Abstract
It is widely known that brassinosteroids (BRs) are involved in various physiological processes during plant growth and development. Roles of BRs have been reported in many plants. However, relevant report is yet not found in carrot. Carrot is a nutrient-rich vegetable from the Apiaceae family. Here, we measured the bioactive contents of BRs at five successive stages and analyzed the expression profiles of genes involved in BR biosynthesis, signaling pathway and catabolism. We found that most biosynthesis regulated genes had higher expression level at the first development stage of carrot and the catabolism gene BAS1/CYP734A1 had significantly high expression level at the first stage in carrot roots and petioles. In addition, we treated carrot plants with exogenous 24-epibrassinolide (24-EBL) and examined the morphological changes after treating. Compared with control plants, carrot plants treated with 24-EBL had higher plant height, more number of petioles and heavier aboveground weight. The expression levels of DcBRI1, DcBZR1, and DcBSU1 in the petioles were significantly up-regulated by treating with exogenous 24-EBL. The expression profiles of DcCYP734A1 were all significantly up-regulated in the three organs when treated with 0.5 mg/L 24-EBL. The elongation of carrot petioles can be promoted by treating with exogenous 24-EBL. These results indicate that BRs playing potential roles during the growth and development of carrot.
Keywords: Brassinosteroids, biosynthesis, signal transduction, gene regulation, 24-epibrassinolide, Daucus carota L.
Introduction
Brassinosteroids (BRs), a kind of sterols that were first discovered and isolated from Brassica napus pollen, have been reported to be involved in many aspects of plant growth and development (Grove et al., 1979; Clouse, 1997). In the growth and development of plant roots, BRs were reported to participate in maintenance of meristem size, lateral root initiation, nodulation in legume species and root hair formation (Bao et al., 2004; Terakado et al., 2005; Hacham et al., 2011; Cheng et al., 2014). Moreover, the roots of BR deficient mutant were found significantly shortened (Müssig et al., 2003). In Arabidopsis, BR was found working cooperatively with auxin in promoting petiole elongation to response the shade stimulus (Kozuka et al., 2010). BRs were also reported to be involved in stem elongation, cell division, vascular differentiation, and leaf bending and epinasty (Thompson et al., 1982; Clouse et al., 1996; Tong and Chu, 2012; Hao et al., 2013). In field production, BRs are believed to improve crop yield and plant tolerance against environmental stress (Divi and Krishna, 2009).
To date, more than 70 kinds of BRs have been identified (Miransari, 2014). Among them, brassinolide (BL) is recognized as the most active form (Fujioka and Sakurai, 1997). With the rapid development of biotechnology, the biosynthesis and signaling pathways of this hormone have been well studied. BL is produced via the early C-6 oxidation pathway and late C-6 oxidation pathway (Noguchi, 2000). Numerous studies have shown that regulation of genes involved in BR biosynthesis pathway can result in alteration in BR accumulation and plant growth (Yoshimitsu et al., 2011; Zhiponova et al., 2013). Some genes, such as DWARF7 (DWF7), DWARF1 (DWF1), DE ETIOLATED2 (DET2), DWARF4 (DWF4), CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM (CPD), CYP85A2, and CYP90C1 have been verified to play roles in BR biosynthesis (Choe et al., 1998, 1999; Noguchi, 1999; Du and Poovaiah, 2005; Ohnishi et al., 2006, 2012; Nole-Wilson et al., 2010). Among them, DWF4 is reported to be a key gene in BR biosynthesis, which may control the putative rate-limiting step in the BR biosynthetic pathway (Choe et al., 1998, 2001).
The homeostasis of BRs is regulated by biosynthesis genes and catabolism genes (Tanaka and Okamoto, 2005). In Arabidopsis, BAS1/CYP734A1 is a major BR inactivating gene (Neff et al., 1996). In many higher plants, the CYP734A paralogs was also found (Thornton et al., 2011). In addition, genes involved in the signaling pathway of BRs have been well investigated in Arabidopsis. Brassinosteroid insensitive 1 (BRI1), BRI1-associated receptor kinase1 (BAK1), BR-signaling kinase1 (BSK1), BRI1 suppressor 1 (BSU1), bridging integrator 2 (BIN2), brassinazole resistant1 (BZR1), and BRI1-EMS-suppressor1 (BES1) were involved in the signaling pathway (Wang and He, 2004; Sun et al., 2010; Yan et al., 2012; Shi et al., 2013a,b; Zhang et al., 2014; Jiang et al., 2015). Among these genes, BRI1 works as the receptor for BRs, BZR1and BES1 are the transcription factors that regulate the growth of plants (Gallegobartolomé et al., 2012; Guo et al., 2013).
Numerous studies have been conducted to determine the function of BRs in plant growth. However, BR accumulation and its potential roles during carrot growth and development remain elusive. Carrot (Daucus carota L.) is a nutrient-rich root crop from the Apiaceae family (Luby et al., 2014; Xu et al., 2014; Wang et al., 2015). It is one of the most economically important members of Apiaceae plants, and the cultivated area of carrot is progressively increasing worldwide (Cavagnaro et al., 2011).
In this study, BR accumulation at five successive growth stages was examined in carrot. Expression profiles of genes involved in BR biosynthesis and signaling pathways were also analyzed. Besides, exogenous 24-epibrassinolide (24-EBL) was applied to carrot plants to study the putative effects of BR on carrot. The results of our work would shed novel insights into studies focusing on hormonal control of plant growth.
Materials and Methods
Plant Material and Exogenous 24-EBL Treatment
‘Kurodagosun’ was selected as the experimental material and cultivated in a climate-control chamber at Nanjing Agricultural University (32°04′N, 118°85′E). Ambient temperature was held at 25°C for 16 h during daytime and 18°C for 8 h in the dark. The mix vermiculite and organic soil (1:1, v/v) was used to cultivated plants. The carrot samples were collected at 20 (stage 1), 40 (stage 2), 60 (stage 3), 75 (stage 4), and 90 (stage 5) days after sowing (DAS). The five stages were classified based on dates and morphological characteristics. The roots, petioles, and leaf blades were separately collected at different stages, and stored at -80°C for molecular research.
To investigate the potential effect of BR on carrot, 40 DAS carrot plants were treated with 24-EBL. The 24-EBL (≥ 85 %, Sigma–Aldrich) was dissolved in ethanol and then diluted by distilled water to make mother liquor. Four concentrations of exogenous 24-EBL (0, 0.1, 0.5, and 1 mg/L) were used, respectively. Plants treated with water alone were used as control (CK). Treatment was carried out every 2 days, five times in total. Afterward, the plants were allowed to grow for another 20 days and harvested for morphology determination.
Assay of Bioactive BR Levels
Samples were ground in a mortar with 10 mL of 80% methanol extraction solution containing 1 mM butylated hydroxytoluene. The mixture was incubated for 4 h at 4°C. The samples were then centrifuged for 10 min at 3500 g. The supernatants were filtered through a C18-Sep-Pak cartridge (Waters, Milford, MA, United States), and the efflux was collected and dried with N2. The mixture was dissolved in 2 mL of PBS containing 0.1 % (v/v) Tween 20 and 0.1% (w/v) gelatin (pH 7.5). The samples were analyzed via indirect enzyme-linked immunosorbent assay according to the methods described previously (Swaczynov et al., 2007; Pradko et al., 2014). Briefly: (1) The calibrating samples (epibrassinolide, CAS: 72962-43-7) or test samples (150 μL per well) were put in wells of the plate with the immobilized antibodies. Plates were placed at 37°C for 30 min. Then, removed the liquid from the wells and washed plates four times with washing buffer. (2) The Horseradish peroxidase (HRP)-conjugate (150 μL) was placed in the wells and placed at 37°C for 30 min. Then, removed the liquid from the wells and washed plates four times with washing buffer. (3) Added TMB solution (containing H2O2) to the wells and placed the plates at 37°C for 20 min. (4) Quenched the reaction by adding 2 mol/L H2SO4 (50 μL) into each well. (5) Measured optical absorbance at 450 nm. (6) Calculated the concentration according to the calibration curve. The epibrassinolide and other related brassinolide analogs were measured in this study.
Total RNA Isolation
An RNA extraction kit (Tiangen, Beijing, China) was used to extract the total RNA of carrot roots, leaf blades, and petioles according to the manufacturer’s instructions. cDNA was synthesized using a PrimerScript RT reagent kit (TaKaRa, Dalian, China). The cDNA was diluted 15-fold for quantitative reverse transcription PCR (qRT-PCR) analysis.
qRT-PCR Analysis
Genes involved in BR biosynthesis and signal transduction pathway were selected from CarrotDB1 (Xu, 2014). The primers of each gene were designed via Primer Premier 5.0 software, and were displayed in Table 1. qRT-PCR was performed in a real-time PCR detection system (Bio-Rad, Hercules, CA, United States). The cycling conditions were maintained as follows: 94°C for 30 s, 40 cycles at 94°C for 10 s, 58°C for 20 s, and 61 cycles at 65°C for 10 s to create a melting curve. The experiments were performed with three independent biological replicates. Under normal growing condition, the DcACTIN gene was selected as the internal control to analyze gene expression and data of DcDWF4 in carrot petioles at 90 DAS were chosen as a calibrator for gene expression analysis (Tian et al., 2015). By contrast, in the analysis of gene expression after application of 24-EBL, the DcTUB gene was selected as the internal control. The expression of DcTUB gene was more stable under abnormal growth conditions.
Table 1.
Oligonucleotide sequences of genes involved in the biosynthesis, catabolism, and signaling pathway.
| Gene name | Oligonucleotide sequences (5′→3′) |
|---|---|
| DcDWF4 | Forward primer: AAACGCTAAGGCTGGGCAATGT |
| Reverse primer: GCACGGCTGCAATCACTGGAA | |
| DcCPD | Forward primer: TCCCACCTACCGTAAAGCCATT |
| Reverse primer: ATCATTCTTCCTCTCCCCTCCT | |
| DcDET2 | Forward primer: TCAACGCCTCTCTCCTCACTCT |
| Reverse primer: CCAGCCCTTACGGTGGTGTTT | |
| DcCYP90C1 | Forward primer: GGTATGGCGAAAACAGGAGAGA |
| Reverse primer: AACACGAGATAGAGTGGCAGGG | |
| DcCYP85A2 | Forward primer: AGCCACTTACATTTAATCCCTG |
| Reverse primer: TCTCCTCCAACTTCTTCCCATC | |
| DcDWF5 | Forward primer: AGATGGTGGTGAAGGAGGAGAA |
| Reverse primer: CACGCAGTCATAGTGGGTTTTG | |
| DcDWF1 | Forward primer: TCCGACCTTTTCTACGCTATTC |
| Reverse primer: TGTAACCCTGTGCCAGTTCTTT | |
| DcBAK1 | Forward primer: TAGCACCCGAGTACCTATCCAC |
| Reverse primer: AGCAACATCACATCGTCATCAT | |
| DcBRI1 | Forward primer: GAAAAGGAGGAAGAAGAAGGAA |
| Reverse primer: CAAGAAGATCTGCAAAAGTGAG | |
| DcBSK1 | Forward primer: TGGCTGCTCTAGCTTAGTCTCC |
| Reverse primer: TTCCTTAGTTGCTCCAATGTGT | |
| DcBZR1 | Forward primer: AGTTTCCAGCCATCGCCGTCCC |
| Reverse primer: CTGCTCCCCAGCCCAATCCTCC | |
| DcBSU1 | Forward primer: TCAGCTTTTTAACTATCTTCCA |
| Reverse primer: ACCTTCTACACTATCATTTTCT | |
| DcBIN2 | Forward primer: TTGTTCCCACTGTTTCTTTGATG |
| Reverse primer: TGTAGATAGCCCACTTTGTCTCC | |
| DcACTIN | Forward primer: AGAAGCACCACTGAATCCTAAGGC |
| Reverse primer: GCATACACCATCACCAGAGTCCAA | |
| DcTUB | Forward primer: CGGTATTGTGTTGGACTCTGGTGAT |
| Reverse primer: CAGCAAGGTCAAGACGGAGTATGG | |
| DcCYP734A1 | Forward primer: TGCCATTCAGCCTTGGAGTA |
| Reverse primer: TGCGGATAAAGAAGCATCAACACC |
Statistical Analysis
Difference in the BRs bioactive content and the expression levels of genes in different organs during carrot development were detected by Duncan’s multiple-range test at a 0.05 probability level. Student’s t-test was used to identify the differences under different concentration treatments at the 0.05 significance.
Results
Plant Growth Analysis
The plant materials were sampled at five stages (20–, 40–, 60–, 75–, and 90 DAS) (Figure 1). The root was white at the first stage, and turned orange at the second stage. The weight of roots, petioles, and leaf blades remarkably increased between stages 2 to 3. The root continued to enlarge, and this trend was maintained at the remaining stages.
FIGURE 1.

Growth status of carrots from five different developmental stages in this research. (A) Stage1, 20 days after sowing, (B) stage2, 40 days after sowing, (C) stage3, 60 days after sowing, (D) stage4, 75 days after sowing, (E) stage5, 90 days after sowing. Black lines in the lower right corner of each plant represent 2 cm in that pixel.
Changes in Bioactive BR Contents
The levels of bioactive BRs in different tissues at five different stages were measured (Figure 2). BR accumulation differed in different carrot tissues and at different developmental stages.
FIGURE 2.
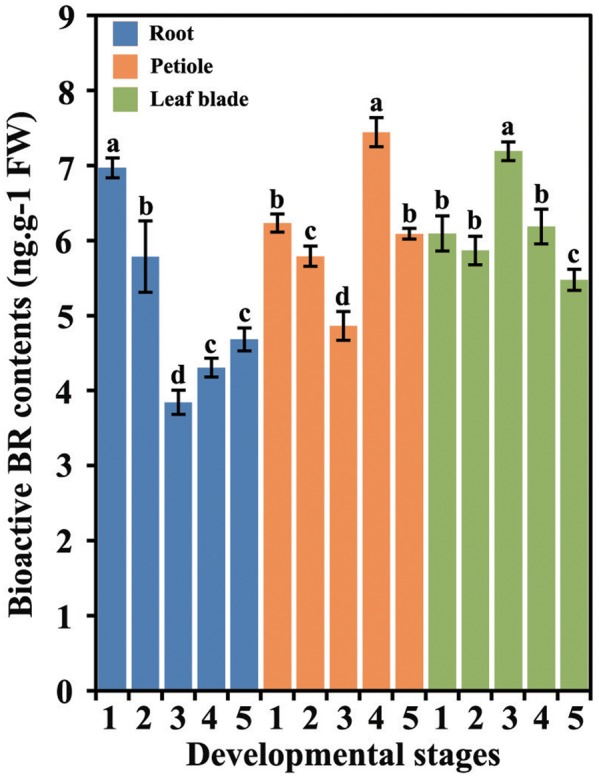
Bioactive BR levels in different tissues during carrot growth and development. Error bars represent standard deviation (SD). Columns with the same letter are not significantly different (P < 0.05).
In the roots, the levels of bioactive BRs were highest at first stage followed by a decrease until stage 3. The BRs levels underwent a slight increase at stage 4 and were constant until the last stage. In the petioles, the highest level of BRs was detected at 75 DAS (stage 4), whereas the lowest level was found at 60 DAS (stage 3). In the leaf blades, the level of BRs was peaked at the third stage.
Effects of 24-EBL on the Growth and Development of Carrot
To determine the potential roles of BR during the development of carrot, 40 DAS (stage 2) carrot plants were treated with four different concentrations of exogenous 24-EBL (0, 0.1, 0.5, and 1 mg/L) (Figure 3). Six physiological indexes including total plant height, root length, root diameter, root weight, aboveground weight, and number of petioles were selected for comparison among the treatments. Under different concentrations of 24-EBL treatments, the root length, root diameter, and root weight did not show obvious difference compared with control. However, 24-EBL obviously increased the total plant height of carrot (Figures 4A,B). When carrot plants were treated with 0.5 mg/L 24-EBL, the aboveground weight of carrot was significantly increased compared to the control (Figure 4B). Similarly, carrot plants exposed to 0.5 mg/L 24-EBL displayed increased number of petioles when compared with control plants (Figure 4C).
FIGURE 3.

Growth status of carrots treated with exogenous BL. 40 days after sowing plants were treated with different concentrations of BL (0, 0.1, 0.5, and 1 mg/L). White lines in the lower right corner of each plant represent 5 cm.
FIGURE 4.
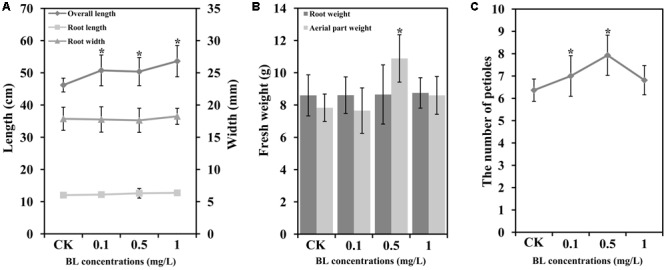
Exogenous BL induces petiole growth in carrot. 40 days after sowing plants were treated with different concentrations of BL (0, 0.1, 0.5 and 1 mg/L). (A) Root length, overall length, and root width; (B) root weight and aerial part (petioles and leaf blades) weight; (C) the number of petioles. Student’s t-test was used to identify the differences under different treatments (P < 0.05; ∗, control versus treatment). Error bars represent standard deviation (SD).
Expression Profiles of Genes Involved in BR Biosynthesis and Catabolism
DcDWF7, DcDWF1, DcDET2, DcDWF4, DcCPD, DcCYP85A2, DcCYP90C1, and DcCYP734A1 were recognized as genes involved in BR biosynthesis and catabolism and their expression levels in carrot roots, petioles and leaf blades at five developmental stages were measured by qRT-PCR (Figure 5) (Noguchi, 2000; Choe, 2010).
FIGURE 5.
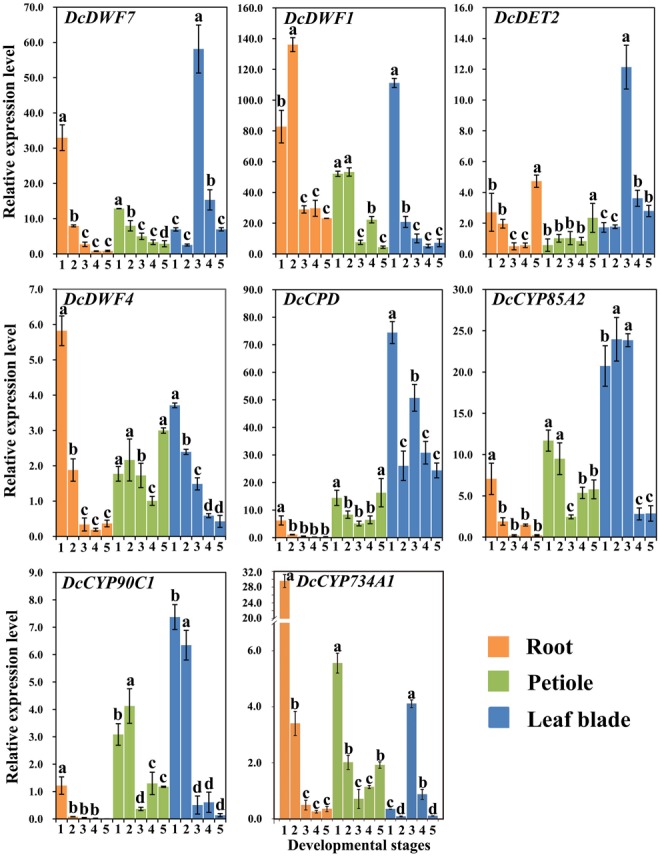
Expression profiles of genes involved in the BR biosynthesis and catabolism pathway at different stages and in different organs. Error bars represent standard deviation (SD). Columns with the same letter are not significantly different (P < 0.05).
In the Roots
DcDWF4, a key gene in BR biosynthesis, showed the highest expression level at stage 1 and the lowest level at stage 4. DcCPD and DcCYP90C1 peaked at stage 1 followed by a decrease. Afterward, the highest level of DcDET2 appeared at stage 5. DcDWF1 showed higher expression as compared with other genes. DcCYP734A1 is a catabolism gene of BRs and showed significantly higher expression level at stage 1 compared with other stages.
In the Petioles
The minimum expression level of the five genes (i.e., DcDWF4, DcCPD, DcDET2, DcCYP90C1, and DcCYP85A2) was observed at stage 3 or 4. The five genes exhibited similar expression trends. DcDWF7 and DcDWF1 were lowly expressed at the last stage. Transcription level of DcDWF1 was relatively higher than that of others. The expression level of catabolism gene DcCYP734A1 peaked at stage 1.
In the Leaf Blades
DcDWF7, DcDET2, and DcCYP85A2 were highly expressed at third stage. DcDWF1, DcDWF4, and DcCYP90C1 showed a similar expression pattern which peaked at first stage and gradually decreased afterwards. In the leaf blades, DcCYP734A1 gene showed the highest expression level at stage 3.
Expression Profiles of Genes Involved in the BR Signaling Pathway
In this study, transcript levels of DcBAK1, DcBRI1, DcBSK1, DcBZR1, DcBSU1, and DcBIN2, which have been reported to be involved in the BR signaling pathway, were analyzed using qRT-PCR (Belkhadir et al., 2006; Wang et al., 2006; Gendron and Wang, 2007). The expression profiles are shown in Figure 6.
FIGURE 6.
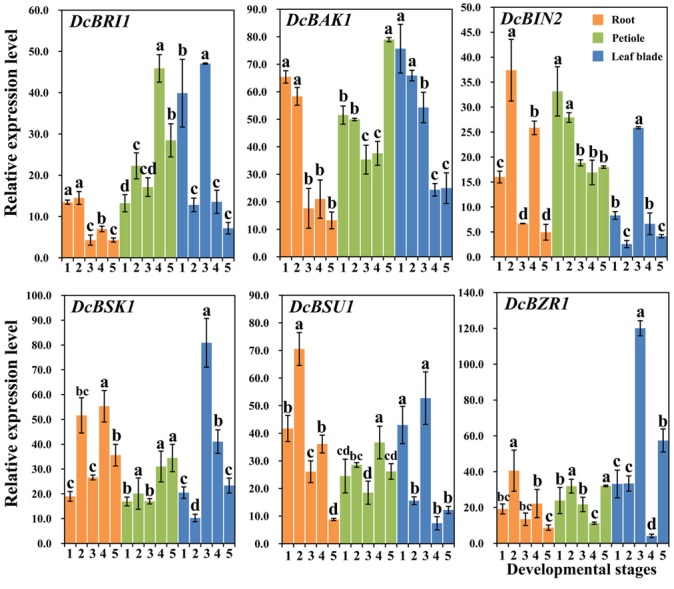
Expression profiles of genes involved in the BR signaling pathway at different stages and in different organs. Error bars represent standard deviation (SD). Columns with the same letter are not significantly different (P < 0.05).
In the Roots
DcBRI1 and DcBAK1 showed higher expression levels at the first two stages and relatively lower expression levels at later stages. The expression levels of DcBSK1, DcBIN2, DcBSU1, and DcBZR1 increased at second stage and declined at the third stage, and then increased at stage 4 and decreased at stage 5.
In the Petioles
DcBRI1 and DcBSU1 were highly expressed at the fourth stage, whereas the lowest expression level of DcBZR1 was detected at this stage. DcBAK1, DcBSK1, DcBZR1 showed highest expression level at the last stage. The highest transcription of DcBIN2 was observed at the first stage, followed by a continuous decrease at the remaining stages.
In the Leaf Blades
DcBRI1, DcBSK1, DcBSU1, DcBZR1, and DcBIN2 showed higher expression levels at stage 3. DcBAK1 showed relatively stable expression level at the first two stages, followed by a obvious decrease at the later stages.
Effects of 24-EBL Application on the Expression Level of BR Biosynthetic and Catabolism Genes
To study the effect of 24-EBL on BR biosynthesis, we investigated the expression level of related genes. The expression levels of these genes under different BL concentrations were measured by quantitative reverse transcription PCR (qRT-PCR). These genes were strongly regulated by exogenous BL treatment (Figure 7).
FIGURE 7.
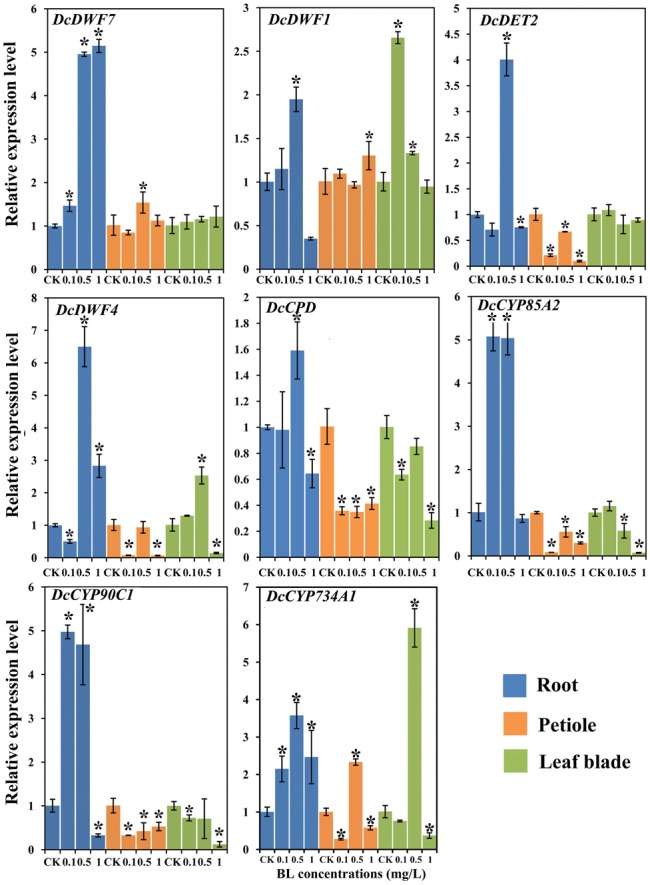
Expression profiles of genes involved in the BR biosynthesis and catabolism pathway under different BL concentrations (0, 0.1, 0.5, and 1 mg/L) and in different organs. Error bars represent standard deviation (SD). Student’s t-test was used to identify the differences under different concentration treatments (P < 0.05; ∗, control versus treatment).
In the Roots
All the genes examined were significantly up-regulated at the 0.5 mg/L. In the presence of 1 mg/L 24-EBL, DcDWF1, DcCPD, and DcCP90C1 showed lower expression levels as compared with control plants. When compared with control group, DcDWF4 expression level was significantly reduced in the roots treated with 0.1 mg/L BL, whereas transcription of DcCYP85A2 and DcCYP90C1 was increased.
In the Petioles
The expression levels of DcDET2, DcDWF4, DcCPD, DcCYP85A2, and DcCYP90C1 were significantly down-regulated after application of exogenous BL. In the expression profile of DcDWF7, expression level did not show significant change with 0.1 and 1 mg/L 24-EBL, but the expression level was significantly up-regulated with 0.5 mg/L. The catabolism gene DcCYP734A1 was significantly up-regulated under the treatment of 0.5 mg/L 24-EBL.
In the Leaf Blades
The expression levels of DcCPD, DcCYP90C1, and DcCYP85A2 was down-regulated after application of 24-EBL. Meanwhile, the expression levels of DcDWF7 and DcDET2 did not show significant change. However, DcDWF1 showed up-regulation trend with 0.1 and 0.5 mg/L and DcDWF4 was up-regulated by 0.5 mg/L 24-EBL. The expression level of the catabolism gene DcCYP734A1 was significantly up-regulated under the treatment of 0.5 mg/L 24-EBL.
Effects of 24-EBL Treatment on the Expression Profiles of BR Response Genes
To further investigate the effect of 24-EBL application on the expression levels of BR response genes, six genes were selected and their expression profiles were evaluated by qRT-PCR. Most genes were strongly regulated after the treatment (Figure 8).
FIGURE 8.
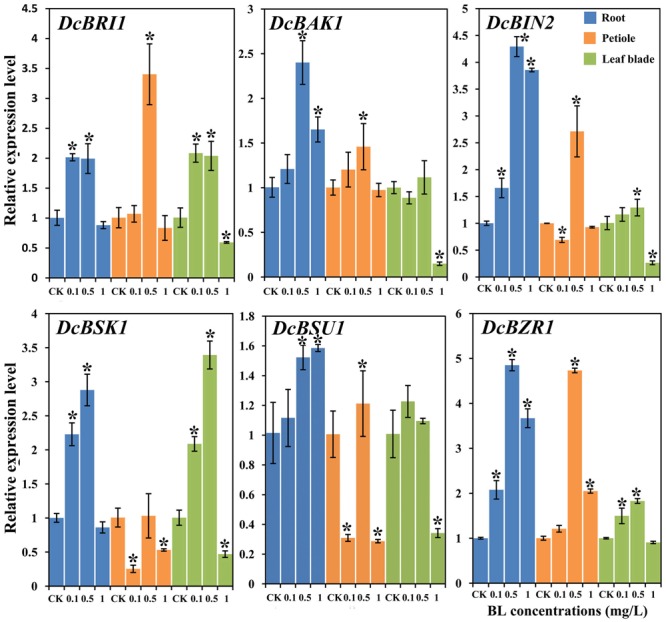
Expression profiles of genes involved in the BR signaling pathway under different BL concentrations (0, 0.1, 0.5, and 1 mg/L) and in different organs. Error bars represent standard deviation (SD). Student’s t-test was used to identify the differences under different concentration treatments (P < 0.05; ∗, control versus treatment).
In the Roots
The expression levels of all genes was increased in the presence of 0.5 mg/L BL. DcBRI1, and DcBSK1 expression levels was up-regulated in the plants treated with 0.1 and 0.5 mg/L BL. However, their expression levels in the presence of 1.0 mg/L showed no significant difference as compared with control group. DcBAK1, DcBIN2, DcBSU1, and DcBZR1 were up-regulated at all the three concentrations 24-EBL levels.
In the Petioles
Most genes showed significant up-regulation at the concentration of 0.5 mg/L, whereas DcBSK1 expression level showed no significant change. The expression levels of DcBSK1 and DcBSU1 was down-regulated when treated with 0.1 and 1 mg/L. Transcription of DcBZR1 was significantly increased in the presence of 1 mg/L BL.
In the Leaf Blades
Most genes (except DcBAK1 and DcBSU1) were up-regulated in the presence of 0.5 mg/L BL. When treated with 1 mg/L BL, the expression level of most genes significantly decreased, whereas DcBZR1 showed no obvious change. DcBAK1 and DcBIN2 expression levels in plants treated with 0.1 and 0.5 mg/L BL did not differ significantly from control.
Discussion
Phytohormones are very important factors affecting almost all the processes of plant growth (Singh and Savaldigoldstein, 2015). Brassinosteroids are a class of phytohormones with various functions in plants. They have been reported to play important roles in many developmental processes including primary root extension and lateral root formation, seed germination, cell and stem elongation, abiotic stress responses (Roddick and Guan, 1991; Clouse et al., 1996; Li and Chory, 1997; Bao et al., 2004; Sharma et al., 2015). Functions of BRs have been documented in many plants, such as, tomato, cucumber, rice, and maize (Montoya et al., 2005; Fu et al., 2008; Kir et al., 2015). However, relevant study regarding BRs in carrot is rare.
The precise role of a specific hormone on plant growth and development is not only a result of its available levels, but also relates to the plant species, and the target organ (Singh and Savaldigoldstein, 2015). In addition, the roles of a specific hormone also depend on the presence of other phytohormone and plant growth regulators (Nemhauser et al., 2006). BRs, acting as a growth-promoting hormone were reported to be most actively biosynthesized in actively growing tissues and function at the sites of synthesis. In Arabidopsis, the highest and the second highest level of endogenous BRs were observed in apical shoots and developing siliques which were actively developing organs, respectively (Shimada et al., 2003; Montoya et al., 2005).
In carrots, the content of bioactive BR levels at different stages and in different organs keeps changing (Figure 2). These results suggested that the roles of BRs were different at different developing stages. At stage1, the main function of BRs may be promoting the formation of roots. At stages 3 and 4, BRs may mainly promote the development of leaf blades and petioles, respectively. Here, the content of bioactive BRs was detected by ELISA. ELISA is an indirect approach for measuring BRs. According to previous reports, high performance liquid chromatography (HPLC), high performance liquid chromatography-mass spectrometry (HPLC-MS), and ELISA were compared in measuring BRs and they determined that ELISA can facilitate the determination of brassinosteroid phytohormones in plant (Pradko et al., 2014). Besides, based on the simple, rapid, fidelity and cost-effective ELISA method, many endogenous hormones in higher plant were detected, such as IAA, gibberellin (GA), and methyl jasmonate (MeJA) (Chen et al., 2009; Wang et al., 2015, 2016; Wu et al., 2016).
According to previous studies, BR plays important role in leaf growth and development. The well-known example is that exogenous BR can affect the bending of the lamina joint of rice (Mori et al., 2002). In rice, Dwarf1 gene was found to encode GTP-binding protein. The phenotype of its recessive mutant is dwarf (Li and Chory, 1997; Ross et al., 1997; Azpiroz et al., 1998). According to previous reports, the dwarf phenotype of dwarf mutants was associated with brassinosteroids and gibberellins (Hong et al., 2002). BR-deficient dwarf 1(brd1) mutant is a BR-related dwarf plant and the elongation of stem and leaves of which were reported to be repressed in rice (Hong et al., 2002). By contrast, exogenous BL stimulated the unrolling of leaf blades in wheat (Wada et al., 2014). In Arabidopsis, BR-deficient mutants appeared to have stunted shoot and small leaves (Nakaya et al., 2002). Here, the expression levels of genes involved in BR biosynthesis in leaf blades were higher than that in the root at the last three stages. The catabolism gene DcCYP734A1 has the same expression trend in leaf blades of carrot plants. In addition, the fresh weight of aerial part of treated carrot plants (0.5 mg/L) was significantly higher than that of control. After treating by 24-EBL, genes DcDWF4, DcCPD, and DcCYP85A2 in BRs biosynthesis pathway were down-regulated and catabolism gene DcCYP734A1 was up-regulated at the concentration of 0.5 mg/L in the leaf blades. These results suggested that BRs may play important roles in carrot leaf blade growth.
BRs regulate plant growth and development through promoting cell elongation and division (Fridman and Savaldi-Goldstein, 2013). In Arabidopsis, the size and intercellular spaces of matured leaves of deetiolated2 (det2) and dwarf1 (dwf1) mutants were smaller than those of wild plants. These phenotypes of det2 and dwf1 mutants were demonstrated to be the result of inhibited elongation and division of cells in Arabidopsis (Nakaya et al., 2002). Similar results were also observed in rice (Hong et al., 2002). Plant height is an important index for plant growth. BR was reported to be a crucial factor for plant height (Hong et al., 2002). In soybean, GmBRI1b (Glyma04g39160) was a putative BR receptor and was found high similar to those of AtBRI1 and pea PsBRI1. The petioles of GmBRI1b over-expression lines were longer than that of wild plants (Peng et al., 2016). Many BR-deficient mutants, such as dwarf4 (dwf4), and dwf7-1 in Arabidopsis, dwarf1-1 in sorghum, exhibited dwarf phenotypes due to deficiency in BR biosynthesis (Choe and Feldmann, 1998; Cheon et al., 2010; Fridman and Savaldi-Goldstein, 2013; Petti et al., 2015). Here, the biosynthesis genes and catabolism genes showed higher expression levels at the first two stages which suggested that BRs may be involved in actively growing of petioles. In petioles of carrot plants, after being treated by exogenous BL, expression levels of most biosynthesis genes were inhibited. However, the catabolism gene DcCYP734A1 was significantly up-regulated treated by exogenous BL at the concentration of 0.5 mg/L 24-EBL. Most signal transduction genes were also up-regulated when treated by 0.5 mg/L 24-EBL. Carrot plants treated by 24-EBL had increased height and aerial part weight. Meanwhile, the number of petioles in carrot plants increased after application of exogenous 24-EBL. All the above results suggested that application of exogenous 24-EBL promoted the growth of carrot petioles.
Carrot is a kind of root vegetable crop. The root of carrot is a good source of phytochemicals, such as carotenoids, anthocyanins, and other phenolic compounds. Till now, the brassinosteroid’s roles involved in carrot root growth and development are not clear. In Arabidopsis, application of exogenous brassinolide was reported to promote root elongation and lateral root germination (Gonzálezgarcía et al., 2011; Singh and Savaldigoldstein, 2015). However, BRs plays different roles in regulating nodule formation in different plant species. BRs can actively regulate the nodule numbers in pea, but function as an inhibitor in soybean nodule formation (Ferguson et al., 2005; Terakado et al., 2005). During the development of carrot, all biosynthesis and catabolism genes showed higher expression levels at the first two stages in root which suggested that BRs may be necessary for the early stage of carrot root growth and development. However, the expression levels of signal transduction genes do not show similar trend. After treatment, all the biosynthesis, catabolism and signal transduction genes were up-regulated. However, the root length and diameter of carrot plants treated with exogenous 24-EBL did not show significant changes in our study. Symons and his colleagues demonstrated that the long-distance transport of BRs cannot be detected in pea (Symons and Reid, 2004). In tomato, the long-distance transport of BRs is also lacking (Montoya et al., 2005). Moreover, BRs were thought to regulate root meristem size maintenance depending on their action site (Wei and Li, 2016). In Arabidopsis, the meristem size of root was reported to be controlled by BR signaling in the root epidermis (Gonzálezgarcía et al., 2011). In high plant, the extensive interactions were existed among different hormonal signaling pathways. Many kinds of hormones were involved in the regulation on plant growth and development (Nemhauser et al., 2006). In rice, the signaling pathways of auxin, ABA and GA3 controlling cell elongation are affected by other pathway involving brassinosteroids (Ephritikhine et al., 1999). In Arabidopsis, the growth of hypocotyls was reported to be controlled by auxin, ethylene and brassinosteroids (De et al., 2005). Ethylene is an important signal for the development of roots and was reported to enhance its inhibition of Arabidopsis roots by up-regulating auxin biosynthesis (Swarup et al., 2007). Moreover, the ethylene level in the primary roots of maize was also increased after application of exogenous brassinolide (Sun et al., 2002). In carrot plant, the growth and development of roots maybe controlled by BRs, and other plant hormones, auxin, ABA, GA3, and ethylene, and so on.
Phytohormone is important for plant, but the concentration of treatment should be appropriate. In Arabidopsis, low concentration of BL can promote the growth of root, but high concentrations will decrease it (Gonzálezgarcía et al., 2011). In present study, 24-EBL at 0.5 mg/L was proved to have a positive impact on carrot petiole growth. In conclusion, this study supports the idea that BRs play an important role in regulating plant height. Carrot plants exposed to exogenous 24-EBL were relatively higher than those treated with water alone. Exogenous 24-EBL can speed up the growth of petioles in carrots.
Author Contributions
Conceived and designed the experiments: A-SX, FQ. Performed the experiments: FQ, G-LW, FW, Z-SX. Analyzed the data: FQ, G-LW. Contributed reagents/materials/analysis tools: A-SX. Wrote the paper: FQ. Revised the paper: A-SX, G-LW. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research was supported by the New Century Excellent Talents in University (NCET-11-0670); Jiangsu Natural Science Foundation (BK20130027); Priority Academic Program Development of Jiangsu Higher Education Institutions.
Abbreviations
- 24-EBL
24-epibrassinolide
- ACTIN
Actin 1 gene
- BAK1
BRI1-associated receptor kinase1
- BES1
BRI1-EMS-suppressor1
- BIN2
bridging integrator 2
- BRI1
Brassinosteroid insensitive 1
- BRs
brassinosteroids
- BSK1
BR-signaling kinase1
- BSU1
BRI1 suppressor 1
- BZR1
brassinazole resistant1
- CPD
CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM
- DAS
days after sowing
- DET2
DE ETIOLATED2
- DWF1
DWARF1
- DWF4
DWARF4
- DWF7
DWARF7
- TUB
Tubulin beta-7 gene
- qRT-PCR
quantitative reverse transcription PCR
Footnotes
References
- Azpiroz R., Wu Y., Locascio J. C., Feldmann K. A. (1998). An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10 219–230. 10.1105/tpc.10.2.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao F., Shen J., Brady S. R., Muday G. K., Asami T., Yang Z. (2004). Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 134 1624–1631. 10.1104/pp.103.036897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y., Wang X., Chory J. (2006). Arabidopsis brassinosteroid signaling pathway. Sci. STKE 2006:cm5 10.1126/stke.3642006cm5 [DOI] [PubMed] [Google Scholar]
- Cavagnaro P. F., Chung S. M., Manin S., Yildiz M., Ali A., Alessandro M. S., et al. (2011). Microsatellite isolation and marker development in carrot - genomic distribution, linkage mapping, genetic diversity analysis and marker transferability across Apiaceae. BMC Genomics 12:386 10.1186/1471-2164-12-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Qi W. B., Reiter R. J., Wei W., Wang B. M. (2009). Exogenously applied melatonin stimulates root growth and raises endogenous indoleacetic acid in roots of etiolated seedlings of Brassica juncea. J. Plant Growth Regul. 166 324–328. 10.1016/j.jplph.2008.06.002 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Zhu W., Chen Y., Ito S., Asami T., Wang X. (2014). Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. eLife 3:e02525 10.7554/eLife.02525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon J., Park S. Y., Schulz B., Choe S. (2010). Arabidopsis brassinosteroid biosynthetic mutant dwarf7-1 exhibits slower rates of cell division and shoot induction. BMC Plant Biol. 10:270 10.1186/1471-2229-10-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S. (2010). “Brassinosteroid biosynthesis and metabolism,” in Plant Hormones: Biosynthesis, Signal Transduction, Action ed. Davies P. J. (Dordrecht: Springer; ) 156–178. 10.1007/978-1-4020-2686-7_8 [DOI] [Google Scholar]
- Choe S., Dilkes B. P., Fujioka S., Takatsuto S., Sakurai A., Feldmann K. A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10 231–243. 10.1105/tpc.10.2.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S., Feldmann K. A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S., Fujioka S., Noguchi T., Takatsuto S., Yoshida S., Feldmann K. A. (2001). Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 26 573–582. 10.1046/j.1365-313x.2001.01055.x [DOI] [PubMed] [Google Scholar]
- Choe S., Noguchi T., Fujioka S., Takatsuto S., Tissier C. P. (1999). The Arabidopsis dwf7/ste1 mutant is defective in the sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 11 207–221. 10.2307/3870851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S. D. (1997). Molecular genetic analysis of brassinosteroid action. Physiol. Plant. 100 702–709. 10.1111/j.1399-3054.1997.tb03077.x [DOI] [Google Scholar]
- Clouse S. D., Langford M., McMorris T. C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111 671–678. 10.1104/pp.111.3.671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De G. L., Vandenbussche F., Tietz O., Palme K., Van D. S. D. (2005). Auxin, ethylene and brassinosteroids: tripartite control of growth in the Arabidopsis hypocotyl. Plant Cell Physiol. 46 827–836. 10.1093/pcp/pci111 [DOI] [PubMed] [Google Scholar]
- Divi U. K., Krishna P. (2009). Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. N. Biotechnol. 26 131–136. 10.1016/j.nbt.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Du L., Poovaiah B. W. (2005). Ca2+/calmodulin is critical for brassinosteroid biosynthesis and plant growth. Nature 437 741–745. 10.1038/nature03973 [DOI] [PubMed] [Google Scholar]
- Ephritikhine G., Fellner M., Vannini C., Lapous D., Barbier-Brygoo H. (1999). The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J. Cell Mol. Biol. 18 303–314. 10.1046/j.1365-313X.1999.00454.x [DOI] [PubMed] [Google Scholar]
- Ferguson B. J., Ross J. J., Reid J. B. (2005). Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol. 138 2396–2405. 10.1104/pp.105.062414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman Y., Savaldi-Goldstein S. (2013). Brassinosteroids in growth control: how, when and where. Plant Sci. Int. J. Exp. Plant Biol. 209 24–31. 10.1016/j.plantsci.2013.04.002 [DOI] [PubMed] [Google Scholar]
- Fu Q. M., Mao W. H., Shi K., Zhou Y. H., Asami T., Yu J. Q. (2008). A role of brassinosteroids in early fruit development in cucumber. J. Exp. Bot. 59 2299–2308. 10.1093/jxb/ern093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S., Sakurai A. (1997). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 100 710–715. [DOI] [PubMed] [Google Scholar]
- Gallegobartolomé J., Minguet E. G., Grauenguix F., Abbas M., Locascio A., Thomas S. G., et al. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109 13446–13451. 10.1073/pnas.1119992109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron J. M., Wang Z. Y. (2007). Multiple mechanisms modulate brassinosteroid signaling. Curr. Opin. Plant Biol. 10 436–441. 10.1016/j.pbi.2007.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzálezgarcía M. P., Vilarrasablasi J., Zhiponova M., Divol F., Moragarcía S., Russinova E., et al. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138 849–859. 10.1242/dev.057331 [DOI] [PubMed] [Google Scholar]
- Grove M. D., Spencer G. F., Rohwedder W. K., Mandava N., Worley J. F., Warthen J. D., et al. (1979). Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281 216–217. 10.1038/281216a0 [DOI] [Google Scholar]
- Guo R., Qian H., Shen W., Liu L., Zhang M., Cai C., et al. (2013). BZR1 and BES1 participate in regulation of glucosinolate biosynthesis by brassinosteroids in Arabidopsis. J. Exp. Bot. 64 2401–2412. 10.1093/jxb/ert094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y., Holland N., Butterfield C., Ubeda-Tomas S., Bennett M. J., Chory J., et al. (2011). Brassinosteroid perception in the epidermis controls root meristem size. Development 138 839–848. 10.1242/dev.061804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J., Yin Y., Fei S. Z. (2013). Brassinosteroid signaling network: implications on yield and stress tolerance. Plant Cell Rep. 32 1017–1030. 10.1007/s00299-013-1438-x [DOI] [PubMed] [Google Scholar]
- Hong Z., Ueguchi-Tanaka M., Shimizu-Sato S., Inukai Y., Fujioka S., Shimada Y., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32 495–508. 10.1046/j.1365-313X.2002.01438.x [DOI] [PubMed] [Google Scholar]
- Jiang J., Zhang C., Wang X. (2015). A recently evolved isoform of the transcription factor BES1 promotes brassinosteroid signaling and development in Arabidopsis thaliana. Plant Cell 27 361–374. 10.1105/tpc.114.133678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kir G., Ye H., Nelissen H., Neelakandan A. K., Kusnandar A. S., Luo A., et al. (2015). RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiol. 169 826–839. 10.1104/pp.15.00367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuka T., Kobayashi J., Horiguchi G., Demura T., Sakakibara H., Tsukaya H., et al. (2010). Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol. 153 1608–1618. 10.1104/pp.110.156802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 929–938. 10.1016/S0092-8674(00)80357-8 [DOI] [PubMed] [Google Scholar]
- Luby C. H., Maeda H. A., Goldman I. (2014). Genetic and phenological variation of tocochromanol (vitamin E) content in wild (Daucus carota L. var. carota) and domesticated carrot (D. carota L. var. sativa). Hortic. Res. 1:14015 10.1038/hortres.2014.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miransari M. (2014). “Plant, mycorrhizal fungi, and bacterial network,” in Plant Signaling: Understanding the Molecular Crosstalk eds Hakim K., Rehman R., Tahir I. (New Delhi: Springer; ) 315–325. [Google Scholar]
- Montoya T., Nomura T., Yokota T., Farrar K., Harrison K., Jones J. G. D., et al. (2005). Patterns of Dwarf expression and brassinosteroid accumulation in tomato reveal the importance of brassinosteroid synthesis during fruit development. Plant J. 42 262–269. 10.1111/j.1365-313X.2005.02376.x [DOI] [PubMed] [Google Scholar]
- Mori M., Nomura T., Ooka H., Ishizaka M., Yokota T., Sugimoto K., et al. (2002). Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 130 1152–1161. 10.1104/pp.007179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig C., Shin G. H., Altmann T. (2003). Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 133 1261–1271. 10.1104/pp.103.028662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M., Tsukaya H., Murakami N., Kato M. (2002). Brassinosteroids control the proliferation of leaf cells of Arabidopsis thaliana. Plant Cell Physiol. 43 239–244. 10.1093/pcp/pcf024 [DOI] [PubMed] [Google Scholar]
- Neff M. M., Nguyen S. M., Malancharuvil E. J., Fujioka S., Noguchi T., Seto H., et al. (1996). BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 96:15316 10.1073/pnas.96.26.15316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J. L., Hong F., Chory J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475. 10.1016/j.cell [DOI] [PubMed] [Google Scholar]
- Noguchi T. (1999). Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-one to (24R)-24-methyl-5alpha-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 120 833–839. 10.1104/pp.120.3.833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T. (2000). Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol. 124 201–210. 10.1104/pp.124.1.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S., Rueschhoff E. E., Bhatti H., Franks R. G. (2010). Synergistic disruptions in seuss cyp85A2 double mutants reveal a role for brassinolide synthesis during gynoecium and ovule development. BMC Plant Biol. 10:198 10.1186/1471-2229-10-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T., Godza B., Watanabe B., Fujioka S., Hategan L., Ide K., et al. (2012). CYP90A1/CPD, a brassinosteroid biosynthetic cytochrome P450 of Arabidopsis, catalyzes C-3 oxidation. J. Biol. Chem. 287 31551–31560. 10.1074/jbc.M112.392720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T., Szatmari A. M., Watanabe B., Fujita S., Bancos S., Koncz C., et al. (2006). C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18 3275–3288. 10.1105/tpc.106.045443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S., Tao P., Xu F., Wu A., Huo W., Wang J. (2016). Functional characterization of soybean Glyma04g39610 as a brassinosteroid receptor gene and evolutionary analysis of soybean brassinosteroid receptors. Int. J. Mol. Sci. 17:897 10.3390/ijms17060897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti C., Hirano K., Stork J., Debolt S. (2015). Mapping of a cellulose-deficient mutant named dwarf1-1 in Sorghum bicolor to the green revolution gene gibberellin20-oxidase reveals a positive regulatory association between gibberellin and cellulose biosynthesis. Plant Physiol. 169 705–716. 10.1104/pp.15.00928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradko A. G., Litvinovskaya R. P., Sauchuk A. L., Drach S. V., Baranovsky A. V., Zhabinskii V. N., et al. (2014). A new ELISA for quantification of brassinosteroids in plants. Steroids 97 78–86. 10.1016/j.steroids.2014.08.022 [DOI] [PubMed] [Google Scholar]
- Roddick J. G., Guan M. (1991). “Brassinosteroids and Root Development,” in Brassinosteroids: Chemistry, Bioactivity, and Application, ACS Symposium Series 474 eds Cutler H. G., Yokota T., Adam G. (Washington, DC: American Chemical Society; ) 231–245. [Google Scholar]
- Ross J. J., Murfet I. C., Reid J. B. (1997). Gibberellin mutants. Physiol. Plant. 100 550–560. 10.1034/j.1399-3054.1997.1000317.x [DOI] [Google Scholar]
- Sharma I., Bhardwaj R., Pati P. K. (2015). Exogenous application of 28-homobrassinolide modulates the dynamics of salt and pesticides induced stress responses in an elite rice variety Pusa basmati-1. J. Plant Growth Regul. 34 509–518. 10.1007/s00344-015-9486-9 [DOI] [Google Scholar]
- Shi H., Shen Q., Qi Y., Yan H., Nie H., Chen Y., et al. (2013a). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25 1143–1157. 10.1105/tpc.112.107904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Yan H., Li J., Tang D. (2013b). BSK1, a receptor-like cytoplasmic kinase, involved in both BR signaling and innate immunity in Arabidopsis. Plant Signal. Behav. 8:e24996 10.4161/psb.24996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y., Goda H., Nakamura A., Takatsuto S., Fujioka S., Yoshida S. (2003). Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol. 131 287–297. 10.1104/pp.013029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. P., Savaldigoldstein S. (2015). Growth control: brassinosteroid activity gets context. J. Exp. Bot. 66 1123–1132. 10.1093/jxb/erv026 [DOI] [PubMed] [Google Scholar]
- Sun H. L., Chang S. C., Lee J. S., Kim S. K., Kim S. Y. (2002). Brassinosteroids affect ethylene production in the primary roots of maize (Zea mays L.). J. Plant Biol. 45 148–153. 10.1007/BF03030307 [DOI] [Google Scholar]
- Sun Y., Fan X. Y., Cao D. M., Tang W., He K., Zhu J. Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19 765–777. 10.1016/j.devcel.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaczynov J., Hauserova N. E., Fuksova K., Sisa M., Kohout L., Strnad M. (2007). New techniques for the estimation of naturally occurring brassinosteroids. J. Plant Growth Regul. 26 1–14. 10.1007/s00344-006-0045-2 [DOI] [Google Scholar]
- Swarup R., Perry P., Hagenbeek D., Van Der Straeten D., Beemster G. T., Sandberg G., et al. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19 2186–2196. 10.1105/tpc.107.052100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons G. M., Reid J. B. (2004). Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol. 135 2196–2206. 10.1104/pp.104.043034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Okamoto S. (2005). Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 138 1117–1125. 10.1104/pp.104.058040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakado J., Fujihara S., Goto S., Kuratani R., Suzuki Y., Yoshida S., et al. (2005). Systemic effect of a brassinosteroid on root nodule formation in soybean as revealed by the application of brassinolide and brassinazole. Soil Sci. Plant Nutr. 51 389–395. 10.1111/j.1747-0765.2005.tb00044.x [DOI] [Google Scholar]
- Thompson M. J., Meudt W. J., Mandava N. B., Dutky S. R., Lusby W. R., Spaulding D. W. (1982). Synthesis of brassinosteroids and relationship of structure to plant growth-promoting effects. Steroids 39 89–105. 10.1016/0039-128X(82)90129-5 [DOI] [PubMed] [Google Scholar]
- Thornton L. E., Peng H., Neff M. M. (2011). Rice CYP734A cytochrome P450s inactivate brassinosteroids in Arabidopsis. Planta 234 1151–1162. 10.1007/s00425-011-1464-2 [DOI] [PubMed] [Google Scholar]
- Tian C., Jiang Q., Wang F., Wang G. L., Xu Z. S., Xiong A. S. (2015). Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PLoS ONE 10:e0117569 10.1371/journal.pone.0117569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Chu C. (2012). Brassinosteroid signaling and application in rice. J. Genet. Genomics 39 3–9. 10.1016/j.jgg.2011.12.001 [DOI] [PubMed] [Google Scholar]
- Wada K., Kondo H., Marumo S. (2014). A simple bioassay for brassinosteroids: a wheat leaf-unrolling test. Agric. Biol. Chem. 49 2249–2251. 10.1271/bbb1961.49.2249 [DOI] [Google Scholar]
- Wang G. L., Huang W., Li M., Xu Z. S., Wang F., Xiong A. S. (2016). Expression profiles of genes involved in jasmonic acid biosynthesis and signaling during growth and development of carrot. Acta Biochim. Biophys. Sin. 48 795–803. 10.1093/abbs/gmw058 [DOI] [PubMed] [Google Scholar]
- Wang G. L., Xiong F., Que F., Xu Z. S., Wang F., Xiong A. S. (2015). Morphological characteristics, anatomical structure, and gene expression: novel insights into gibberellin biosynthesis and perception during carrot growth and development. Hortic. Res. 2:15028 10.1038/hortres.2015.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Y., He J. X. (2004). Brassinosteroid signal transduction – choices of signals and receptors. Trends Plant Sci. 9 91–96. 10.1016/j.tplants.2003.12.009 [DOI] [PubMed] [Google Scholar]
- Wang Z. Y., Wang Q. M., Chong K., Wang F. R., Wang L., Bai M. Y., et al. (2006). The brassinosteroid signal transduction pathway. Cell Res. 16 427–434. 10.1038/sj.cr.7310054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z. Y., Li J. (2016). Brassinosteroids regulate root growth, development, and symbiosis. Mol. Plant 9 86–100. 10.1016/j.molp.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Wu X. J., Wang G. L., Song X., Xu Z. S., Wang F., Xiong A. S. (2016). Regulation of auxin accumulation and perception at different developmental stages in carrot. Plant Growth Regul. 80 243–251. 10.1007/s10725-016-0161-3 [DOI] [Google Scholar]
- Xu Z. S. (2014). CarrotDB: a genomic and transcriptomic database for carrot. Database 2014 1229–1245. 10.1093/database/bau096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. S., Huang Y., Wang F., Song X., Wang G. L., Xiong A. S. (2014). Transcript profiling of structural genes involved in cyanidin-based anthocyanin biosynthesis between purple and non-purple carrot (Daucus carota L.) cultivars reveals distinct patterns. BMC Plant Biol. 14:262 10.1186/s12870-014-0262-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Ma Y., Liu D., Wei X., Sun Y., Chen X., et al. (2012). Structural basis for the impact of phosphorylation on the activation of plant receptor-like kinase BAK1. Cell Res. 22 1304–1308. 10.1038/cr.2012.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimitsu Y., Tanaka K., Fukuda W., Asami T., Yoshida S., Hayashi K., et al. (2011). Transcription of DWARF4 plays a crucial role in auxin- regulated root elongation in addition to brassinosteroid homeostasis in Arabidopsis thaliana. PLoS ONE 6:e23851 10.1371/journal.pone.0023851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ye H., Guo H., Johnson A., Zhang M., Lin H., et al. (2014). Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in Arabidopsis. Plant J. 77 59–70. 10.1111/tpj.12368 [DOI] [PubMed] [Google Scholar]
- Zhiponova M. K., Vanhoutte I., Boudolf V., Betti C., Dhondt S., Coppens F., et al. (2013). Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phytol. 197 490–502. 10.1111/nph.12036 [DOI] [PubMed] [Google Scholar]


