in Table 1. The qRT-PCR reaction consisted of an initial denaturation at 94 °C and 40 amplifications cycles of 5 s at 94 °C and 60 s at 64 °C. The target gene transcription level was normalized internally to that of 16S rRNA gene for its transcription during overall growth stages was relatively stable.
Table 1.
Primers used in this study
| Name | Sequences (5′–3′) | Application |
|---|---|---|
| 16S rRNA_L1 | AGCAGCGGAGCATGTGGCTT | 16S rRNA gene transcription |
| 16S rRNA_R1 | TGCGCTCGTTGCGGGACTTA | |
| pro-smtg-L1 | CATGTCGAGGGACAGGAACA | pro-smtg gene transcription |
| pro-smtg-R1 | TTGCGGAACTTGCTCTCGTA | |
| pro-smtg-FP | CGGAATTCATGCCGTCCGCAGGC | pro-smtg gene amplification |
| pro-smtg-RP | CCCAAGCTTTCACGGCCAGCCCTG |
Cloning of pro-smtg gene from S. mobaraensis mutants
Genomic DNAs were obtained from S. mobaraensis mutants and the wild-type strain. The primers used for the PCR are listed in Table 1. The PCR procedure was set as follows: (95 °C, 5 min) 1 cycle; (94 °C, 1 min; 65 °C, 30 s; 72 °C, 90 s) 30 cycles; and (72 °C, 10 min) 1 cycle. The amplified PCR products (1200 bp) were ligated into plasmid pMD-19T and sequenced for alignment to that of the wild-type strain.
Results and discussion
Mutation and screening of the mutants
A well-controlled lethal rate is fundamental for effective mutation and screening of the mutants. The lethal rates of S. mobaraensis with respect to various treatment times are shown in Fig. 2, which indicated that the lethal rate increased to 66.5, 93.1, and 99.2%, respectively, after treated for 30, 40, and 50 s. When the samples were treated for 50 s or even longer, no spores could survive. According to previous reports (Guo et al. 2011; Hua et al. 2010), a lethal rate of 90% was considered appropriate. Besides, keeping the lethal rate high is necessary for the effective mutation and selection of mutant strains. Therefore, the ARTP treatment time applied in this study was determined as 40 s.
Fig. 2.
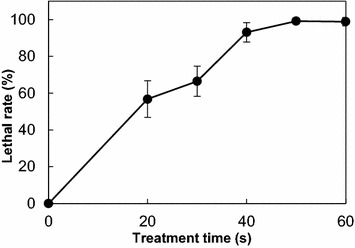
Lethal rate of S. mobaraensis by ARTP mutagenesis
After eight rounds of iterative ARTP mutagenesis, 501 mutants in total were selected for tube-fermentation screening, as shown in Additional file 1: Figure S1. To investigate the accumulative effect on TGase production by iterative ARTP mutagenesis, the mutation and screening results for each round were collected and compared. The statistic results shown in Table 2 and Fig. 3 indicated that the proportion of positive mutants was scaled up with the increase of iterative rounds. Moreover, the TGase production of the best mutant for each round presented a fluctuant improving trend. Thus, the increase of iterative rounds might exhibit an accumulative effect on TGase production of the mutants.
Table 2.
Mutation and screening results for each round of ARTP mutagenesis
| Iterative round | R M (%) | R P (%) | Average of mutants |
|---|---|---|---|
| WT | – | – | 0.56 ± 0.08 |
| 1 | 32.1 | 6.4 | 0.44 ± 0.19 |
| 2 | 48.8 | 36.6 | 0.69 ± 0.33 |
| 3 | 45.7 | 32.6 | 0.71 ± 0.35 |
| 4 | 34.8 | 19.1 | 0.58 ± 0.23 |
| 5 | 56.8 | 54.6 | 0.81 ± 0.31 |
| 6 | 62.5 | 50.0 | 0.75 ± 0.28 |
| 7 | 74.5 | 68.1 | 0.98 ± 0.41 |
| 8 | 69.6 | 52.2 | 0.83 ± 0.43 |
Fig. 3.
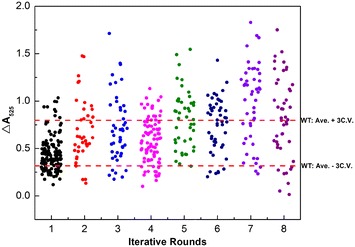
Distribution of screening results for each round of ARTP mutagenesis. , which is proportional to the TGase activity
TGase production of the mutants
Shake flask fermentation was conducted to verify the potentially positive mutants identified in the tube fermentation. Finally, four mutants, Sm5-V1, Sm6-V13, Sm2-V10, and Sm7-V12, were identified in the ARTP breeding process, and the TGase production of these mutants is shown in Table 3. The highest TGase production reached 5.85 U/mL, which represented a 27% increase as compared with the wild-type strain.
Table 3.
TGase production of S. mobaraensis wild-type and ARTP mutants
| Strain | TGase titer (U/mL) | Relative production (%) |
|---|---|---|
| WT | 4.60 ± 0.17 | 100 ± 4 |
| Sm5-V1 | 5.85 ± 0.21 | 127 ± 5 |
| Sm6-V13 | 5.72 ± 0.16 | 124 ± 4 |
| Sm2-V10 | 5.68 ± 0.19 | 124 ± 4 |
| Sm7-V12 | 5.49 ± 0.13 | 119 ± 3 |
The genetic stability is one of the key performance factors for a promising industrial strain because it reflects the potential mutations at the gene level (Ren et al. 2017). The genetic stability test results shown in Additional file 1:Figure S2 indicated that the TGase production of the identified mutants still remained stable after eight rounds of subcultures. These results proved that ARTP mutagenesis is a promising mutation breeding tool for industrial applications.
Pro-smtg gene expression level of the wild-type and mutant strains
The enhanced TGase production caused by ARTP mutagenesis may be attributed to two effects. A direct effect will happen if the structural gene (pro-smtg) of TGase zymogen is altered, leading to the change in the specific activity of the protein. Indirect effect happens when the relevant genes regulating the expression of TGase zymogen are altered.
Firstly, the pro-smtg gene nucleotide sequences of the four mutants showed no differences from the original sequence of the native enzyme from the wild-type strain, as indicated by gene cloning and sequence alignment shown in Additional file 1: Figure S3. However, qRT-PCR results shown in Fig. 4 indicated that the pro-smtg expression levels of the four mutants were remarkably improved as compared to that of the parental strain, which might lead to the increase in TGase production. The analysis from transcription level conducted in this study is of great importance for further investigation of metabolic changes caused by ARTP mutagenesis.
Fig. 4.
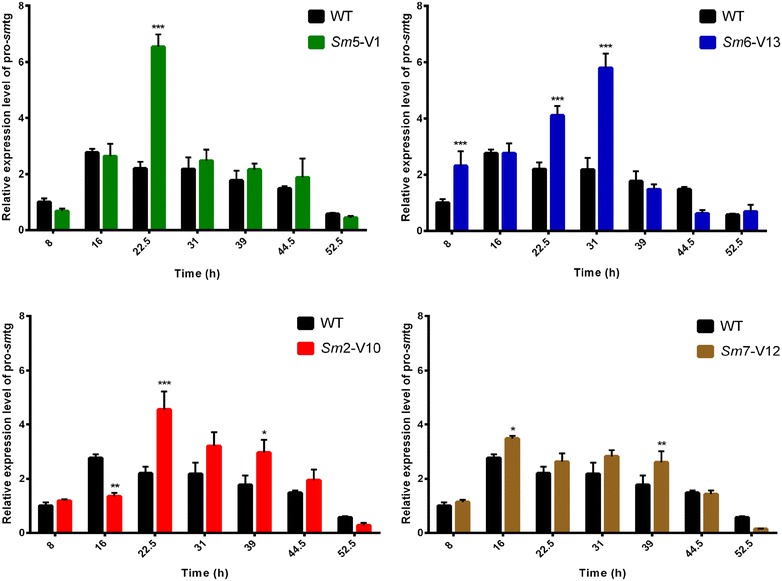
Transcription analysis of pro-smtg gene by qRT-PCR. The pro-smtg transcription levels were detected after flask fermentation for different times in wild-type strain (WT) and mutants Sm5-V1, Sm6-V13, Sm2-V10, and Sm7-V12, which were normalized to that of 16S rRNA gene by the method. The experiments were performed in three replications
Conclusions
Iterative ARTP mutagenesis was applied to improve the TGase production in S. mobaraensis as a novel and efficient mutation tool. As a result, four mutants, Sm5-V1, Sm6-V13, Sm2-V10, and Sm7-V12, with increased TGase production by 27, 24, 24, and 19%, respectively, were identified after eight rounds of iterative mutagenesis. The best mutant Sm5-V1 showed an enhanced TGase production of 5.85 U/mL. In addition, the results of sequence alignment and qRT-PCR revealed that the expression of TGase zymogen gene increased obviously under the ARTP treatment, while the gene sequence remained unchanged. The analysis of the target gene sequence as well as transcription level conducted in this study would provide a valuable guidance for further researches and applications of ARTP mutagenesis.
Authors’ contributions
YJ wrote the experiment; YJ and CZ conducted the experiment; YS and HL helped to revise the manuscript; YJ, CL, and JX conceived the research; JP and YB provided experimental materials and guidance. All authors read and approved the final manuscript.
Acknowledgements
The authors are thankful to the Prof. Yonghong Wang’s research group for providing the ARTP mutation breeding system as well as the operative skills to perform this research work.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential competing interest.
Availability of data and materials
All data generated or analyzed during this study are included in the main manuscript file.
Consent for publication
The authors approved the consent for publishing the manuscript.
Ethics approval and consent to participate
All the authors have read and agreed the ethics for publishing the manuscript.
Funding
This work is financially sponsored by the National Natural Science Foundation of China (No. 21536004), Shanghai Commission of Science and Technology (No. 15JC1400403), and Shanghai Pujiang Program (No. 15PJ1401200).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ARTP
atmospheric and room-temperature plasma
- TGase
transglutaminase
- MTG
microbial transglutaminases
- RT-PCR
reverse transcription PCR
- qRT-PCR
quantitative real-time PCR
Additional file
Additional file 1. Additional information.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s40643-017-0168-2) contains supplementary material, which is available to authorized users.
Contributor Information
Ying Jiang, Email: jiangyingzt@126.com.
Yue-Peng Shang, Email: shangyuepeng415@163.com.
Hao Li, Email: lihao59@sina.com.
Chao Zhang, Email: chaozhangecust@163.com.
Jiang Pan, Email: panjiang@ecust.edu.cn.
Yun-Peng Bai, Email: ybai@ecust.edu.cn.
Chun-Xiu Li, Phone: +86-21-6425-2498, Email: chunxiuli@ecust.edu.cn.
Jian-He Xu, Phone: +86-21-6425-2498, Email: jianhexu@ecust.edu.cn.
References
- Duran R, Junqua M, Schmitter JM, Gancet C, Goulas P. Purification, characterisation, and gene cloning of transglutaminase from Streptoverticillium cinnamoneum CBS 683.68. Biochimie. 1998;80:313–319. doi: 10.1016/S0300-9084(98)80073-4. [DOI] [PubMed] [Google Scholar]
- Grossowicz N, Wainfan E, Borek E, Waelsch H. The enzymatic formation of hydroxamic acids from glutamine and asparagine. J Biol Chem. 1950;187:111–125. [PubMed] [Google Scholar]
- Guo T, Tang Y, Xi YL, He AY, Sun BJ, Wu H, Liang DF, Jiang M, Ouyang PK. Clostridium beijerinckii mutant obtained by atmospheric pressure glow discharge producing high proportions of butanol and solvent yields. Biotechnol Lett. 2011;33:2379–2383. doi: 10.1007/s10529-011-0702-9. [DOI] [PubMed] [Google Scholar]
- Hua XF, Wang J, Wu ZJ, Zhang HX, Li HP, Xing XH, Liu Z. A salt tolerant Enterobacter cloacae mutant for bioaugmentation of petroleum- and salt-contaminated soil. Biochem Eng J. 2010;49:201–206. doi: 10.1016/j.bej.2009.12.014. [DOI] [Google Scholar]
- Kobayashi K, Suzuki SI, Izawa Y, Yokozeki K, Miwa K, Yamanaka S. Transglutaminase in sporulating cells of Bacillus subtilis. J Gen Appl Microbiol. 1998;44:85–91. doi: 10.2323/jgam.44.85. [DOI] [PubMed] [Google Scholar]
- Kumar AK. UV mutagenesis treatment for improved production of endoglucanase and β-glucosidase from newly isolated thermotolerant actinomycetes, Streptomyces griseoaurantiacus. Bioresour Bioprocess. 2015;2:22. doi: 10.1186/s40643-015-0052-x. [DOI] [Google Scholar]
- Laroussi M. Low temperature plasma-based sterilization: overview and state-of-the-art. Plasma Process Polym. 2005;2:391–400. doi: 10.1002/ppap.200400078. [DOI] [Google Scholar]
- Lu Y, Wang LY, Ma K, Li G, Zhang C, Zhao HX, Lai QH, Li HP, Xing XH. Characteristics of hydrogen production of an Enterobacter aerogenes mutant generated by a new atmospheric and room temperature plasma (ARTP) Biochem Eng J. 2011;55:17–22. doi: 10.1016/j.bej.2011.02.020. [DOI] [Google Scholar]
- Martins IM, Matos M, Costa R, Silva F, Pascoal A, Estevinho LM, Choupina AB. Transglutaminase: recent achievements and new sources. Appl Microbiol Biotechnol. 2014;98:6957–6964. doi: 10.1007/s00253-014-5894-1. [DOI] [PubMed] [Google Scholar]
- Marx CK, Hertel TC, Pietzsch M. Purification and activation of a recombinant histidine-tagged pro-transglutaminase after soluble expression in Escherichia coli and partial characterization of the active enzyme. Enzyme Microb Technol. 2008;42:568–575. doi: 10.1016/j.enzmictec.2008.03.003. [DOI] [Google Scholar]
- Ren F, Chen L, Tong QY. Highly improved acarbose production of Actinomyces through the combination of ARTP and penicillin susceptible mutant screening. World J Microbiol Biotechnol. 2017;33:16. doi: 10.1007/s11274-016-2156-7. [DOI] [PubMed] [Google Scholar]
- Salis B, Spinetti G, Scaramuzza S, Bossi M, Jotti GS, Tonon G, Crobu D, Schrepfer R. High-level expression of a recombinant active microbial transglutaminase in Escherichia coli. BMC Biotechnol. 2015;15:84. doi: 10.1186/s12896-015-0202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares LHB, Assmann F, Ayub MAZ. Purification and properties of a transglutaminase produced by a Bacillus circulans strain isolated from the Amazon environment. Biotechnol Appl Biochem. 2003;37:295–299. doi: 10.1042/BA20020110. [DOI] [PubMed] [Google Scholar]
- Tan J, Chu J, Wang YH, Zhuang YP, Zhang SL. High-throughput system for screening of Monascus purpureus high-yield strain in pigment production. Bioresour Bioprocess. 2014;1:16. doi: 10.1186/s40643-014-0016-6. [DOI] [Google Scholar]
- Wang Q, Feng LR, Wei L, Li HG, Wang L, Zhou Y, Yu XB. Mutation breeding of lycopene-producing strain Blakeslea Trispora by a novel atmospheric and room temperature plasma (ARTP) Appl Biochem Biotechnol. 2014;174:452–460. doi: 10.1007/s12010-014-0998-8. [DOI] [PubMed] [Google Scholar]
- Xu F, Jin HJ, Li HM, Tao L, Wang JP, Lv J, Chen SF. Genome shuffling of Trichoderma viride for enhanced cellulase production. Ann Microbiol. 2012;62:509–515. doi: 10.1007/s13213-011-0284-8. [DOI] [Google Scholar]
- Yokoyama K, Nio N, Kikuchi Y. Properties and applications of microbial transglutaminase. Appl Microbiol Biotechnol. 2004;64:447–454. doi: 10.1007/s00253-003-1539-5. [DOI] [PubMed] [Google Scholar]
- Yurimoto H, Yamane M, Kikuchi Y, Matsui H, Kato N, Sakai Y. The pro-peptide of Streptomyces mobaraensis transglutaminase functions in cis and in trans to mediate efficient secretion of active enzyme from methylotrophic yeasts. Biosci Biotechnol Biochem. 2004;68:2058–2069. doi: 10.1271/bbb.68.2058. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang XF, Li HP, Wang LY, Zhang C, Xing XH, Bao CY. Atmospheric and room temperature plasma (ARTP) as a new powerful mutagenesis tool. Appl Microbiol Biotechnol. 2014;98:5387–5396. doi: 10.1007/s00253-014-5755-y. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang C, Zhou QQ, Zhang XF, Wang LY, Chang HB, Li HP, Oda Y, Xing XH. Quantitative evaluation of DNA damage and mutation rate by atmospheric and room-temperature plasma (ARTP) and conventional mutagenesis. Appl Microbiol Biotechnol. 2015;99:5639–5646. doi: 10.1007/s00253-015-6678-y. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Rinzema A, Tramper J, Bol J. Microbial transglutaminase—a review of its production and application in food processing. Appl Microbiol Biotechnol. 1995;44:277–282. doi: 10.1007/BF00169916. [DOI] [Google Scholar]
- Zotzel J, Keller P, Fuchsbauer HL. Transglutaminase from Streptomyces mobaraensis is activated by an endogenous metalloprotease. Eur J Biochem. 2003;270:3214–3222. doi: 10.1046/j.1432-1033.2003.03703.x. [DOI] [PubMed] [Google Scholar]
- Zotzel J, Pasternack R, Pelzer C, Ziegert D, Mainusch M, Fuchsbauer HL. Activated transglutaminase from Streptomyces mobaraensis is processed by a tripeptidyl aminopeptidase in the final step. Eur J Biochem. 2003;270:4149–4155. doi: 10.1046/j.1432-1033.2003.03809.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in the main manuscript file.


