Abstract
Mitochondrial DNA (mtDNA) encodes RNAs and proteins critical for cell function. In human cells, 100–1000s mtDNA copies are replicated asynchronously, packaged into protein-DNA nucleoids, and distributed within a dynamic mitochondrial network. The mechanisms that govern how nucleoids are chosen for replication and distribution are not understood. Mitochondrial distribution depends on division, which occurs at endoplasmic reticulum (ER)-mitochondria contact sites. These sites were spatially linked to a subset of nucleoids selectively marked by mtDNA polymerase and engaged in mtDNA synthesis, events that occurred upstream of mitochondrial constriction and division machine assembly. Our data suggest that ER tubules proximal to nucleoids were necessary but not sufficient for mtDNA synthesis. Thus, ER-mitochondria contacts coordinate licensing of mtDNA synthesis with division to distribute newly replicated nucleoids to daughter mitochondria.
Main Text
Mutations in mitochondrial DNA (mtDNA) genes and in nuclear genes that control mtDNA maintenance cause mitochondrial dysfunction and are linked to human disease and aging, underscoring the functional importance of mtDNA (1–4). Nucleoids are the protein-DNA structures used for mtDNA inheritance in cells (5). Accurate maintenance of mtDNA requires replication, repair, packaging and, distribution of the mitochondrial nucleoid at the cellular level. In mammalian cells, mitochondrial DNA replication is mediated by a nuclear-encoded replisome comprised of a polymerase gamma holoenzyme, containing a catalytic subunit (POLG1) and a processivity subunit (POLG2) (6–10). In addition to the polymerase complex, the replisome contains the helicase Twinkle and a mitochondrial-specific single stranded DNA binding protein, which together facilitate the formation of a single stranded DNA replication template (11, 12). Within cells, mtDNA is packaged into a nucleoid by TFAM, a nuclear-encoded DNA bending protein, which also plays a role in mtDNA replication and transcription (13–18).
Although the molecular players involved in mtDNA replication and packaging have been described, the mechanisms underlying the spatial regulation of mtDNA replication and intracellular nucleoid distribution have been elusive. This is in part due to the fact that mtDNA is present in cells in multiple copies and the spatiotemporal regulation of its replication and distribution is relaxed in comparison to the nuclear genome (19). Indeed, mtDNA replication occurs asynchronously with the cell cycle and within post-mitotic tissues such as the brain and muscle (20, 21). Nucleoids are evenly distributed within mitochondria and constrained in their motility (5, 22). Mitochondrial distribution is in large part dependent on cytoskeletal-based motility and on mitochondrial division(23), mediated in mammalian cells by DRP1, a cytosolic dynamin-related GTPase that forms assemblies around mitochondria to facilitate membrane scission (24, 25). DRP1 recruitment and assembly occur at sites of endoplasmic reticulum (ER)-mitochondrial contact, where mitochondrial constriction is also observed (26). Perturbation of mitochondrial division in both yeast and mammalian cells causes nucleoid aggregation, mtDNA deletions and mtDNA depletion, suggesting a fundamental and functional link between mitochondrial division and mtDNA maintenance (27–29). In yeast, ER-linked division sites, marked by the fungal specific ER-mitochondria encounter structure (ERMES) complex, are spatially linked to nucleoids, further suggesting a role for ER-mitochondria contacts in mtDNA maintenance(30). Here we asked whether ER-mitochondria contact sites function to couple mtDNA replication with mitochondrial division for the purpose of distributing newly replicated mtDNA in human cells.
ER-mitochondria contacts and ER-associated mitochondrial division are spatially linked to nucleoids in mammalian cells
We asked if ER-associated mitochondrial division (ERMD) events are spatially linked to mitochondrial nucleoids in human cells by simultaneously imaging mitochondria, nucleoids and the ER network at high spatial and temporal resolution using spinning disk confocal microscopy. Osteosarcoma cells (U2OS) were transiently transfected with Green Fluorescent Protein (GFP)-tagged TFAM (TFAM-GFP), a well-characterized marker of the total nucleoid population, mitochondrial matrix targeted Blue Fluorescent Protein (mito-BFP) and ER targeted mRuby (mRuby-KDEL). TFAM-GFP labeled foci were evenly spaced within mitochondria as previously described for nucleoids in other cell types [Fig. 1A](5, 16, 18). TFAM-GFP labeled nucleoids were also localized adjacent to points where ER tubules crossed over mitochondria in a perpendicular fashion [Fig. 1A, right panel], and a subset of nucleoids remained stably linked to ER-mitochondria contacts over time, despite ER network remodeling and mitochondrial motility [Fig. 1B, arrow]. We further assessed the spatial link between ER-mitochondria contacts and nucleoids by determining the Pearson correlation coefficient of mRuby-KDEL and TFAM-GFP fluorescence intensity along linescans of mitochondria imaged in live U2OS cells (n=58). Consistent with our observations, this analysis indicated a highly significant enrichment of ER signal specifically within 17 pixels (approximately 1 μm) laterally adjacent to nucleoids [Pearsons’ R=0.59; Fig. S1A]. Thus, in general, nucleoids are spatially linked to ER-mitochondria contacts in human cells.
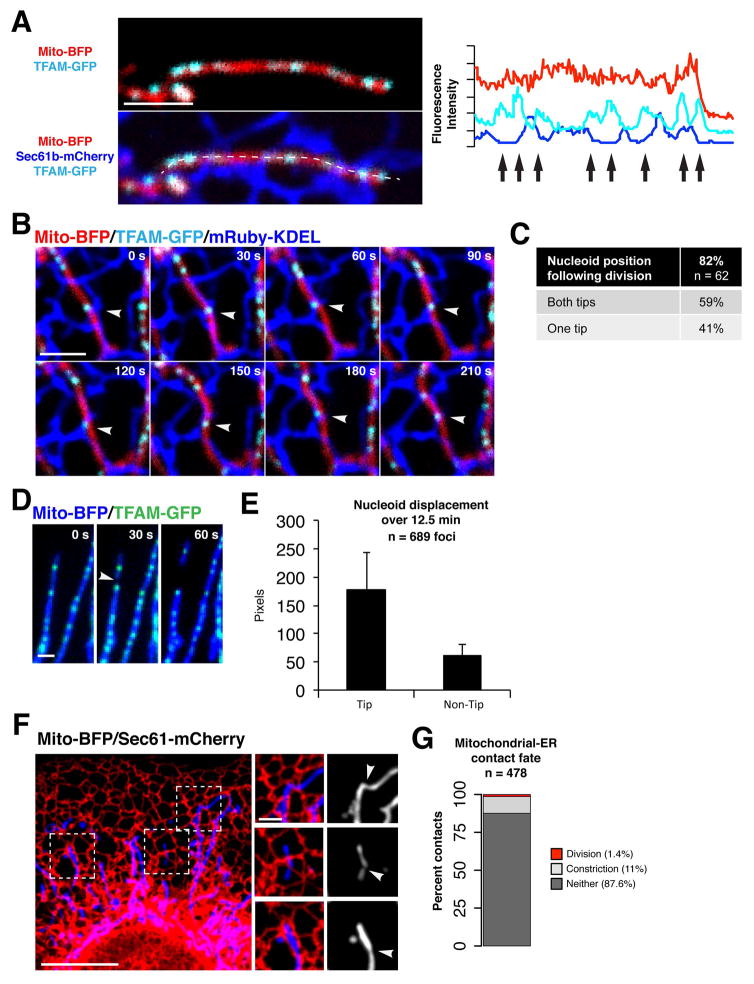
Figure 1. Mitochondrial DNA nucleoids are spatially linked to mitochondria-ER contacts in human cells.
(A) Left panels show a merged image of a live U2OS cell expressing mito-BFP, TFAM-GFP and Sec61b-mCherry (ER). Right panel shows the pixel intensity of mito-BFP, TFAM-GFP and Sec61b-mCherry from a line scan drawn along the mitochondrial tubule (dotted line), arrows indicate nucleoid positions. (B) Time-lapse images of a U2OS cell expressing mito-BFP, TFAM-GFP and mRuby-KDEL (ER), a single plane is shown. Arrow indicates a site of persistent co-localization between a TFAM-GFP labeled nucleoid and an ER-mitochondria contact. (C) Number of mitochondrial divisions in U2OS cells spatially linked to TFAM-GFP labeled nucleoids, from 43 cells. (D) Time-lapse images of mitochondrial division (marked by arrow) spatially linked to a TFAM-labeled nucleoid focus in a U2OS cell. (E) Graph indicating the displacement of picogreen-labeled nucleoids in live Cos7 cells over 12.5 minutes as a function of their intra-mitochondrial position. Data are mean +/− SD. (F) Merged image of a live U2OS cell expressing mito-BFP and Sec61b-mCherry (ER). Right panels show examples of mitochondrial constrictions co-localized with ER tubules. (G) Graph indicating the percentage of persistent mitochondrial-ER co-localizations that become sites of mitochondrial constriction or division over 5 minutes in live U2OS cells. Scale bars: (A, B, D, F inset) 2 um; (F) 10 um.
Accordingly, we observed that a majority of ERMD events (82%, n=62) were spatially linked to nucleoids (within 1 μm distance), resulting in their localization at mitochondrial tips following division [Fig. 1C and D, arrow]. We also performed time lapse imaging of nucleoids and mitochondria in Cos7 primate cells labeled with the selective vital dyes, Picogreen DNA stain and Mitotracker Red, respectively. Retrospective nucleoid tracking over time revealed that nucleoids at mitochondrial tips had been displaced a greater distance on average (3X greater over a time period of 12.5 min.) than intra-mitochondrial nucleoids, suggesting that the subset of nucleoids linked to ERMD sites are preferentially distributed within cells [Fig. 1E].
ER-mitochondria contacts are not rate limiting for mitochondrial division
The high density of ER and mitochondrial networks in mammalian cells suggested that ER-mitochondria contact sites may not be rate-limiting for ERMD. To test this, we defined and quantified persistent ER-mitochondria contacts in cells by time-lapsed imaging of mitochondria and ER for 5 minutes at 15 sec intervals using mito-BFP and mRuby-KDEL, respectively, in U2OS cells. ER and mitochondria were segmented in time-lapse images by thresholding and regions of overlap between the organelles were identified and tracked over 5 min (see Fig. S1B, arrows). Most regions of ER-mitochondria co-localization were transient, but approximately 100 distinct regions per cell were identified in which the ER and mitochondria persistently co-localized over the duration of imaging [Fig. S1C]. We followed the fate of persistent ER-mitochondria co-localized regions in relation to mitochondrial division over the duration of imaging. At a small fraction of these regions, mitochondrial constrictions were observed and/or developed [Fig. 1F, arrows]. An even smaller fraction of ER-mitochondria co-localized regions was linked to division events, consistent with published observations indicating that ER-linked mitochondrial constriction precedes division [Fig. 1G](26). Thus, although there are many ER-mitochondria contacts, only a subset are destined to be linked to ERMD. Given that ER-mitochondria contacts are also linked spatially to nucleoids in general [Fig. S1A], we considered whether a functionally specialized subset of nucleoids marks ERMD events in cells.
Nucleoids engaged in mtDNA synthesis mark nascent mitochondrial division sites at ER-mitochondria contacts
It has been proposed that functional sub-populations of nucleoids co-exist in mammalian cells, distinguished on the basis of mtDNA replication and/or transcription status(31, 32). In yeast, a sub-population of nucleoids are spatially linked to ERMES foci, which in turn mark a fraction of mitochondrial division sites(30, 33). Thus, we tested whether in mammalian cells mtDNA synthesis is specifically coupled to ERMD, which might help to ensure the segregation of nascent mtDNA to daughter mitochondria.
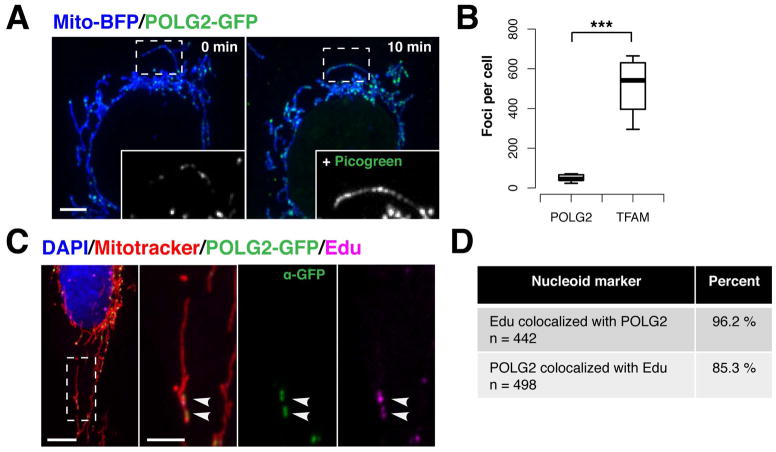
To visualize nucleoids engaged in mtDNA synthesis in cells, we employed POLG2-GFP, a fluorescently tagged version of the processivity subunit of the human mitochondrial DNA polymerase holoenzyme, whose enzymatic properties and affinity for mtDNA are indistinguishable from untagged POLG2 in vitro (34). 16 hours after transfection of live U2OS cells, POLG2-GFP labeled a subset of the total nucleoid population within mitochondria, which was revealed by the addition of Picogreen DNA stain and subsequent re-imaging [Fig. 2A and Fig. S2A]. Consistent with this, comparison of the total number of nucleoids labeled with TFAM-GFP to the number of POLG2-GFP labeled nucleoids in live U2OS cells indicated that there were significantly less POLG2-GFP foci per cell (9.4% of total nucleoids) [Fig. 2B]. To test whether the POLG2-labeled nucleoids were selectively engaged in mtDNA synthesis, live cells expressing POLG2-GFP and labeled with Mitotracker Red, were incubated with the nucleotide thymidine analog 5-ethynyl–2-deoxyuridine (EdU) for 1 hour, which was subsequently visualized in fixed cells with AlexaFluor647 using copper click chemistry(35). The vast majority of POLG2-GFP foci (as detected via α-GFP-AlexaFluor488) co-localized with detectable EdU incorporation at nucleoids within mitochondria (96% from 15 cells) [Fig. 2C and D]. Consistently, there was no significant difference between the number of EdU or POLG2-GFP foci per cell in either live or fixed cells [Fig. S2B]. We also assessed whether expression of POLG2-GFP perturbed mtDNA maintenance by examining mtDNA copynumber by quantitative Polymerase Chain Reaction (qPCR). We detected no difference in mtDNA levels between cells expressing POLG2-GFP versus mock-transfected cells [Fig. S2C]. In contrast, and as previously shown, a significant increase in mtDNA copy number was observed in cells overexpressing TFAM-GFP, indicating that U2OS cells were capable of modulating mtDNA synthesis during our experiments (13, 36). Thus, exogenously expressed POLG2-GFP is recruited to the endogenous mtDNA replisome providing a sensitive and highly specific marker for monitoring mtDNA synthesis in live cells.
Figure 2. POLG2-GFP is specifically recruited to replicating nucleoids in live cells.
(A) A live U2OS cell expressing mito-BFP and POLG2-GFP was imaged (left panel), then stained with Picogreen DNA dye and re-imaged ten minutes later (right panel). Inset is 488 channel signal intensity in an example mitochondrion, left panel, and the same organelle after Picogreen staining, right panel. Magnified 2X. (B) The number of mitochondrial POLG2-GFP (n= total 441 foci from 10 cells) or TFAM-GFP foci (n=5,182 foci from 10 cells) per U2OS cell (***p<0.001, two-tailed t test). Data are mean +/− SD. (C) Representative image of a U2OS cell expressing POLG2-GFP and pulse-labeled with 50mM EdU, fixed and stained with DAPI (DNA, blue), Mitotracker (mitochondria, red), anti-GFP AlexaFluor488 conjugate antibody (POLG2-GFP, green), and ClickIt Edu-AlexaFluor647 (nascent DNA, magenta). (D) Observed co-localization between mitochondrial POLG2-GFP and EdU foci in fixed U2OS cells labeled as in C, from 15 cells. Scale bars: (A, C) 10 um, (C inset) 5 um.
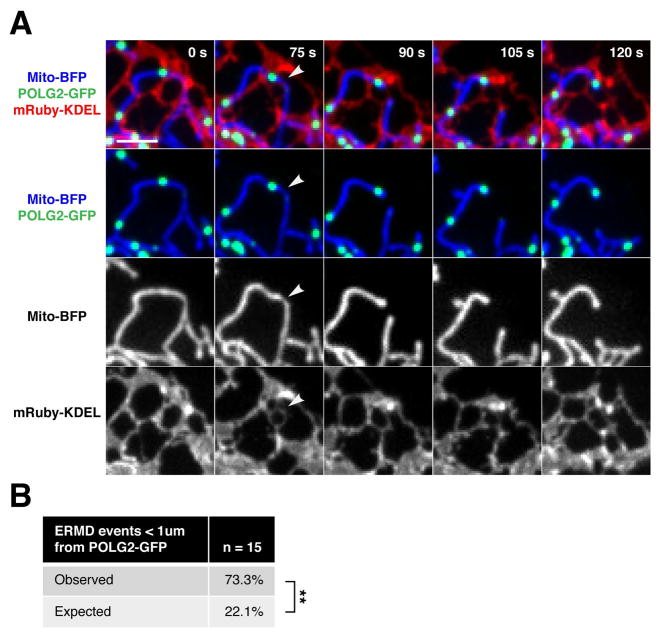
With this tool in hand, we asked whether nucleoids engaged in mtDNA synthesis, labeled faithfully by POLG2-GFP, were spatially linked to ERMD in a selective manner. As shown in a representative time-lapse series [Fig. 3A], POLG2-GFP–labeled nucleoids were indeed spatially linked to mitochondrial division at ER-mitochondria contact sites in live U2OS cells, as labeled by mRuby-KDEL and mito-BFP markers, respectively. A majority of ERMD events were linked to POLG2 labeled nucleoids (within 1 μm) and occurred at an over three-fold higher rate in cells than expected from random chance at persistent ER-mitochondria contacts [Fig. 3B]. In addition, a comparable proportion of ERMD events were linked to nucleoids in cells expressing either the general nucleoid marker TFAM-GFP, or the replication-specific marker POLG2-GFP [Fig. 1C and 3B, 82% v 73%, p-val =0.71]. Thus, nucleoid-linked ERMD events seem to occur predominantly at nucleoids engaged in mtDNA synthesis.
Figure 3. Replicating nucleoids mark sites of ER-mediated division.
(A) Representative time-lapse images of a U2OS cell expressing mito-BFP, mRuby-KDEL (ER) and POLG2-GFP demonstrating mitochondrial division at a mitochondrial-ER contact site spatially linked to POLG2-labeled nucleoid. (B) Percentage of ERMD events (marked by mRuby-KDEL and mito-BFP as in E) in live U2OS cells that occurred within one micron of a POLG2-GFP labeled nucleoid (n = 15 events from 7 cells, **p<.01, Fisher’s exact test). Significantly more division events occurred at pre-existing POLG2-GFP foci (73.3%) than expected by random chance at stable ER-mitochondria contacts (22.1%). Scale bar: 2um.
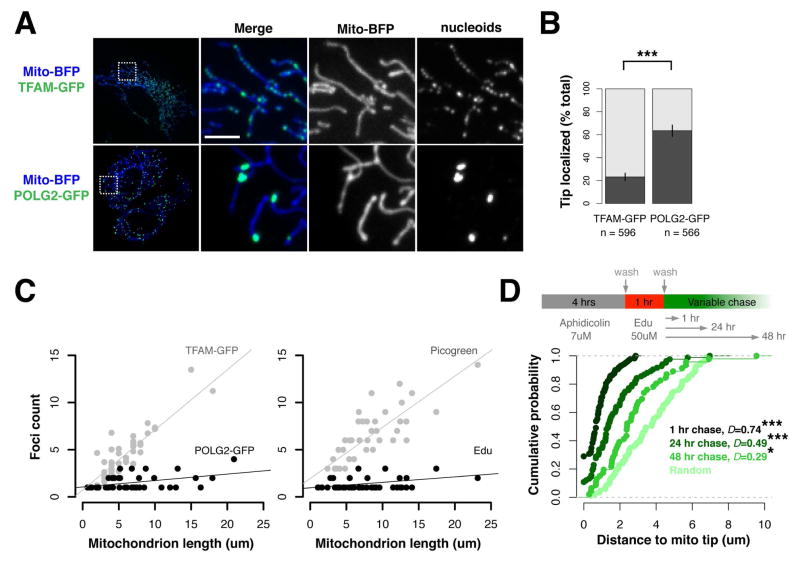
To further test the idea that nucleoid-linked ERMD events occur predominantly at nucleoids actively engaged in mtDNA synthesis, we exploited the observation that POLG2-GFP nucleoids localized at the tips of daughter mitochondria as a consequence of ERMD [Fig. 3A]. Steady state analysis of nucleoid position in U2OS cells revealed a highly significant spatial enrichment of POLG2-GFP labeled nucleoids within 1 micron of mitochondrial tips, as compared to the total TFAM-GFP labeled nucleoid population [Fig. 4A and B], despite the nearly tenfold greater number of TFAM-GFP labeled nucleoids detected per cell [Fig. 2B]. To validate that our observations were generally reflective of mammalian mtDNA segregation, we analyzed two additional cell lines: non-cancerous ARPE19 retinal epithelial cells and Cos7 primate fibroblasts. As in U2OS cells, POLG2-GFP-labeled nucleoids were enriched at mitochondrial tips in these additional lines, where they were also co-localized with focal EdU labeling in fixed cells [Fig. S3, A and B]. In addition, we reasoned that if mtDNA replication was selectively linked to ERMD, the number of POLG2-GFP and/or EdU-labeled nucleoids per mitochondrion would be constrained to the number of mitochondrial tips created by mitochondrial division, as opposed to scaling to the total length of the organelle. Thus, we examined the distribution of replicating and total nucleoids per mitochondrion in live U2OS cells (labeled with TFAM-GFP or POLG2-GFP) and in fixed ARPE19 cells (labeled with Picogreen and EdU) [Fig. 4C, left and right panels, respectively]. Indeed, consistent with the relatively uniform distribution of nucleoids within mitochondria [Fig. 1A], individual organelles contained numerous TFAM-GFP or Picogreen labeled nucleoids and the total number of nucleoids per mitochondrion was highly correlated with the length of the organelle [Fig. 4C]. In contrast, the number of POLG2-GFP or EdU labeled nucleoids per organelle was not well correlated with mitochondrial length and in a majority of instances there was a clear constraint on the number of EdU or POLG2-GFP labeled nucleoids to 2 or less per organelle, regardless of organelle length [Fig. 4C]. Closer examination of outlier EdU or POLG2 labeled nucleoids in mitochondria that contained greater that the median number of nucleoids revealed that these organelles were either branched, and consequently possessed a greater number of tips with associated EdU or POLG2 labeled nucleoids, or were unbranched and contained an additional internally localized pair of EdU foci [Fig. S4]. Internal EdU foci pairs were closely spaced, suggestive of a segregation intermediate. Thus, in mammalian cells, a majority of ERMD events are spatially linked to the subset of nucleoids that are actively engaged in mtDNA replication, consistent with a role for ERMD in the coordinated segregation of nascent mitochondrial genomes.
Figure 4. Nascent mtDNA is segregated to daughter mitochondria by division.
(A) Representative image of live U2OS cells expressing mito-BFP and TFAM-GFP, (top panels) or POLG2-GFP (bottom panels). (B) Significant spatial enrichment of the total population of POLG2-GFP foci within one micron of mitochondrial tips, as compared to TFAM-GFP foci (in dark grey) in live U2OS cells as labeled and imaged in (A); ***p<.001, two-tailed t test. (C) Left panel: In live U2OS cells, the number of TFAM-GFP foci per mitochondrion, but not POLG2-GFP foci, is correlated with mitochondrion length. Linear regression with best-fit line is shown. For TFAM-GFP, adjusted R2 = 0.86, Pearson’s R = 0.92 (****p<.0001). For POLG2-GFP, adjusted R2= 0.15, Pearson’s R= 0.41 (n.s.). Right panel: In fixed Arpe19 cells, the number of Picogreen foci per mitochondrion, but not Edu foci, is correlated with mitochondrion length. Linear regression as in A. For Picogreen, adjusted R2= 0.68, Pearson’s R=0.82 (****p<.0001). For Edu, adjusted R2 =0.07, Pearson’s R= 0.31 (n.s.). (D) Top panel, diagram of Edu pulse-chase experiments in Arpe19 cells. Bottom, empirical cumulative distribution analysis of Edu focus position along mitochondria, demonstrating depletion of pulse-labeled nascent mtDNA from mitochondrial tips over time, towards a simulated random distribution (***p<.001, **p<.01, Kolmogorov-Smirnov test). Scale bar: 5 um.
To rigorously test this model and determine the fate of replicating nucleoids linked to ERMD, we performed an EdU pulse chase analysis of mtDNA in cells under native conditions, in the absence of POLG2-GFP expression [Fig. 4D, upper panel]. We pulse labeled ARPE19 cells with EdU for 1 hour under conditions where the replication of nuclear DNA was inhibited, and chased for 1, 24 or 48 hours and subsequently analyzed the position of EdU-labeled nucleoids relative to mitochondrial tips after fixation. Consistent with our previous observations [Figs. 3A–B, and 4A–B], EdU nucleoids detected during the pulse and after the 1 hour chase were highly enriched within 1 micron of mitochondrial tips [Fig. 4D and Fig. S5A–C, representative images]. Quantification revealed that the positioning of EdU-labeled nucleoids relative to tips progressively decreased after the 24 and 48 hours chase, towards a random distribution [Fig. 4D]. Thus, replicating nucleoids are indeed both spatially and temporally linked to mitochondrial division and following the completion of mtDNA replication, nascent mtDNA is segregated into daughter mitochondria and subsequently distributed away from preceding division sites.
The mtDNA replisome is an early marker of nascent mitochondrial division sites
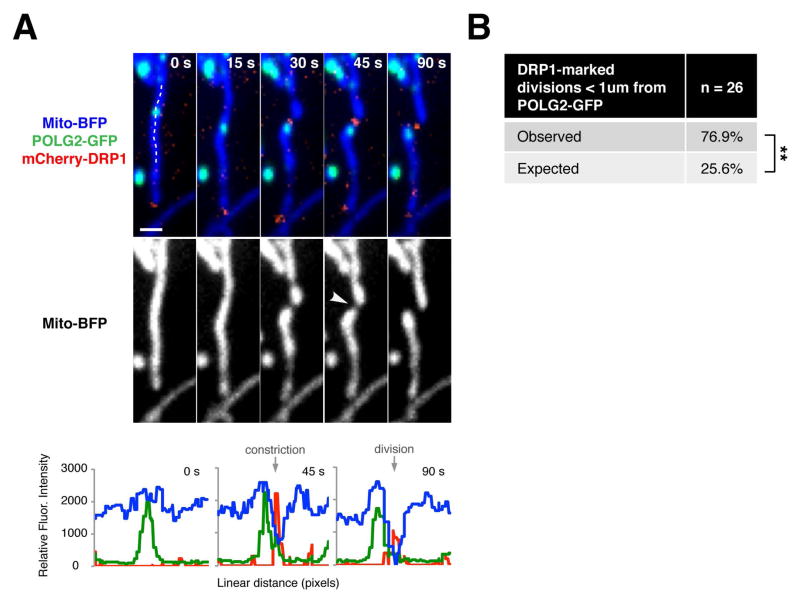
To gain further insight into the relationship between ERMD and replicating nucleoids, we further examined the temporal relationship of active mtDNA synthesis to known mitochondrial division events: ER-linked mitochondrial constriction and recruitment of the mitochondrial division dynamin, DRP1 (26). We performed time-lapse microscopy to assess the relationship of POLG2-GFP labeled nucleoids to mitochondrial division events in U2OS cells co-expressing mito-BFP, and either mCherry-DRP1 or mRuby-KDEL. Consistent with our previous observations, a majority of mitochondrial division events marked by DRP1 assemblies occurred within 1 micron of a POLG2-GFP focus [Fig. 5A–B]. To further define the spatial link between replicating nucleoids and DRP1-marked division sites in live cells, we quantified the distance from the center of each POLG2-GFP focus to the position of matrix marker discontinuity, for 26 division events. We observed that division sites were enriched within a zone greater than 200 nm, but less than 400 nm, from a POLG2-GFP focus [Fig. S4]. In every division event marked by mCherry-DRP1 (100%, n=26), POLG2-GFP-labeled nucleoids marked a future division site prior to mitochondrial constriction and also preceded the recruitment of mCherry-DRP1 to mitochondrial constrictions [Fig. 5B]. Indeed, for some division events, POLG2-GFP was detected at a future division site over 15 minutes prior to the recruitment of mCherry-DRP1. Thus, replication of mtDNA precedes known events linked to mitochondrial division.
Figure 5. Replicating nucleoids mark division sites prior to mitochondrial constriction or DRP1 recruitment.
(A) Top panel, time-lapse images of a U2OS cell expressing mito-BFP, mCherry-DRP1 and POLG2-GFP. Arrow indicates site of division. Bottom panel, linescan drawn along the mitochondrial tubule to show relative fluorescence intensity of mitochondria, DRP1 division machinery and POLG2 signal for timepoints t = 0s (pre-constriction), t = 45s (post-constriction), and t = 90s (post-division). Scale bar: 1 um. (B) Table indicating the percentage of mitochondrial divisions (marked by mCherry-DRP1 as in A) that occur within one micron of a POLG2-GFP focus, in live U2OS cells (n = 26 events from 22 cells, **p<.01, Fisher’s exact test).
ER structure is required for mtDNA replication licensing and nucleoid distribution
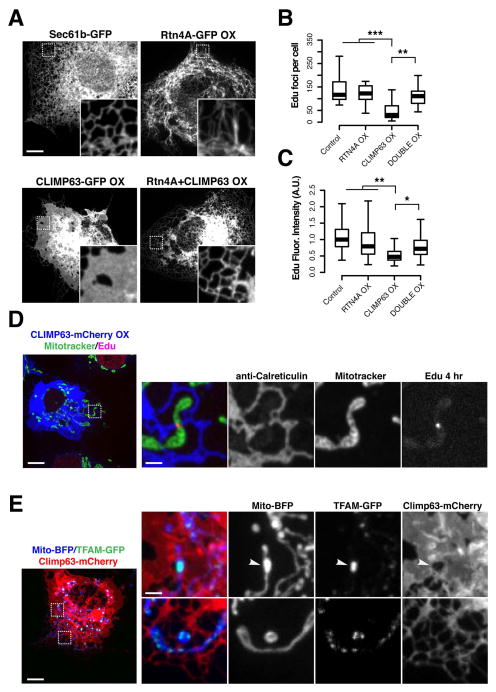
The spatio-temporal relationship between replicating nucleoids and nascent ERMD sites prompted us to further examine the role of the ER in mtDNA replication and nucleoid distribution in cells. In addition to the nuclear envelope, the peripheral ER forms a dense and dynamic network of interconnected tubules and sheet-like structures(37–39). Members of the conserved reticulon protein family (RTN1, RTN2, RTN3, and RTN4/Nogo) are thought to be key factors in determining the ratio of ER tubules and sheet like structures by localizing to and stabilizing highly curved ER tubules and edges of ER sheet-like structures (40, 41). The coiled-coil membrane protein CLIMP63 is enriched in sheet-like ER structures where is it is thought to maintain the lumenal distance between ER membrane bilayers(41–43). Overexpression of the ER shaping proteins RTN4 or CLIMP63 in mammalian cells shifts the proportion of tubules to sheet-like structures or sheet-like structures to tubules, respectively(40, 41, 44). Thus, we used transient over-expression of CLIMP63 or RTN4A in Cos7 cells to manipulate ER structure and examine its potential role in nucleoid replication and distribution. As expected, acute overexpression of fluorescently tagged RTN4A or CLIMP63 caused a dramatic increase in the proportion of tubular ER or sheet-like ER structures respectively, as compared to control cells expressing SEC61-mCherry, an ER membrane marker with no known role in membrane morphogenesis [Fig. 4A] (40). In contrast and consistent with previous observations, ER morphology in cells simultaneously overexpressing CLIMP63-GFP and RTN4A-GFP was similar to control cells, consistent with a dynamic balance between the tubules and sheet-like structures within ER networks [Fig. 6A](41).
Figure 6. ER tubules license mtDNA synthesis and are required for nucleoid distribution.
(A) ER network morphologies in representative Cos7 cells expressing fluorescently tagged ER membrane proteins: Sec61b-GFP, upper left; RTN4A-GFP, upper right; CLIMP63-GFP over-expression, lower left; or Rtn4A-GFP and Climp63-mCherry double over-expression, lower right. Inset regions magnified 5X. (B) Quantification of the number of mitochondrial EdU foci per fixed Cos7 cell following a 4 hour pulse of 50mM EdU, in cells labeled with Mitotracker Red and indicated ER markers, n = 15+ cells per condition (***p<.001, **p<.01, two-tailed t test). (C) Quantification of fluorescence intensity of mitochondrial Edu foci, n = 600+ foci from 15+ cells per condition. (D) Image of fixed Cos7 cell over-expressing CLIMP63-mCherry (ER) following a 4 hour pulse of 50mM EdU in cells labeled with Mitotracker Red. (E) Image of a live Cos7 cell expressing mito-BFP (mitochondria), TFAM-GFP (nucleoids), and over-expressing CLIMP63-mCherry (ER). Left, full field view of entire cell; right, examples of aggregated nucleoids associated with sheet-like ER (top panels) and distributed nucleoids associated with reticular ER (bottom panels). Scale bars: (A) 10 um, (D, E) 10 um, (D inset, E inset) 2 um.
In Cos7 labeled with Mitotracker Red and overexpressing CLIMP63 or RTN4 or both, EdU pulse labeling of mtDNA was used to assess the number of nucleoids engaged in mtDNA synthesis within a four hour window. Post-fixation, EdU-labeled nucleoids were detected in cells using AlexaFluor647 and total nucleoids and the ER were visualized using Picogreen staining and indirect immunofluorescence with anti-Calreticulin antibody, respectively. Relative to control cells, there was a significant reduction in the number of EdU-labeled nucleoids in the cell population overexpressing CLIMP63-mCherry, but not RTN4A-GFP [Fig. 6B]. In addition, the fluorescence intensity of AlexaFluor647 at EdU-labeled nucleoids was significantly reduced in CLIMP63-overexpressing cells depleted of tubular ER in comparison to control and RTN4A-GFP overexpressing cells [Fig. 6C]. This observation indicates that the reduced number of apparent EdU foci in CLIMP63 overexpressing cells was not due to aggregation of replicating nucleoids. Moreover, in cells simultaneously overexpressing CLIMP63-mCherry and RTN4A-GFP, the number and fluorescence intensity of EdU labeled nucleoids and ER morphology was similar to control cells, suggesting that the nucleoid phenotypes in CLIMP63 overexpressing cells were a consequence of changes in ER structure and not CLIMP63 expression per se [Fig. 6B–C]. Analysis of mtDNA copy number by qPCR indicated that there was no significant difference in mtDNA content in cells transiently overexpressing CLIMP63-mCherry, Rtn4A-GFP or both CLIMP63-mCherry and Rtn4A-GFP relative to control cells [Fig. S6A]. This observation suggests that the transient reduction of the proportion of ER tubules in cells acutely disrupted mtDNA synthesis, as opposed to causing mtDNA loss or mtDNA EdU labeling resistance [Fig. 6B]. ER morphological phenotypes were not completely penetrant within the total population of cells overexpressing CLIMP63-GFP. We observed some cells with mosaic intracellular phenotypes in which the severity of ER tubule depletion varied spatially within the cytoplasm [Fig. 6D–E]. In this context, rare EdU-labeled nucleoids were detected even in highly overexpressing CLIMP63 cells and, in every case, were spatially linked to tubular ER co-localized with mitochondria [Fig. 6D, full field views in Fig. S6B]. Consistently, in live Cos7 cells overexpressing CLIMP63-mCherry and expressing POLG2-GFP and mito-BFP, POLG2-GFP labeled nucleoids were observed adjacent to residual ER tubules co-localized with mitochondria [Fig. S6C]; these sites subsequently marked mitochondrial constriction and ultimately division events. In addition, we observed a reduction in the number of POLG2-GFP foci in cells overexpressing CLIMP63 as compared to control cells expressing the ER marker Sec61b-mCherry [Fig. S6C]. Thus, ER structure, and in particular tubular ER-mitochondria contacts, is necessary but not sufficient for homeostatic mtDNA replication.
Given the significant spatial link observed between ER-mitochondria contacts and nucleoid position [Fig. S1A], we also examined the overall distribution of the total nucleoid population in cells under conditions of perturbed ER structure. In a majority of cells, overexpression of CLIMP63-mCherry, but not RTN4A-GFP, caused a significant increase in the area of resolvable TFAM-GFP foci, consistent with nucleoid aggregation and hence disturbed distribution [Fig. 6E and S7A–B]. Further examination of a subset of CLIMP63-overexpressing cells that contained normally distributed nucleoids revealed that these cells contained ER tubules co-localized with mitochondria at positions adjacent to TFAM-GFP labeled nucleoids [Fig. S7C]. In addition, at the subcellular level, depletion of ER tubules was highly correlated with nucleoid aggregation [Fig. 6E]. In cells simultaneously overexpressing CLIMP63-mCherry and RTN4A-GFP, the ER morphology and nucleoid distribution was similar to control cells [Fig. S7A]. Thus, normal ER structure is required for intracellular nucleoid distribution tubular ER-mitochondria contacts may play a role in the facilitation of nucleoid segregation within the mitochondrial network of mammalian cells. These findings demonstrate critical functional interdependencies between mitochondrial and ER dynamics and mitochondrial genome maintenance.
Discussion
Our data indicate that within mitochondrial nucleoids in mammalian cells, homeostatic mtDNA synthesis is spatially linked to a small subset of ER-mitochondria contacts that are selectively coupled to mitochondrial division. We also observed that nucleoids at mitochondrial tips, which are the products of mitochondrial division, exhibit preferential motility within cells as compared to intra-mitochondrial nucleoids. We propose that the successive events of mtDNA replication, mitochondrial division and mitochondrial motility are intimately linked, and function together as part of a programmed process that ensures the accurate distribution of mtDNA within cells. Such a process may be especially important for highly polarized cells, such as neurons, whose long axonal processes likely depend on the transport and amplification of mitochondria and associated mtDNA derived from the cell body. It will be important to determine the fundamental molecular mechanisms linking mtDNA replication initiation to mitochondrial division. Our analyses suggest that contacts between the ER and mitochondria are required to license mtDNA replication. Increasing evidence implicates inter-organellar membrane contacts in the formation of membrane microdomains with specialized lipid and protein composition(45). In this capacity, ER-mitochondria contacts could function to facilitate the creation of a spatially defined platform within and on mitochondria that selectively recruits components required for the initiation of mtDNA replication, such as POLG1, POLG2 or other components of the mtDNA replisome. Supporting this idea are recent biochemical observations suggesting that mtDNA, and mtDNA replisome proteins, associate with cholesterol-rich membrane structures that would be predicted to have raft-like properties(46). Our findings also raise the question of how mtDNA replication, which occurs inside mitochondria, is coordinated with division events associated with the outer surface of the organelle. Perhaps, in addition to contact sites between mitochondria and the ER, there are intra-mitochondrial spatial determinants that contribute to division site placement.
Our findings connect ER structure with mtDNA maintenance. This connection has implications for understanding the cellular pathology underlying human diseases and suggests that for human diseases linked to defects ER morphogenesis, pathogenesis could be a consequence of mitochondrial dysfunction (47, 48).
Methods
Plasmids
All fluorescent constructs have been previously described. Mito-BFP and mCherry-Drp1 (26), mCherry-SEC61b (49), mCherry-CLIMP63 (50), GFP-CLIMP63, and RTN4A-GFP (44) were gifts from Gia Voeltz. mRuby-KDEL was a gift from Joerg Wiedenmann (51). Human POLG2-GFP was a gift from William Copeland (34). Human TFAM-GFP was a gift from Mikhail Alexeyev (52).
Mammalian cell growth and transfection
U2OS, COS7 and ARPE19 cells (ATCC) were grown in high glucose DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were seeded at ~0.5×105 cells per mL in 35mm glass-bottom dishes (Mattek) 24 hours prior to transient transfection and 40 hours prior to imaging. Plasmid transfections were performed for 4 hours in serum- and antibiotic-free DMEM with 3 ul FuGENE6 reagent (Millipore) per dish. 16 hours later, cells were imaged in Fluorobrite DMEM (ThermoFisher) containing 10% FBS.
Cell fixation, antibodies and immunofluorescence
Cells were seeded as described above; 24 hours later, cells were stained with 500nM Mitotracker Red CMX Ros (ThermoFisher), and where indicated, a 1:1000 dilution of Picogreen DNA stain (ThermoFisher) for 15 min at 37 C. Cells were rinsed once in complete media and twice in warm PBS and fixed in 4% paraformaldehyde in PBS pH 7.4 for 20 minutes at room temperature. Dishes were washed twice in PBS and permeabilized in 0.1% TritonX-100 for 20 minutes. Dishes were blocked in 3% BSA PBS solution for 1 hour at room temperature. Primary antibodies were added at 1:1000 dilution in PBST (PBS pH 7.4, 1% BSA, 0.1% Tween-20) overnight at 4 C, rinsed twice in PBS and incubated with Alexa-Fluor conjugated secondary antibodies at 1:2000 dilution in PBST for 1 hour. Antibodies used: mouse anti-GFP AlexaFluor 488 conjugate (ThermoFisher), donkey anti-rabbit AlexaFluor 405 conjugate (ThermoFisher), anti-GFP (clone N86/8, Neuromabs), rabbit Anti-Calreticulin (2907, Abcam).
Edu incorporation was detected via Click-It Edu AlexaFluor 647 Labeling Kit (C10640, ThermoFisher) according to manufacturer instructions with minor deviations. Briefly, cells were incubated in 7 uM aphidicolin (A4487, Sigma) for 4 hours in complete media prior to a pulse of 50 uM Edu. The Edu pulse was followed by a chase in Edu-free complete DMEM for 1, 24 or 48 hours as described in the main text. For all Edu labeling experiments, cells were fixed and stained while sub-confluent, during logarithmic growth.
Spinning-disk Confocal Microscopy
Live cell imaging was performed using the spinning-disk module of an inverted objective fluorescence microscope (Marianas SDC Real Time 3D Confocal-TIRF; intelligent Imaging Innovations) with 100X, 1.46 NA objective. Either a Photometrics QuantiEM EMCCD, or Hamamatsu Orca Flash 4.0 sCMOS camera was used, depending on the experiment. Images were captured with Slidebook (Intelligent Imaging Innovations); if necessary, linear adjustments were made with ImageJ (NIH). Morphological and quantification analyses were performed in Nikon Elements-Advanced Research (Nikon) as described below. Scale bars were generated using Slidebook.
Mitochondrial division proximity analysis
To predict the frequency that mitochondrial division could occur at POLG2-GFP marked ER-mitochondrial contacts by random chance, we counted the total number of persistent contacts from each mitochondrion containing a POLG2-GFP focus in the frames leading up to each division event, considering each to be a potential future division site. We then compared the number of ERMD events that occurred within 1 micron of the POLG2-GFP to the total number of persistent contacts, and averaged over all events. For example, a mitochondrion with three persistent contacts and one division event would have a per-contact expected frequency of 1/3.
MtDNA copynumber and quantitative PCR analyses
Total DNA was isolated from U2OS cells using the DNeasy Blood and Tissue Kit (Qiagen). Quantitative PCR was carried out using SsoAdvanced Universal Probes Supermix (Biorad). Mitochondrial DNA copy number of control, and TFAM-, CLIMP63-, RTN4A- and double-overexpression osteosarcoma cells was performed as in (11) using primer sequences described therein. Briefly, mtDNA copynumber was normalized to nuclear DNA by amplifying a ~132 nucleotide fragment of cytochrome b (cyt b) as compared to a similarly sized fragment of the single copy nuclear gene, APP. The delta delta Ct method was used in calculations of fold change.
Statistical analyses, plotting, and modeling
All statistical analyses and plotting were performed in R version 3.2.0 within the RStudio development environment, version 0.99.441. To model the distribution of Edu foci in fixed ARPE19 cells, we used the Empirical Cumulative Distribution Function ecdf(). Random simulated data was generated with the runif() function using 83 observations and a range of 0–7, consistent with our real data from the first biological replicate of the 1 hour Edu pulse with 1 hour chase. To recreate the random dataset shown in Figure 3, employ the set.seed() function as follows:
>set.seed(100)#random number generator, initial state
>randomDistToTip<-runif(83, min=0, max=7)#generate random dataset
>R<-ecdf(randomDistToTip)#calculate ecdf
>plot(R)#plot random dataset
Supplementary Material
Acknowledgments
We thank members of the Nunnari lab for discussion, and Dr. Michael Paddy and the UC Davis MCB Microscopy facility for shared equipment and helpful suggestions. This work is supported by NIH grants R01GM106019 and R01GM097432 (to J. Nunnari), an NIH Ruth L. Kirschstein Postdoctoral Fellowship F32GM113388 (to S. C. Lewis), and a UC Davis Provost’s Undergraduate Research Fellowship (to L. Uchiyama). J. Nunnari is on the Advisory Board of Mitobridge, and declares no financial interest related to this work. The other authors declare that no competing interests exist. All data are included in the main manuscript and Supplementary Materials.
Footnotes
References and Notes
- 1.Copeland WC. Defects of mitochondrial DNA replication. Journal of Child Neurology. 2014 Sep;29:1216. doi: 10.1177/0883073814537380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012 Apr 16;148:1145. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004 Jun 27;429:417. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 4.Kujoth GC, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005 Jul 15;309:481. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 5.Garrido N, et al. Composition and dynamics of human mitochondrial nucleoids. Molecular Biology of the Cell. 2003 May;14:1583. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ropp PA, Copeland WC. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics. 1996 Sep 15;36:449. doi: 10.1006/geno.1996.0490. [DOI] [PubMed] [Google Scholar]
- 7.Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. The Journal of Biological Chemistry. 1999 Dec 31;274:38197. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- 8.Kaguni LS. DNA polymerase γ, the mitochondrial replicase. Annual Review of Biochemistry. 2004 Jul;73:293. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 9.Wanrooij S, Goffart S, Pohjoismäki JLO, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Research. 2007;35:3238. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wanrooij S, Falkenberg M. The human mitochondrial replication fork in health and disease. Biochimica et Biophysica Acta. 2010 Aug;1797:1378. doi: 10.1016/j.bbabio.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Tyynismaa H, et al. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Human Molecular Genetics. 2004 Dec;24:3219. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 12.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO Journal. 2004 Jul 16;23:2423. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alam TI, et al. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Research. 2003 Apr 15;31:1640. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufman BA, et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Molecular Biology of the Cell. 2007 Sep;18:3225. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ngo HB, Kaiser JT, Chan DC. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nature Structural & Molecular Biology. 2011 Nov;18:1290. doi: 10.1038/nsmb.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kukat C, et al. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proceedings of the National Academy of Sciences. 2011 Aug 16;108:13534. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kukat C, et al. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proceedings of the National Academy of Sciences. 2015 Sep 08;112:11288. doi: 10.1073/pnas.1512131112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown TA, et al. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Molecular and Cellular Biology. 2011 Dec;31:4994. doi: 10.1128/MCB.05694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birky CW., Jr The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annual Review of Genetics. 2001;35:125. doi: 10.1146/annurev.genet.35.102401.090231. [DOI] [PubMed] [Google Scholar]
- 20.Pohjoismäki JLO, et al. Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. The Journal of Biological Chemistry. 2009 Aug 07;284:21446. doi: 10.1074/jbc.M109.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnusson J, Orth M, Lestienne P, Taanman JW. Replication of mitochondrial DNA occurs throughout the mitochondria of cultured human cells. Experimental Cell Research. 2003 Sep 10;289:133. doi: 10.1016/s0014-4827(03)00249-0. [DOI] [PubMed] [Google Scholar]
- 22.Nunnari J, et al. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Molecular Biology of the Cell. 1997 Jul 01;8:1233. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labbé K, Murley A. J. (to S. C. Lewis), and a UC Davis functions of mitochondrial behavior. Annual Review of Cell and Developmental Biology. 2014;30:357. doi: 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- 24.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Molecular Biology of the Cell. 2001 Aug;12:2245. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. The Journal of Cell Biology. 1998 Oct 19;143:351. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science. 2011 Oct 21;334:358. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ban-Ishihara R, Ishihara T, Sasaki N, Mihara K, Ishihara N. Dynamics of nucleoid structure regulated by mitochondrial fission contributes to cristae reformation and release of cytochrome c. Proceedings of the National Academy of Sciences. 2013 Jul;110:11863. doi: 10.1073/pnas.1301951110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishihara T, et al. Dynamics of mtDNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Molecular and Cellular Biology. 2014 Oct 27;35:211. doi: 10.1128/MCB.01054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itoh K, Tamura Y, Iijima M, Sesaki H. Effects of Fcj1-Mos1 and mitochondrial division on aggregation of mitochondrial DNA nucleoids and organelle morphology. Molecular Biology of the Cell. 2013 Jul;24:1842. doi: 10.1091/mbc.E13-03-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murley A, et al. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iborra FJ, Kimura H, Cook PR. The functional organization of mitochondrial genomes in human cells. BMC Biology. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajala N, Gerhold JM, Martinsson P, Klymov A, Spelbrink JN. Replication factors transiently associate with mtDNA at the mitochondrial inner membrane to facilitate replication. Nucleic Acids Research. 2014 Feb;42:952. doi: 10.1093/nar/gkt988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meeusen S, Nunnari J. Evidence for a two membrane–spanning autonomous mitochondrial DNA replisome. The Journal of Cell Biology. 2003 doi: 10.1083/jcb.200304040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young MJ, Humble MM, DeBalsi KL, Sun KY, Copeland WC. POLG2 disease variants: analyses reveal a dominant negative heterodimer, altered mitochondrial localization and impaired respiratory capacity. Human Molecular Genetics. 2015 Sep 15;24:5184. doi: 10.1093/hmg/ddv240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proceedings of the National Academy of Sciences. 2008 Mar 19;105:2415. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanki T, et al. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004 Nov;24:9823. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu J, Prinz WA, Rapoport TA. Weaving the web of ER tubules. Cell. 2011 Dec 09;147:1226. doi: 10.1016/j.cell.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voeltz GK, Rolls MM, Rapoport TA. Structural organization of the endoplasmic reticulum. EMBO Reports. 2002 Oct;3:944. doi: 10.1093/embo-reports/kvf202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West M, Zurek N, Hoenger A, Voeltz GK. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. The Journal of Cell Biology. 2011 May 18;193:333. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006 Mar 10;124:573. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 41.Shibata Y, et al. Mechanisms determining the morphology of the peripheral ER. Cell. 2010 Nov 24;143:774. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klopfenstein DR, et al. Subdomain-specific localization of CLIMP-63 (p63) in the endoplasmic reticulum is mediated by its luminal alpha-helical segment. The Journal of Cell Biology. 2001 Jul 11;153:1287. doi: 10.1083/jcb.153.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui-Wang T, et al. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell. 2012 Jan 20;148:309. doi: 10.1016/j.cell.2011.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shibata Y, et al. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. The Journal of Biological Chemistry. 2008 Jul 04;283:18892. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrew JN, Murley C. The emerging network of mitochondria-organelle contacts. Molecular Cell. 2016;61 doi: 10.1016/j.molcel.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerhold JM, et al. Human Mitochondrial DNA-Protein Complexes Attach to a Cholesterol-Rich Membrane Structure. Scientific Reports. 2015;5:15292. doi: 10.1038/srep15292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blackstone C. Cellular pathways of hereditary spastic paraplegia. Annual Review of Neuroscience. 2012;35:25. doi: 10.1146/annurev-neuro-062111-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rugarli EI, Langer T. Translating m-AAA protease function in mitochondria to hereditary spastic paraplegia. Trends in Molecular Medicine. 2006 Jun;12:262. doi: 10.1016/j.molmed.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 49.Zurek N, Sparks L, Voeltz G. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 2011 Jan;1:28. doi: 10.1111/j.1600-0854.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.English AR, Zurek N, Voeltz G. Peripheral ER structure and function. Current Opinion in Cell Biology. 2009 Aug;4:596. doi: 10.1016/j.ceb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kredel S, et al. mRuby, a bright monomeric fluorescent protein for labeling of cellular structures. PLoS ONE. 2009 Feb;2:4391. doi: 10.1371/journal.pone.0004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pastukh V, Shokolenko I, Wang B, Wilson G, Alexeyev M. Human mitochondrial transcription factor A possesses multiple subcellular targeting signals. FEBS J. 2007 Dec;274:6488. doi: 10.1111/j.1742-4658.2007.06167.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.