Abstract
The emergence of endomembrane systems was a pivotal event in the evolution of the eukaryotic cell. Two ancient and fundamental, but evolutionarily distinct, eukaryotic endomembrane systems are mitochondria and the endoplasmic reticulum (ER). Both of these compartments are actively shaped into an extended reticular network, which enables them to communicate with each other and with other organelles distributed throughout the cell. Active protein-, lipid-, and ion-meditated interorganellar communication between mitochondria and the ER occur at points of contact at a distance of 10–30 nm between the two organelles. Recent advances, made primarily in budding yeast, have begun to describe the molecular features and mechanisms that underlie ER-mitochondria contact site formation and function. These and other studies have revealed that mitochondria make contacts with multiple organelles that possess functions beyond lipid and ion exchange. A more general model has emerged in which ion and lipid transport at interorganellar contacts sites also serve to form specialized microdomains that coordinate diverse activities, such as mitochondrial dynamics with cell stress signaling pathways. Here we highlight advances demonstrating the functional integration of activities at mitochondrial contact sites and speculate on how the functional outputs of different types of contact sites are somehow coordinated with each other and non-mitochondrial membrane contacts and pathways in the cell.
Mitochondrial contact sites function in lipid transport
Mitochondria retain features of their bacterial ancestors, including a genome. Thus, in contrast to other organelles that are born at the ER, such as the Golgi, mitochondria cannot be created de novo in cells. However, they are not autonomous. Their replication is dependent on the delivery of essential building blocks, which include a majority of their proteome, encoded by the nuclear genome, and their lipids. While mitochondria house biochemical pathways to make phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL), the respective precursors of these pathways, phosphatidylserine (PS) and phosphatidic acid (PA), must be imported from other organelles. In addition, mitochondria lack the ability to synthesize phosphatidylcholine (PC), the major bilayer-forming structural lipid, phosphatidylinositol (PI), and the raft-forming lipids, sterols and sphingolipids. The endoplasmic reticulum is the primary site of lipid biosynthesis. However, as mitochondria are not connected to the secretory pathway, the essential influx of lipids from the ER instead occurs through non-vesicular means.
Elegant biochemical studies demonstrated that ER-derived PS serves as a precursor for PE synthesis within mitochondria, which can in turn be exported to other organelles or to the ER where it is used at to fuel the synthesis of PC. The enzymes that mediate these reactions were found to be enriched in a distinct biochemically isolated membrane fraction, which contained ER associated with outer mitochondrial membranes termed the MAM, which also has raft-like features. From this and many other studies, a model emerged in which both lipid and ion transport between mitochondria and the ER occur via regions of close intermembrane apposition. Lipid transport at regions of contact may be direct, facilitated by lipid and/or protein mediated membrane curvature, but emerging evidence indicates that it primarily occurs via lipid transporting protein complexes that may also serve to mediate the intracellular distribution of mitochondria by tethering.
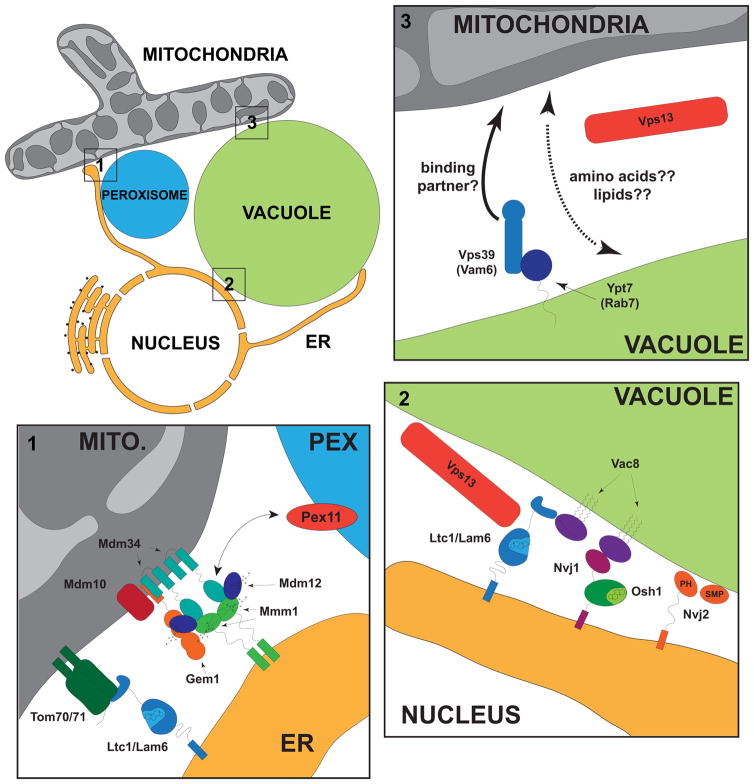
The molecular identity and mechanisms of the protein complexes that facilitate lipid transport between the ER and mitochondria are currently emerging, primarily in the budding yeast. The prototypical contact is mediated by the ER-mitochondria encounter structure (ERMES) complex, identified in a yeast screen for mutants whose growth defects could be suppressed by an artificial ER-mitochondrial tether, immediately implicating ERMES as a physical tether in wild type cells (Kornmann et al., 2009). The core ERMES complex is composed of four interdependent subunits: the integral mitochondrial outer membrane proteins, Mdm34 and Mdm10, the integral ER protein Mmm1, and the soluble cytosolic subunit, Mdm12 (Figure 1). Observed by fluorescence microscopy, ERMES subunits are co-localized in focal structures that are persistently at regions of overlap between the ER and mitochondria, further supporting its role as a tether (Murley et al., 2013). The discovery of this protein complex illuminated the fundamental functional importance of ER-mitochondria contacts: cells without ERMES have severe mitochondrial morphological defects in which the distributed tubular mitochondrial reticulum collapses into large spheres, cells rapidly lose mitochondrial DNA, cannot respire, and they have severe growth defects, even on media containing fermentable carbon sources. The ERMES complex is ancient as it was present in the ancestor of yeast and animals. But ERMES was lost in the animal lineage, suggesting that other proteins or protein complexes have replaced its functions in metazoans.
Figure 1.
Current understanding of proteins localized to contact sites between mitochondria (gray), vacuoles (green), ER (yellow) and peroxisomes (blue) in budding yeast. Detailed depictions of ER-mitochondria-peroxisome, nucleus –vacuole and mitochondria-vacuole contacts are shown in enlargements 1–3, respectively. ERMES and Ltc1 localize independently to ER-mitochondria contacts, shown in the bottom left enlargement. Ltc1 interacts with Tom70 and Tom71 via its GRAM domain (light blue) and is anchored into the ER via its predicted transmembrane domain. In ERMES, Mmm1 (green) homodimers are flanked by single Mdm12 SMP domains (purple) and this tetramer interacts with Mdm34 (turquoise) to potentially transport PC from the ER to mitochondria. The roles and interactions of the core ERMES and peripheral ERMES subunits, Mdm10 and Gem1, respectively, are not known. Peroxisomes are adjacent to ER-mitochondria contacts, possibly thorough an interaction with Mdm34. Enlarged in the top left are vCLAMPs and shown schematically are Vps39 and Ypt7, which are required for their formation, and Vps13. Both Vps39 and Vps13 are enriched in vCLAMP regions and Vps13 may reinforce vCLAMP formation. Whether specific mitochondrial components are required for vCLAMP formation or are enriched at these regions as part of a vCLAMP tether complex is not known. Enlarged in the lower right corner are types of ER-vacuolar contacts. Nuclear-vacuolar junctions (NVJs) are sites at which starvation-induced piecemeal microautophagy of the nuclear envelope occurs and are formed by a physical interaction between Nvj1 and Vac8. NVJs also contain Osh1, a sterol transport protein and the PH and SMP-domain containing protein, Nvj2, which binds to membranes, but its lipid ligands are not well established. Vac8 is also required to localize Ltc1/Lam6 to these sites and to non-NVJ ER-vacuole contacts required for stress induced vacuolar membrane domain formation. The conserved protein, Vps13 is localized to ER-vacuolar contacts in a regulated manner under respiratory growth conditions.
In addition to physically tethering mitochondria and ER, functional studies and, more compellingly, recent biochemical and structural studies of ERMES suggest the primary function of this complex is to transport phospholipids between the two organelles (AhYoung et al., 2015). Three of the four ERMES subunits, the ER localized Mmm1, the mitochondrial localized Mdm34 and the cytosolic Mdm12, possess SMP (synaptotagmin-like mitochondrial-lipid binding protein) domains. The remaining ERMES complex subunit, Mdm10, is a β-barrel protein that can separately interact with the mitochondrial outer membrane SAM (Sorting and Assembly Machinery) core complex, which functions in the insertion of all β-barrel outer membrane proteins. This suggests a role for ERMES, and of contact sites more generally, in the coordination of lipid and protein biogenesis.
SMP domain family members share two features: localization to interorganellar contacts sites and formation of a tubular lipid-binding protein (TULIP) domain structure, which is present in a diverse superfamily of proteins that can bind lipids and, in some cases, function in lipid transport (Schauder et al., 2014; Toulmay and Prinz, 2012). Recent structural analysis of an ERMES sub-complex containing Mmm1 and Mdm12 found that, similar to other SMP proteins, Mmm1 and Mdm12 form TULIP-like structures. The Mmm1 and Mdm12 TULIP domains interact to form a stable crescent-shaped head to tail hetero-tetramer containing two Mdm12 SMP domains flanking an Mmm1 SMP dimer, potentially creating an extended hydrophobic lipid transport cavity (AhYoung et al., 2015). Biochemical studies have shown that the Mmm1-Mdm12 complex preferentially interacts with PC, suggesting that it functions as a primary transporter of ER-derived PC for mitochondria. The crescent-shape of this complex and its position relative to the ER membrane may also serve to drive local membrane curvature and bilayer destabilization to facilitate lipid desorption at the ER and transport to mitochondria through the long central cavity of the tetramer. The mitochondrial SMP subunit of ERMES, Mdm34, was also shown to interact more weakly with the Mmm1-Mdm12 tetramer, suggesting the possibility that Mdm34 functions as the terminal mitochondrial lipid acceptor in the Mmm1-Mdm12 mediated relay. However, from these studies, it remains unclear if and how vectorial transport of PC occurs to mitochondria via ERMES and how other glycerophospholipids, such as PS and PA, reach mitochondria from the ER or other organelles. It is possible that transport of other lipid species is mediated by ERMES also and in particular, Mdm34, whose phospholipid binding specificity has not been determined, or through other lipid transfer proteins, such as proteins in the Osh family that have recently shown to bind PS (Maeda et al., 2013). Another possibility is the conserved ER Membrane Complex (EMC), which is comprised of six integral ER proteins. Deletion of multiple EMC proteins causes a synthetic growth phenotype when paired with mutations that disrupt ERMES and impairs transfer of PS from the ER-to-mitochondria, inferred from the rate at which PS is converted to PE (Lahiri et al., 2014). Although EMC proteins do not accumulate at ER-mitochondria contacts, they interact with the mitochondrial outer membrane protein, Tom5, a component of the mitochondrial protein import machinery. Thus, EMC proteins may function to augment ER-mitochondria tethering for mitochondrial lipid import.
A potential mechanism of sterol transport from the ER to mitochondria was recently revealed by the identification Ltc1/Lam6, a transmembrane ER protein localized to both ER-mitochondria and ER-vacuole contacts sites (Elbaz-Alon et al., 2015; Gatta et al., 2015; Murley et al., 2015) (Figure 1). Ltc1 also likely functions as a physical tether at mitochondria and vacuoles through its interactions with the mitochondrial protein import receptors, Tom71/70 or the membrane anchored vacuolar protein Vac8, respectively. Ltc1/Lam6 specifically catalyzes the transfer of sterol lipids between membranes via a VASt/StART-like domain (Murley et al., 2015). This domain is predicted to form a lipid-binding cavity similar in structure to the StART domains in lipid transport proteins such as CERT, which mediates transport of ceramides from the ER to the Golgi. Thus, similar to ERMES, Ltc1 potentially has the ability to create organelle contacts via its lipid transporting mechanism.
Mitochondrial membrane contacts: a mode of communication for cellular homeostasis
In light of the compelling biochemical evidence indicating a role of ERMES in phospholipids transport from the ER to mitochondria, it was puzzling why functional data supporting ERMES-mediated phospholipid transport from the ER to mitochondria has been less convincing with several groups reporting relatively mild or no defects in ER-mitochondria lipid exchange and/or in mitochondrial lipid homeostasis in cells lacking ERMES function. The basis of this apparent discrepancy is suggested by recent studies, which have found that multiple pathways can potentially sense and bypass ERMES and thus obscure its function(s) (Elbaz-Alon et al., 2014; Honscher et al., 2014; Lang et al., 2015; Tan et al., 2013). Analysis of the features of these alternative pathways suggests that they bypass ERMES by altering lipid homeostasis, consistent with lipid transport as a primary function of this tethering complex. For example, overexpression of the poorly characterized mitochondrial outer and inner integral proteins Mcp1 and Mcp2, respectively, suppresses the mitochondrial lipid and morphology defects associated with deletion of ERMES complex subunits (Tan et al., 2013).
Recent studies suggest that ERMES also can be bypassed by altering the lipid delivery pathway to mitochondria via contacts with organelles other than the ER, including the vacuole/lysosome (Elbaz-Alon et al., 2014; Honscher et al., 2014). The formation of mitochondria-vacuole contacts, termed vCLAMPs (vacuolar and mitochondrial patch), depends on the vacuolar proteins, Vps39, a component of the HOPS tethering complex, and the Rab GTPase, Ypt7 (Elbaz-Alon et al., 2014; Honscher et al., 2014) (Figure 1). Vps39 is enriched at vCLAMP, but whether it also functions as a component of a tether is not known as the identity of a mitochondrial lipid(s) and/or protein(s) component of vCLAMP required for interface formation has yet to be identified. Consistent with redundant roles for ERMES and vCLAMPs, mutations disrupting both contacts render cells non-viable and acute depletion of ERMES and vCLAMPs results in additive defects in mitochondrial lipid import. Overexpression of Vps39 expands the vCLAMP area in cells and can also rescue ERMES associated growth defects.
In addition to functional redundancy, evidence suggests that ERMES and vCLAMP sites are functionally linked and can coordinately respond to cellular needs. Loss of vCLAMP components increases the number of ERMES marked ER-mitochondrial contacts in cells and conversely loss of ERMES expands the area vCLAMP (Elbaz-Alon et al., 2014). The metabolic state of the cell also plays a role in the regulation of ERMES and vCLAMPs, as actively respiring cells have reduced vCLAMPs, corresponding to phosphorylated Vps39, and an increase in ERMES structures (Honscher et al., 2014). The basis of this regulation may involve the highly conserved peripheral membrane protein Vps13, as, similar to vCLAMPs, dominant mutations in Vps13 also suppress loss of ERMES function (Lang et al., 2015). In wild type cells grown in fermentable carbon sources, Vps13 localizes primarily to vCLAMPs, but is also observed at ER-vacuole junctions (Figure 1). Under respiratory conditions, however, the vast majority of Vps13 is localized to regions of ER-vacuole contacts in cells. Dominant mutated versions of Vps13 that suppress loss of ERMES function are detected only at vCLAMPs in cells and fail to localize to ER-vacuole contacts in non-fermentable carbon sources. Thus, although the molecular function of Vps13 is not understood, these observations suggest that Vps13 functions to preferentially stabilize vCLAMPs in cells. Suppression of ERMES defects mediated by dominant mutations in Vps13 also depends on the mitochondrial outer membrane ERMES multicopy suppressor, Mcp1, not the inner membrane suppressor Mcp2, suggesting that Vps13 and Mcp1 function as part of the same pathway (Lang et al., 2015). Thus, Vps13 might function together with Mcp1 at mitochondria to sense and augment lipid import into mitochondria from vacuoles via vCLAMPs in response to metabolic needs. In this context, it is interesting to note that vCLAMPs are also enriched for specific ion and amino acid transporters (Elbaz-Alon et al., 2014). Given that vacuoles are linked to amino acid sensing via regulation of the TORC1 pathway, and that mitochondria and vacuoles are connected metabolically via amino acids and additional metabolites, it is possible that vCLAMP function may extend beyond lipid transport to nutrient sensing and utilization.
The potential for additional cross-talk between mitochondria and vacuoles in yeast also potentially occurs via the sterol transporter, Ltc1 as it has been shown to perform separable functions at ER-mitochondria and ER–vacuole contact sites (Elbaz-Alon et al., 2015; Murley et al., 2015) (Figure 1). At mitochondria, Ltc1 functions with Tom70/71 in parallel with ERMES, as disruption of both contacts results in additive severe growth defects. In this context, it is interesting to note that although Ltc1 and ERMES independently localize to ER-mitochondria contacts, they form focal structures adjacent to one another within cells, potentially sharing an ER-mitochondria contact and contributing non-redundantly to the creation of a membrane domain. Indeed, the amount of sterols present in bulk mitochondria is relatively low and the function of sterol lipids in mitochondria is not well understood, except in the specialized case of steroid hormone production in mammals. However, given the well-established ability of sterols to promote the formation of lipid rafts, Ltc1 may function to facilitate membrane domain formation at ER-mitochondria contacts by enriching these sites with these lipids. Such a membrane domain defined by preferential lipid transport and interorganellar tethering dimensions could further recruit and mature a specialized subset of lipid and protein species to create a platform that triggers additional functions. This idea is reinforced by the observation that Ltc1 function at ER-vacuolar contacts is distinctly required for the formation of sterol-enriched vacuolar membrane domains under stress conditions (Murley et al., 2015).
Although the functions of Ltc1 at mitochondria and vacuoles are separate, shifting the population of Ltc1 to ER–vacuole contacts is sufficient to induce sterol-enriched vacuolar domains under normal growth conditions (Murley et al., 2015). In addition, evidence suggests that Ltc1 may also localize to vCLAMPs under conditions where vCLAMP area is expanded, potentially creating a tripartite ER-mitochondria-vacuole junction and Ltc1 is required for the expansion of vCLAMP sites in response to loss of ERMES (Elbaz-Alon et al., 2015). Thus, as with Vps13, the shared localization of Ltc1 to multiple organellar contacts may serve as a mode of interorganellar communication. The physiological pathways and mechanisms that potentially regulate the relative distribution of Ltc1 at these distinct contacts are not known. It is possible that, similar to the cross talk between ERMES and vCLAMP contacts, coordination between mitochondria and vacuoles via Ltc1 will be important for lipid homeostasis and nutrient sensing and signaling, which has been shown to be important to cellular replicative aging in yeast.
Mitochondrial metabolism might also be linked to peroxisomes by membrane contact sites. In yeast, peroxisomes are major sites of fatty acid β-oxidation, the product of which, acetylcarnitine, can be used to make acetyl-CoA and power the TCA cycle and drive respiration. Two recent studies have found that peroxisomes are juxtaposed to ERMES marked ER-mitochondria contacts and the pyruvate decarboxylase complex in the mitochondrial matrix, which generates the hub metabolite, acetyl-CoA (Cohen et al., 2014; Mattiazzi Usaj et al., 2015). The connection between mitochondria and peroxisomes is mediated, in part, by interactions between the mitochondrial localized ERMES subunit, Mdm34 and Pex11, which has been implicated as a peroxisomal membrane shaping protein. One interesting possibility is that ER-mitochondria-peroxisome contacts are promoted by starvation conditions, in which fatty acids replace sugars as the source of acetyl-CoA. Whether this occurs and how interactions between Mdm34 and Pex11 regulate these contacts is not known.
Beyond lipid transport -- mitochondrial contacts regulate mitochondrial dynamics and distribution
Mitochondria contact sites play roles beyond lipid and ion transport. In both yeast and mammalian cells, sites of contact between ER and mitochondria create hot spots for mitochondrial division (Friedman et al., 2011). Although the molecular basis of ER-associated mitochondrial division (ERMD) is not known in metazoans, in yeast, both ERMES and the peripheral ERMES subunit, Gem1, which is a conserved Miro GTPase that functions in microtubule based mitochondrial motility in metazoans, mark ER contacts destined to become mitochondrial division sites (Murley et al., 2013). Following ERMD, mitochondria rapidly separate from each other in a Gem1-dependent manner, suggesting it plays a similar role in the regulation of mitochondrial motility in yeast. In this capacity, Gem1 might function more directly by regulating turnover of the ERMES complex to remodel ER-mitochondria connections as mitochondrial motility in yeast is actin, not microtubule, dependent.
ERMES is also spatially linked to mtDNA nucleoids in the matrix of mitochondria, but it is not known whether this is a conserved feature of ERMD in metazoan cells (Meeusen and Nunnari, 2003; Murley et al., 2013). The molecular basis for the link between ER-mitochondria contacts and nucleoids is also not known, but one possible candidate is the MICOS complex. MICOS is a conserved six protein-containing complex associated with the inner membrane, which functions to mediate contact sites between the outer and inner mitochondrial membranes, where lipid and protein transport are thought to occur (Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011). In addition, evidence suggests that MICOS serves to control the distribution and copy number of cristae, which are inner membrane invaginations that house respiratory complexes (Friedman et al., 2015). If ER-mitochondria contacts are spatially linked to MICOS, they could function to couple events on the surface of mitochondria – division and Gem1-dependent mediated motility – to mtDNA inside mitochondria to ensure genome distribution throughout mitochondrial networks and into daughter cells.
Work in yeast suggests that mitochondria-organelle contact sites may have a general role in coordinating mitochondrial motility and positioning in collaboration with cytoskeletal factors. In this context, the bud specific protein, Mmr1 functions to both regulate actin based mitochondrial transport via interactions with the myosin motor, Myo2, and to anchor mitochondria to the cortical ER in the growing bud (Itoh et al., 2004; Swayne et al., 2011). Mitochondria are also anchored to the mother cell cortex in budding yeast via interactions with the cortical ER and PM. The mother specific mitochondria-ER cortex anchor, MECA, contains Num1, a BAR domain protein that interacts with the plasma membrane via a PIP2 lipid binding PH domain and Mdm36, which promotes the assembly of Num1 at regions of contact between mitochondria, the plasma membrane and the ER (Lackner et al., 2013). Mmr1 and MECA have opposing functions as loss of either tether can be suppressed by disruption of the other (Klecker et al., 2013). However, it is not clear if and how these structures communicate with one another and the identity of mitochondrial proteins or lipids to mediate tethering is also not known. However, the pervasive role of ER-mitochondria contacts as determinants of mitochondrial behavior and the possibility of communication amongst them raises the interesting idea that the ER functions to control the systems properties of mitochondria.
Mitochondrial contact sites in metazoans
The existence of ER-mitochondria contact sites in metazoan cells is firmly established by electron microscopy and time-lapse fluorescence microscopy, and is also suggested by the biochemical isolation of the MAM fraction, but the molecular basis of ER-mitochondria “tethering” remains poorly understood. In addition to the role of ER-mitochondria contacts in marking mitochondrial division, ER-mitochondrial contacts serve as platforms to coordinate ERMD with apoptosis and innate immunity stress signaling pathways as well as calcium uptake from the ER into mitochondria (reviewed in (Labbe et al., 2014)). Given these critical roles, the molecular identification of mammalian ER-mitochondria tethers and proteins enriched at contacts sites has increasingly become a major focus in cell biology. The dynamic nature of the ER and mitochondria and their interactions with one another make the definitive identification and validation of contact site components technically challenging. Indeed, in mammalian cells, the mitochondrial outer membrane dynamin-related protein MFN2, which directly mediates mitochondrial outer membrane fusion, is found in MAMs and has been proposed to function as an ER-mitochondria tether (de Brito and Scorrano, 2008). In addition to its presence in the MAM fraction, it localizes to discrete foci at regions of proximity between mitochondria and the ER and plays a role in Ca2+ homeostasis, a feature consistent with it being an ER-mitochondria contact site and/or tether component. However, more recent work reports that Mfn2 depletion increases ER-mitochondria contact and suggests that its role in Ca2+ homeostasis is more indirect through destabilization of the mitochondrial calcium uniporter, thus more work is necessary to understand the role of MFN2 in ER-mitochondria contact site biogenesis (Filadi et al., 2015).
Current advances in proximity labeling technology, high-resolution live cell imaging, and electron tomography techniques, have made the rigorous identification of contact site components more tractable. In addition, although ERMES is not conserved in metazoans, Ltc1/Lam6 homologs in mammalian cells GramD1a-c, the EMC proteins, as well as the mitochondrial division dynamin Drp1 and its receptor Mff, known to be enriched at contact sites, may be means for exploring the proteome of ER-mitochondria contact sites in mammalian cells. This approach is promising as it was used to identify Ltc1/Lam6 at ER-mitochondria contacts in yeast.
Outstanding questions
In addition to learning the composition of mitochondrial contacts sites in mammalian cells, there are several outstanding questions in this exciting area of cell biology. What are the signals and mechanisms that enable contacts sites to integrate multiple functions? Do they serve to create membrane domains that function as signaling platforms? How do distinct mitochondrial contact sites communicate with one another and with other interorganellar contacts? Do ER- mitochondria contacts fundamentally serve to distribute mtDNA in cells and if so, how do they couple events outside and inside mitochondria to accomplish this? Answering these questions will provide fundamental insights into cell function and cell dysfunction in disease.
Summary.
Membrane contact sites between mitochondria and other organelles are important for lipid and ion exchange, membrane dynamics and signaling. Recent advances are revealing their molecular features and how different types of mitochondria contacts are coordinated with each other for cell function.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AhYoung AP, Jiang J, Zhang J, Khoi Dang X, Loo JA, Zhou ZH, Egea PF. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E3179–3188. doi: 10.1073/pnas.1422363112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Klug YA, Dimitrov L, Erez Z, Chuartzman SG, Elinger D, Yofe I, Soliman K, Gartner J, Thoms S, et al. Peroxisomes are juxtaposed to strategic sites on mitochondria. Molecular bioSystems. 2014;10:1742–1748. doi: 10.1039/c4mb00001c. [DOI] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y, Eisenberg-Bord M, Shinder V, Stiller SB, Shimoni E, Wiedemann N, Geiger T, Schuldiner M. Lam6 Regulates the Extent of Contacts between Organelles. Cell reports. 2015;12:7–14. doi: 10.1016/j.celrep.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M. A dynamic interface between vacuoles and mitochondria in yeast. Developmental cell. 2014;30:95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Filadi R, Greotti E, Turacchio G, Luini A, Pozzan T, Pizzo P. Mitofusin 2 ablation increases endoplasmic reticulum-mitochondria coupling. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E2174–2181. doi: 10.1073/pnas.1504880112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science (New York, NY) 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Mourier A, Yamada J, McCaffery JM, Nunnari J. MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. eLife. 2015;4 doi: 10.7554/eLife.07739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta AT, Wong LH, Sere YY, Calderon-Norena DM, Cockcroft S, Menon AK, Levine TP. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. eLife. 2015;4 doi: 10.7554/eLife.07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M, Korner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, Neupert W. The mitochondrial contact site complex, a determinant of mitochondrial architecture. The EMBO journal. 2011;30:4356–4370. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, van der Laan M, Cabrera M, Reggiori F, Ungermann C. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Developmental cell. 2014;30:86–94. doi: 10.1016/j.devcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. The Journal of cell biology. 2011;195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Toh EA, Matsui Y. Mmr1p is a mitochondrial factor for Myo2p-dependent inheritance of mitochondria in the budding yeast. The EMBO journal. 2004;23:2520–2530. doi: 10.1038/sj.emboj.7600271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klecker T, Scholz D, Fortsch J, Westermann B. The yeast cell cortical protein Num1 integrates mitochondrial dynamics into cellular architecture. Journal of cell science. 2013;126:2924–2930. doi: 10.1242/jcs.126045. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science (New York, NY) 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe K, Murley A, Nunnari J. Determinants and functions of mitochondrial behavior. Annual review of cell and developmental biology. 2014;30:357–391. doi: 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- Lackner LL, Ping H, Graef M, Murley A, Nunnari J. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E458–467. doi: 10.1073/pnas.1215232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S, Chao JT, Tavassoli S, Wong AK, Choudhary V, Young BP, Loewen CJ, Prinz WA. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS biology. 2014;12:e1001969. doi: 10.1371/journal.pbio.1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AB, John Peter AT, Walter P, Kornmann B. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. The Journal of cell biology. 2015;210:883–890. doi: 10.1083/jcb.201502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Anand K, Chiapparino A, Kumar A, Poletto M, Kaksonen M, Gavin AC. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature. 2013;501:257–261. doi: 10.1038/nature12430. [DOI] [PubMed] [Google Scholar]
- Mattiazzi Usaj M, Brloznik M, Kaferle P, Zitnik M, Wolinski H, Leitner F, Kohlwein SD, Zupan B, Petrovic U. Genome-Wide Localization Study of Yeast Pex11 Identifies Peroxisome-Mitochondria Interactions through the ERMES Complex. Journal of molecular biology. 2015;427:2072–2087. doi: 10.1016/j.jmb.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S, Nunnari J. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. The Journal of cell biology. 2003;163:503–510. doi: 10.1083/jcb.200304040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A, Lackner LL, Osman C, West M, Voeltz GK, Walter P, Nunnari J. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A, Sarsam RD, Toulmay A, Yamada J, Prinz WA, Nunnari J. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. The Journal of cell biology. 2015;209:539–548. doi: 10.1083/jcb.201502033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TT, Lewandowska A, Choi JY, Markgraf DF, Junker M, Bilgin M, Ejsing CS, Voelker DR, Rapoport TA, Shaw JM. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic (Copenhagen, Denmark) 2012;13:880–890. doi: 10.1111/j.1600-0854.2012.01352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder CM, Wu X, Saheki Y, Narayanaswamy P, Torta F, Wenk MR, De Camilli P, Reinisch KM. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 2014;510:552–555. doi: 10.1038/nature13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne TC, Zhou C, Boldogh IR, Charalel JK, McFaline-Figueroa JR, Thoms S, Yang C, Leung G, McInnes J, Erdmann R, et al. Role for cER and Mmr1p in anchorage of mitochondria at sites of polarized surface growth in budding yeast. Current biology: CB. 2011;21:1994–1999. doi: 10.1016/j.cub.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T, Ozbalci C, Brugger B, Rapaport D, Dimmer KS. Mcp1 and Mcp2, two novel proteins involved in mitochondrial lipid homeostasis. Journal of cell science. 2013;126:3563–3574. doi: 10.1242/jcs.121244. [DOI] [PubMed] [Google Scholar]
- Toulmay A, Prinz WA. A conserved membrane-binding domain targets proteins to organelle contact sites. Journal of cell science. 2012;125:49–58. doi: 10.1242/jcs.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Malsburg K, Muller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, Becker T, Loniewska-Lwowska A, Wiese S, Rao S, Milenkovic D, et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Developmental cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]