Abstract
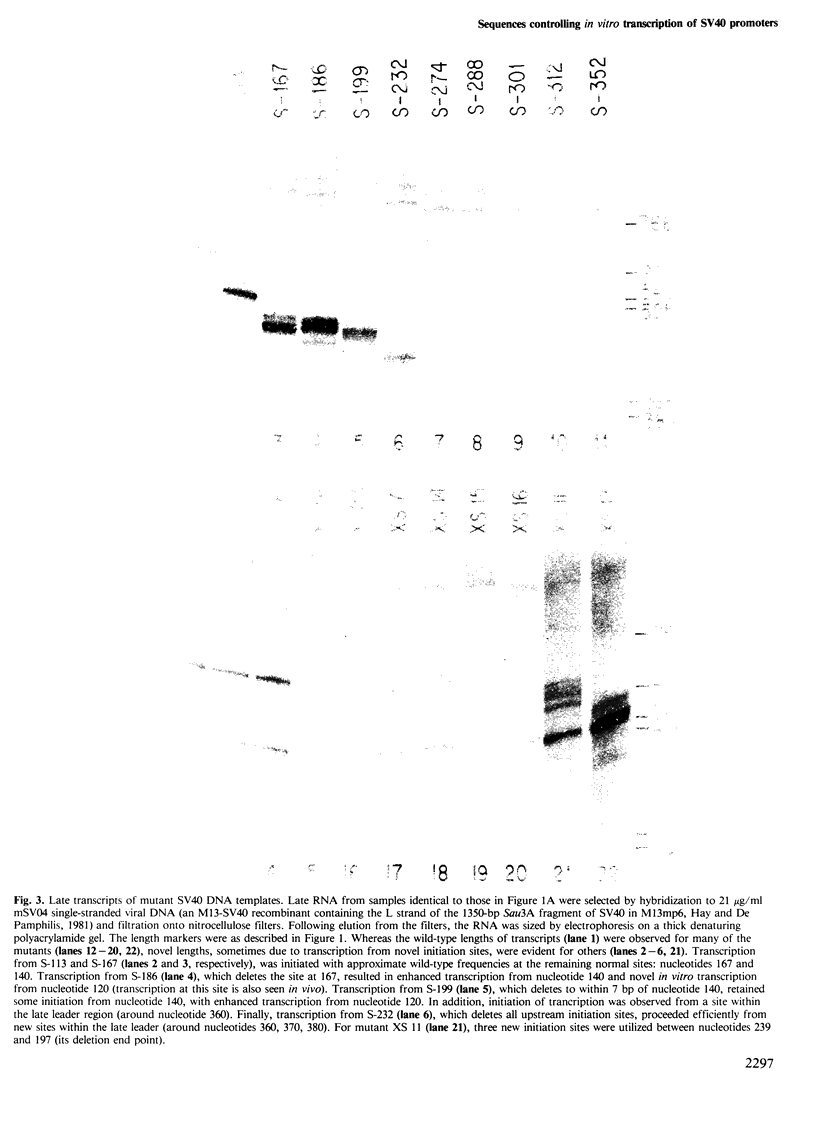
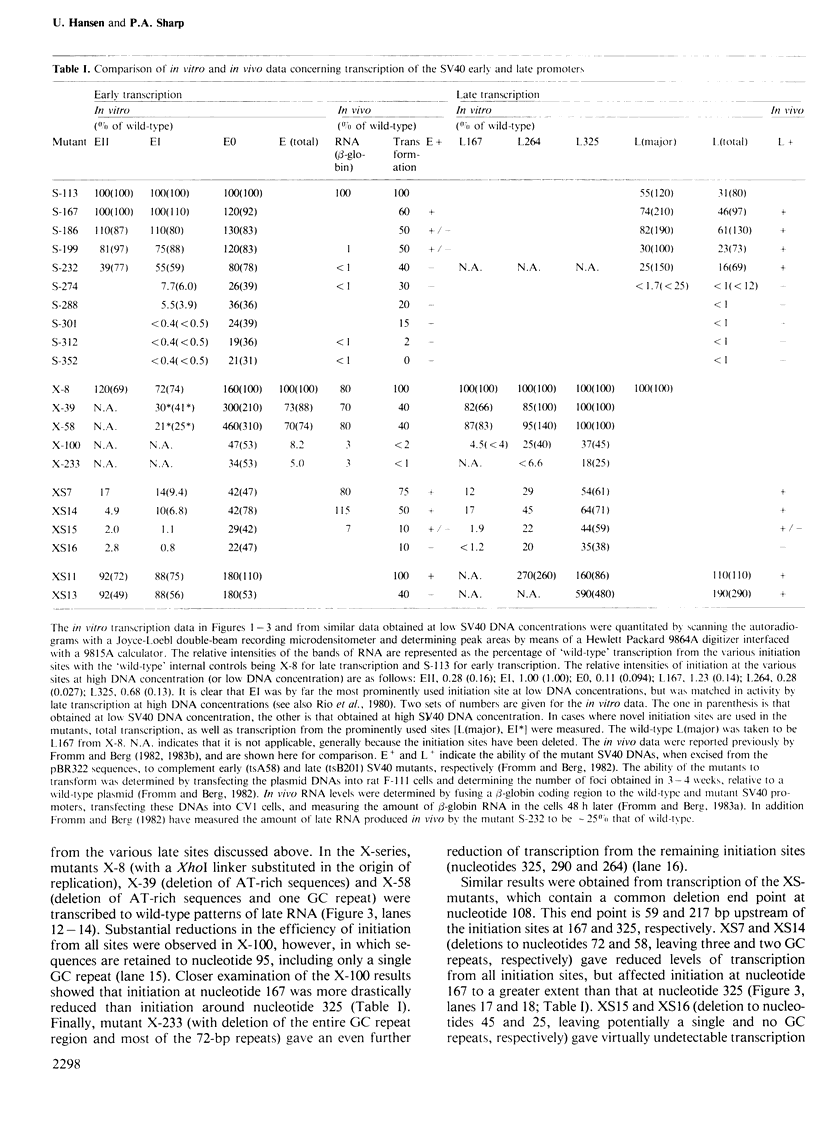
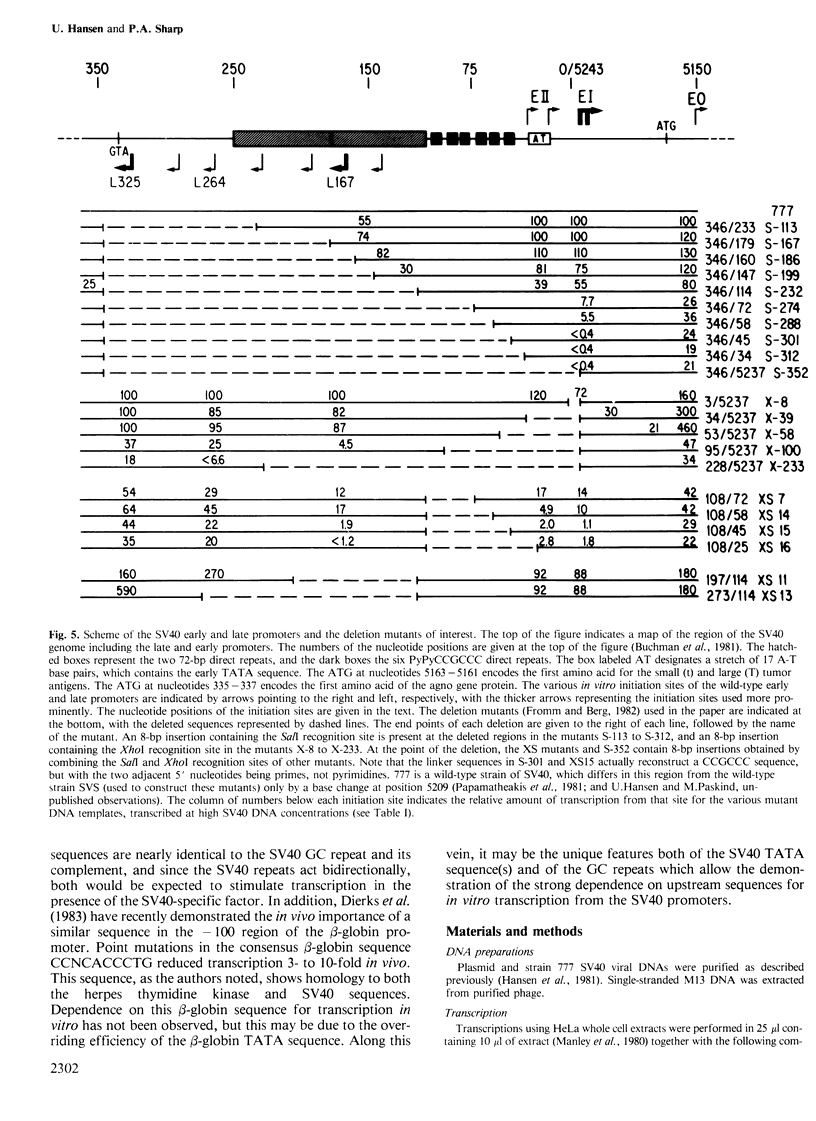
A series of deletion mutants of SV40 were tested for early and late promoter activity in vitro in a transcription extract prepared from HeLa cells. These mutants had previously been characterized for expression in vivo. Transcription in vitro from both the SV40 early and late promoters was strongly dependent on an upstream region of DNA that contains six direct GC repeats. Sequences spanning two or more of these repeats stimulated transcription in a bidirectional fashion, at distances of 50-200 bp. These sequences may function by mediating the activity of a specific transcriptional factor. Little effect on transcription in vitro was observed upon deletion of the 72-bp enhancer elements. With this exception, the sequence dependence of early and late transcription in vitro was similar to that observed previously in vivo, both of the region including the GC repeats and of the early TATA sequence.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Brady J., Radonovich M., Vodkin M., Natarajan V., Thoren M., Das G., Janik J., Salzman N. P. Site-specific base substitution and deletion mutations that enhance or suppress transcription of the SV40 major late RNA. Cell. 1982 Dec;31(3 Pt 2):625–633. doi: 10.1016/0092-8674(82)90318-x. [DOI] [PubMed] [Google Scholar]
- Byrne B. J., Davis M. S., Yamaguchi J., Bergsma D. J., Subramanian K. N. Definition of the simian virus 40 early promoter region and demonstration of a host range bias in the enhancement effect of the simian virus 40 72-base-pair repeat. Proc Natl Acad Sci U S A. 1983 Feb;80(3):721–725. doi: 10.1073/pnas.80.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras R., Fiers W. Initiation of transcription by RNA polymerase II in permeable, SV40-infected or noninfected, CVI cells; evidence for multiple promoters of SV40 late transcription. Nucleic Acids Res. 1981 Jan 24;9(2):215–236. doi: 10.1093/nar/9.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Cochran M. D., Dobkin C., Reiser J., Weissmann C. Three regions upstream from the cap site are required for efficient and accurate transcription of the rabbit beta-globin gene in mouse 3T6 cells. Cell. 1983 Mar;32(3):695–706. doi: 10.1016/0092-8674(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Dierks P., van Ooyen A., Mantei N., Weissmann C. DNA sequences preceding the rabbit beta-globin gene are required for formation in mouse L cells of beta-globin RNA with the correct 5' terminus. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1411–1415. doi: 10.1073/pnas.78.3.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Baty D., Chambon P. The repeated GC-rich motifs upstream from the TATA box are important elements of the SV40 early promoter. Nucleic Acids Res. 1983 Apr 25;11(8):2447–2464. doi: 10.1093/nar/11.8.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M., Berg P. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet. 1982;1(5):457–481. [PubMed] [Google Scholar]
- Fromm M., Berg P. Simian virus 40 early- and late-region promoter functions are enhanced by the 72-base-pair repeat inserted at distant locations and inverted orientations. Mol Cell Biol. 1983 Jun;3(6):991–999. doi: 10.1128/mcb.3.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M., Berg P. Transcription in vivo from SV40 early promoter deletion mutants without repression by large T antigen. J Mol Appl Genet. 1983;2(1):127–135. [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P., Frisque R. J., Gluzman Y. Identification of a promoter component involved in positioning the 5' termini of simian virus 40 early mRNAs. Proc Natl Acad Sci U S A. 1981 Jan;78(1):100–104. doi: 10.1073/pnas.78.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P. Simian virus 40 early mRNA's contain multiple 5' termini upstream and downstream from a Hogness-Goldberg sequence; a shift in 5' termini during the lytic cycle is mediated by large T antigen. J Virol. 1981 Oct;40(1):224–240. doi: 10.1128/jvi.40.1.224-240.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Piatak M., Mertz J. E., Weissman S. M., Lebowitz P. Altered utilization of splice sites and 5' termini in late RNAs produced by leader region mutants of simian virus 40. J Virol. 1982 Nov;44(2):610–624. doi: 10.1128/jvi.44.2.610-624.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Lebowitz P., Weissman S. M. Heterogeneity and 5'-terminal structures of the late RNAs of simian virus 40. J Mol Biol. 1978 Dec 25;126(4):813–846. doi: 10.1016/0022-2836(78)90022-0. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Delimitation of far upstream sequences required for maximal in vitro transcription of an H2A histone gene. Proc Natl Acad Sci U S A. 1982 Jan;79(2):297–301. doi: 10.1073/pnas.79.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7102–7106. doi: 10.1073/pnas.77.12.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld G. C., Rosenthal A., Flavell R. A. Sequence requirements for the transcription of the rabbit beta-globin gene in vivo: the -80 region. Nucleic Acids Res. 1982 Aug 25;10(16):4951–4971. doi: 10.1093/nar/10.16.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld G. C., Shewmaker C. K., Jat P., Flavell R. A. Localization of DNA sequences necessary for transcription of the rabbit beta-globin gene in vitro. Cell. 1981 Jul;25(1):215–226. doi: 10.1016/0092-8674(81)90246-4. [DOI] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman G., Fiers W. Localization of the 5' terminus of late SV40 mRNA. Nucleic Acids Res. 1978 Jul;5(7):2359–2371. doi: 10.1093/nar/5.7.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen U., Tenen D. G., Livingston D. M., Sharp P. A. T antigen repression of SV40 early transcription from two promoters. Cell. 1981 Dec;27(3 Pt 2):603–613. doi: 10.1016/0092-8674(81)90402-5. [DOI] [PubMed] [Google Scholar]
- Hen R., Sassone-Corsi P., Corden J., Gaub M. P., Chambon P. Sequences upstream from the T-A-T-A box are required in vivo and in vitro for efficient transcription from the adenovirus serotype 2 major late promoter. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7132–7136. doi: 10.1073/pnas.79.23.7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S. L., Manley J. L. DNA sequence required for initiation of transcription in vitro from the major late promoter of adenovirus 2. Proc Natl Acad Sci U S A. 1981 Feb;78(2):820–824. doi: 10.1073/pnas.78.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz P., Ghosh P. K. Initiation and regulation of simian virus 40 early transcription in vitro. J Virol. 1982 Feb;41(2):449–461. doi: 10.1128/jvi.41.2.449-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D. J., Chambon P. The SV40 early region TATA box is required for accurate in vitro initiation of transcription. Nature. 1981 Mar 26;290(5804):310–315. doi: 10.1038/290310a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Mellon P., Parker V., Gluzman Y., Maniatis T. Identification of DNA sequences required for transcription of the human alpha 1-globin gene in a new SV40 host-vector system. Cell. 1981 Dec;27(2 Pt 1):279–288. doi: 10.1016/0092-8674(81)90411-6. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Rio D. C., Robbins A. K., Tjian R. SV40 gene expression is modulated by the cooperative binding of T antigen to DNA. Cell. 1981 Aug;25(2):373–384. doi: 10.1016/0092-8674(81)90056-8. [DOI] [PubMed] [Google Scholar]
- Papamatheakis J., Lee T. N., Thayer R. E., Singer M. F. Recurring defective variants of simian virus 40 containing monkey DNA segments. J Virol. 1981 Jan;37(1):295–306. doi: 10.1128/jvi.37.1.295-306.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R. A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell. 1982 Sep;30(2):517–528. doi: 10.1016/0092-8674(82)90249-5. [DOI] [PubMed] [Google Scholar]
- Rio D., Robbins A., Myers R., Tjian R. Regulation of simian virus 40 early transcription in vitro by a purified tumor antigen. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5706–5710. doi: 10.1073/pnas.77.10.5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels M., Fire A., Sharp P. A. Separation and characterization of factors mediating accurate transcription by RNA polymerase II. J Biol Chem. 1982 Dec 10;257(23):14419–14427. [PubMed] [Google Scholar]
- Tsai S. Y., Tsai M. J., O'Malley B. W. Specific 5' flanking sequences are required for faithful initiation of in vitro transcription of the ovalbumin gene. Proc Natl Acad Sci U S A. 1981 Feb;78(2):879–883. doi: 10.1073/pnas.78.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M., Suzuki Y. Faithful transcription initiation of fibroin gene in a homologous cell-free system reveals an enhancing effect of 5' flanking sequence far upstream. Cell. 1981 Nov;27(1 Pt 2):175–182. doi: 10.1016/0092-8674(81)90371-8. [DOI] [PubMed] [Google Scholar]
- Weil P. A., Luse D. S., Segall J., Roeder R. G. Selective and accurate initiation of transcription at the Ad2 major late promotor in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979 Oct;18(2):469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]