Abstract
Yeast MET18, a subunit of the cytosolic iron-sulfur (Fe/S) protein assembly (CIA) machinery which is responsible for the maturation of Fe/S proteins, has been reported to participate in the oxidative stress response. However, the underlying molecular mechanisms remain unclear. In this study, we constructed a MET18/met18Δ heterozygous mutant yeast strain and found that MET18 deficiency in yeast cells impaired oxidative stress resistance as evidenced by increased sensitivity to hydrogen peroxide (H2O2) and cumene hydroperoxide (CHP). Mechanistically, the mRNA levels of catalase A (CTA1) and catalase T (CTT1) as well as the total catalase activity were significantly reduced in MET18-deficient cells. In contrast, overexpression of CTT1 or CTA1 in MET18-deficient cells significantly increased the intracellular catalase activity and enhanced the resistance ability against H2O2 and CHP. In addition, MET18 deficiency diminished the replicative capacity of yeast cells as evidenced by the shortened replicative lifespan, which can be restored by CTT1 overexpression, but not by CTA1, in the MET18-deficient cells. These results suggest that MET18, in a catalase-dependent manner, plays an essential role in enhancing the resistance of yeast cells to oxidative stress and increasing the replicative capacity of yeast cells.
1. Introduction
Oxygen (O2) is crucial for the livelihood of organisms grown aerobically. However, exposure to higher O2 concentration can increase the production of reactive oxygen species (ROS), including hydroxyl radical (OH−), hydrogen peroxide (H2O2), and superoxide anions (O2−), which may exert detrimental effects on cell growth. ROS can also be generated by redox-cycling chemicals and heavy metals in the environment, as well as by the endogenous metabolic process. In addition, the effects of environmental oxygen concentrations on the intracellular ROS production are highly dependent upon the culture medium, the cells themselves, the metabolic state of the cells, and the presence of any antioxidative compound or enzyme and other known and unknown factors [1–3]. It has been shown that culturing cells in air (21% O2) exposes the cells to much higher levels of oxygen than what they are usually submitted to under physiological conditions (2~5% O2) [2]. Cells normally have oxidant defense systems to protect themselves against ROS or oxidative stress. However, the underlying mechanisms remain largely unknown [3, 4].
Yeast MET18, the homologue of methyl-methanesulfonate sensitivity protein 19 (MMS19) in human, is a conserved component of the cytosolic iron-sulfur (Fe/S) protein assembly (CIA) machinery in eukaryotes [5–8]. From yeasts to plant cells to human cells, the CIA machinery is generally located in the cytoplasm and responsible for the maturation of Fe/S proteins through catalyzing the synthesis of Fe/S clusters [9, 10]. In the CIA pathway, MET18 and other subunits, such as CIA1, CIA2, and nuclear architecture-related protein 1 (NAR1), form a CIA complex in which MET18 functions as a delivering agent by directly interacting with the target protein and transferring the Fe/S cluster to the protein, leading to the maturation of Fe/S proteins [11, 12]. MET18 could form complexes with Dos2, Rki1, and Cdc20 in order to participate in various cell processes such as DNA repair, RNA polymerase transcription, chromosome segregation, and maintenance of telomere length, among others [13]. Therefore, MET18 has a number of roles in cell metabolism.
Previous studies have suggested that some components of CIA machinery, CIA2 and NAR1, are associated with yeast response to oxidative stress and thus regulate the replicative lifespan (RLS) of yeast. In CIA2/cia2Δ mutants, the deficiency of CIA2 enhances the sensitivity of yeast to the oxidant cumene hydroperoxide (CHP), resulting in an impaired oxidative stress tolerance and thus a decreased RLS [14, 15]. Similarly, NAR1-deficient yeast cells have been found to be more sensitive to O2−-generating compound paraquat (PQ) and have shorter RLS than wild-type (WT) yeast strain [16]. Any deficiency in the CIA pathway will impact the activities of enzymes involved in ROS and oxidative stress management [13].
Since MET18 is also a component of CIA, we hypothesized that MET18 may play an important role in yeast response to oxidative stress. To confirm this hypothesis, we performed the functional assay for MET18 mutation by constructing a MET18/met18Δ heterozygous mutant yeast strain. Our results demonstrate for the first time that MET18 deficiency may increase the response sensitivity of yeast to oxidative stress and shorten the RLS through suppressing the expression and activity of catalases.
2. Materials and Methods
2.1. Yeast Strains and Culture
The yeast strains used in this study are listed in Table 1. All strains were isogenic to BY4743 and were stored in liquid yeast peptone dextrose (YPD; 1% yeast extract, 2% peptone, and 2% glucose; Oxoid, Basingstoke, UK) medium mixed with equal volume of 50% (v/v) glycerol at −80°C. For all experiments, cells were removed from frozen stock and streaked onto solid YPD plate followed by an incubation for 2~3 days at 30°C. Then the single colonies were picked and grown in liquid YPD until the exponential phase using an orbital shaker where yeast cells were shaken at 30°C and 180 rpm.
Table 1.
Yeast strains in this study.
| Strain name | Genotype | Comments | Source |
|---|---|---|---|
| BY4743 | MATa/MATαhis3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2/lys2Δ0 met15Δ0/met15 ura3Δ0/ura3Δ0 | Wild-type strain | Gift from Matt Kaeberlein |
| GDMUB1001 | BY4743 MET18::URA3 | Deletion of one copy of MET18 in BY4743 | This experiment |
| GDMUB1002 | BY4743 + pAUR123 | BY4743 harboring pAUR123 | This experiment |
| GDMUB1003 | BY4743MET18::URA3 + pAUR123 | GDMUB1001 harboring pAUR123 | This experiment |
| GDMUB1004 | BY4743MET18::URA3 + pAUR123-CTA1 | GDMUB1001 harboring pAUR123-CTA1 | This experiment |
| GDMUB1005 | BY4743MET18::URA3 + pAUR123-CTT1 | GDMUB1001 harboring pAUR123-CTT1 | This experiment |
2.2. MET18/met18Δ Mutants and Plasmids Construction
The MET18/met18Δ mutants were constructed as previously described [14]. Briefly, URA3 cassette was amplified by polymerase chain reaction (PCR) from pRS306 vector using the following primers: 5′-TGT TTT AAC TGG GAA AAA GCG GAA CAA TTG GGC CTT ACA AGA TTG TAC TGA GAG TGC AC-3′ (forward) and 5′-CGT GCT CAT CAA TGT GAA CAA ATT ATT AAA TAC AAG CGT CTG TGC GGT ATT TCA CAC CG-3′ (reverse) (Invitrogen, Shanghai, China). Then the PCR products were transformed into BY4743 to replace one copy of MET18 by homologous recombination, and the transformants were selected on SD-URA medium (Clontech, Mountain View, CA, USA). The heterozygous MET18/met18Δ cells were verified by PCR using the following primers: 5′-TGT GGC TGT CGT TTC GTG G-3′ (forward) and 5′-TAC AGT TTC CAC TGC GAA CAC A-3′ (reverse) (Invitrogen, Shanghai, China).
The entire coding regions of catalase A (CTA1) and catalase T (CTT1) were amplified from genomic DNA of BY4743 using primers flanked by Sac I and Xba I restriction sites (CTT1 primers: 5′-TAT GTC GAC ATG AAC GTG TTC GGT AAA AAA G-3′ (forward) and 5′-GCG TCT AGA TTA ATT GGC ACT TGC AAT GG-3′ (reverse); CTA1 primers: 5′-ATA GTC GAC ATG TCG AAA TTG GGA CAA GA-3′ (forward) and 5′-CGC TCT AGA TCA AAA TTT GGA GTT ACT CG-3′ (reverse)) followed by digestion with Sac I and Xba I (Takara, Otsu, Shiga, Japan) and ligation into pAUR123 vector (Takara, Otsu, Shiga, Japan) to generate pAUR123-CTT1 and pAUR123-CTA1 expressing CTT1 and CTA1, respectively. Subsequently, pAUR123-CTT1 or pAUR123-CTA1 was transformed into MET18/met18Δ mutants. The empty pAUR123 vector was used as a negative control. The transformants were selected on YPD or SD-URA media containing aureobasidin A (Sigma-Aldrich, St. Louis, MO, USA) and then verified by PCR using the following primers: 5′-TCT GCA CAA TAT TTC AAG C-3′ (forward) and 5′-TTC GTT TTA AAA CCT AAG AGT CAC-3′ (reverse).
2.3. Real-Time PCR
Total RNA was isolated from yeast cells using a Yeast RNAiso Kit (Takara, Otsu, Shiga, Japan) followed by cDNA synthesis using a Transcriptor First-Strand cDNA Synthesis Kit with gDNA Eraser (Takara, Otsu, Shiga, Japan). Real-time PCR was performed using SYBR® Premix Ex Taq™ (Takara, Otsu, Shiga, Japan) and the primers are shown in Table 2. Data were normalized to the internal control PRP8 [14].
Table 2.
The real-time PCR primers used in this study.
| Gene | Primers | Sequence |
|---|---|---|
| PRP8 | Forward | TCATGGCTGCGTCTGAAGTA |
| Reverse | GGCTCAAACCCTTCCGATAG | |
| MET18 | Forward | TGCTGGAAGTTGTCGTTGC |
| Reverse | TCGTTTTTGGAGAGGTGGTC | |
| CTT1 | Forward | GATTCCGTTCTACAAGCCAGAC |
| Reverse | GGAGTATGGACATCCCAAGTTTC | |
| CTA1 | Forward | CCAACAGGACAGACCCATTC |
| Reverse | TTACCCAAAACGCGGTAGAG |
2.4. Spot Assay
The sensitivity of yeast to oxidant stressors, including H2O2, CHP, and PQ (Sigma-Aldrich, St. Louis, MO, USA), was determined by spot assay. Yeast cells were cultured to exponential phase (OD600 = 2.0) in liquid YPD followed by a 10-fold dilution with sterile water. Four additional 10-fold serial dilutions were performed, and 5 μL of each dilution was inoculated onto solid YPD plates supplemented with indicated stressors. The plates were incubated at 30°C for 2~5 days for colony formation.
2.5. Enzyme Activity Assay and Determination of Intracellular H2O2 Level
Yeast cells in exponential phase were harvested and washed twice with cold sterile water followed by resuspension in lysis buffer containing acid-washed glass beads and 20 cycles of 10 s vortexing plus 20 s cooling [14]. After centrifugation at 12000 rpm for 15 min, the supernatants were collected and used for an enzyme activity assay and determination of intracellular H2O2 levels. The protein concentration was determined using the BCA protein Assay Kit (Sangon Biotech, Shanghai, China) following the manufacturer's instruction. The total superoxide dismutase (SOD) activity and the catalase activity were determined using a Total Cellular SOD Activity Kit (Dojindo Molecular Technologies, Rockville, MD, USA) and a Catalase Assay Kit (Beyotime Biotechnology, Shanghai, China), respectively. The intracellular H2O2 level was determined using an H2O2 Assay Kit (Beyotime Biotechnology, Shanghai, China).
2.6. Replicative Lifespan (RLS) Assay
RLS assay was performed as previously described [15]. Briefly, cells were thawed from frozen stocks and grown on freshly made YPD plate for 2 days and then patched onto the second YPD plate and incubated for an additional 24 h. Cells were then restruck on the third YPD plate and incubated for about 12 h, and then the virgin buds were isolated and used for RLS analysis.
2.7. Statistical Analysis
All experiments were repeated at least three times. Data were expressed as the mean ± standard deviation (SD). Statistical significance was assessed by calculating p values using a two-tailed Student's t-test or Wilcoxon rank-sum test. A value of p < 0.05 was considered statistically significant.
3. Results
3.1. MET18 Deficiency Enhances the Cellular Response of Yeast to Oxidative Stress
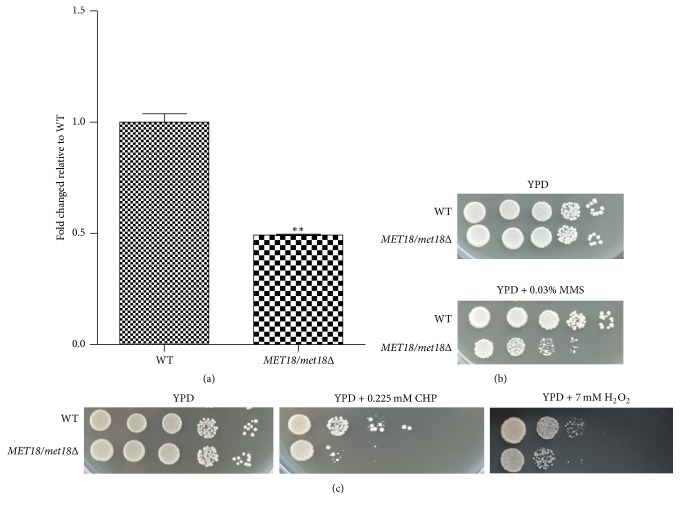
To investigate the role of MET18 in yeast response to oxidative stress, we constructed a MET18/met18Δ mutant strain which was confirmed by a real-time PCR detection of MET18- and URA3-specific PCR products. As shown in Figure 1(a), the mRNA level of MET18 was decreased by more than 50% in MET18-deficient cells compared with WT cells, indicating that the expression of MET18 mRNA in MET18/met18Δ mutants was successfully suppressed by the replacement of one copy of MET18 with URA3 gene.
Figure 1.
Effects of MET18 deficiency on yeast response to oxidative stress. (a) mRNA expression of MET18 under unstressed condition was determined by real-time PCR. (b) Tenfold dilution series of WT and MET18/met18Δ cells were spotted on YPD plates with or without 0.03% MMS and then incubated at 30°C for 2~3 days to detect the cellular response to MMS. (c) Tenfold dilution series of WT and MET18/met18Δ cells were spotted on YPD plates with 0.225 mM CHP or 7 mM H2O2 and incubated at 30°C for 2~5 days to detect the cellular response to CHP and H2O2. Results shown are representative of three independent experiments, ∗∗p < 0.01 versus WT cells (n = 3).
We then sought to examine the cellular response of MET18/met18Δ mutants to oxidative stressors, including H2O2 and CHP. A DNA-damaging agent methyl-methanesulfonate (MMS) was used as a positive control [17]. As shown in Figures 1(b) and 1(c), the significant decreases in yeast growth in response to MMS, H2O2, and CHP were observed in MET18/met18Δ mutants, compared with those in WT cells, suggesting that MET18 deficiency may increase the oxidative stress response in yeast cells.
3.2. MET18 Deficiency Decreases Catalase Activity While Increasing Intracellular H2O2 Levels
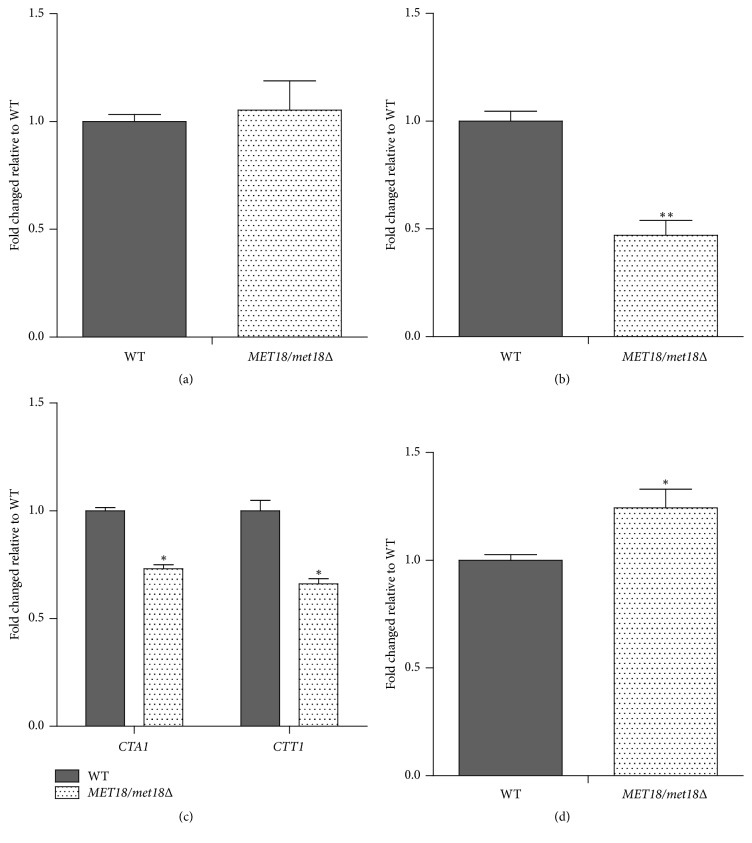
To further examine whether MET18 is involved in enzymatic antioxidant defense against oxidative stress, we determined the activities of SOD and catalases which are responsible for ROS scavenging [18, 19]. As shown in Figures 2(a) and 2(b), the activity of total SOD in MET18/met18Δ mutants was comparable to that in WT strain, whereas the total catalase activity was significantly decreased by more than 50% in MET18-deficient cells compared with WT strain. Meantime, the mRNA expression of both CTT1 and CTA1 was significantly downregulated in MET18-deficient cells (Figure 2(c)). Conversely, H2O2 levels in MET18/met18Δ mutants were higher than those in WT strain (Figure 2(d)), suggesting an increased oxidative stress in MET18/met18Δ mutants. These results indicate that MET18 deficiency may downregulate antioxidant defense enzymes, leading to the increased oxidative stress.
Figure 2.
Effects of MET18 deficiency on enzymatic defense system against oxidative stress. Total SOD activity (a), catalase activity (b), CTA1 and CTT1 mRNA levels (c), and intracellular H2O2 levels (d) under unstressed condition were determined and compared between WT and MET18/met18Δ cells. Results shown are representative of three independent experiments. ∗p < 0.05, ∗∗p < 0.01 versus WT cells (n = 3).
3.3. Either CTT1 or CTA1 Restores the MET18 Deficiency-Suppressed Oxidative Stress Resistance in Yeast
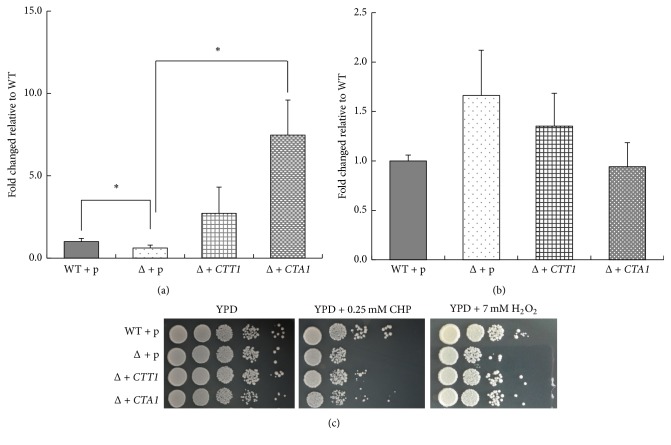
To investigate if the increased oxidative stress response in MET18/met18Δ mutants is attributable to inhibition of catalases, pAUR123-CTT1 or pAUR123-CTA1 was transfected into MET18/met18Δ mutants to generate CTT1- or CTA1-overexpressing mutants. As shown in Figure 3(a), the catalase activity in MET18/met18Δ mutants overexpressing CTT1 (Δ + CTT1) or CTA1 (Δ + CTA1) was significantly increased as compared with the empty vector-transfected WT strain (WT + p) or the empty vector-transfected mutant strain (Δ + p). Furthermore, CTT1- or CTA1-overexpression also reduced the intracellular levels of H2O2 (Figure 3(b)), consistent with our findings (Figure 2). Importantly, CTT1- or CTA1-overexpression greatly enhanced the resistance of MET18/met18Δ mutants against H2O2 and CHP (Figure 3(c)), leading to an increased cell growth. In contrast, CTT1- or CTA1-overexpression did not appear to promote cell growth under unstressed conditions (Figure 3(c)), suggesting the important role of catalases in oxidative stress response of MET18/met18Δ mutants. These results indicate that inhibition of CTT1 or CTA1 activity may be a major mechanism underlying the impaired oxidative response in MET18-deficient yeasts.
Figure 3.
Effects of CTT1 or CTA1 on oxidative stress resistance in MET18/met18Δ cells. Catalase activity (a) and the intracellular H2O2 levels (b) under unstressed conditions were determined and compared among pAUR123-transfected WT strain (WT + p), pAUR123-, and CTT1- and CTA1-transfected MET18/met18Δ mutants ((Δ + p), (Δ + CTT1), and (Δ + CTA1), resp.). ∗p < 0.05 versus WT cells (n = 3). (c) Tenfold dilution series of WT + p, Δ + p, Δ + CTT1, or Δ + CTA1 were spotted on YPD plates with or without the stressors, as indicated, and incubated at 30°C for 2~3 days to detect the cellular response. Results shown are representative of three independent experiments.
3.4. CTT1, but Not CTA1, Restores the Lifespan of MET18-Deficient Yeast
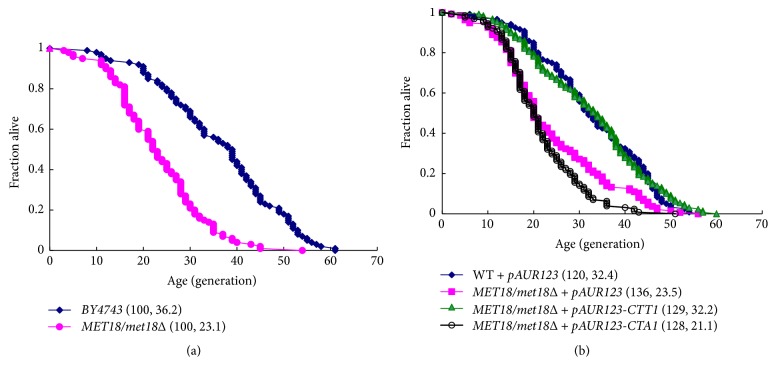
To further examine if MET18 has effects on yeast aging, we performed a RLS assay in WT strains and MET18/met18Δ mutants. The results showed that MET18/met18Δ mutants had a shortened RLS compared with the WT strain BY4743 (mean lifespan: 23.1 versus 36.2) (Figure 4(a)). Of note, overexpression of CTT1 rescued MET18 deficiency-induced RLS defects, whereas overexpression of CTA1 did not appear to affect RLS in MET18/met18Δ mutants (Figure 4(b)), suggesting that MET18 deficiency leads to shortened RLS or aging in yeast, at least in part, through inhibition of catalases.
Figure 4.
Effect of CTT1 or CTA1 on RLS in MET18/met18Δ mutants. (a) RLS of WT and MET18/met18Δ cells; (b) RLS of WT and MET18/met18Δ cells transfected with pAUR123, CTT1, or CTA1. Results shown are representative of three independent experiments.
4. Discussion
In the present study, we constructed a MET18/met18Δ mutant using the WT BY4743 yeast strain and examined the response of MET18-deficient yeast to oxidative stress caused by H2O2 and CHP. Our results suggest that MET18 deficiency increases the cellular response of yeast to H2O2 and CHP and shortens RLS through downregulation of catalases CTT1 and CTA1.
It has been reported that MET18 participates in oxidative stress responses [20, 21]; however, the underlying mechanism remains unknown. Oxidative stress is termed as a disruption of cellular redox balance due to the failure of oxidant defense system to scavenge excess ROS which can be generated from normal cellular metabolism in living organisms and is a potential damage to cells [22]. In our study, the total catalase activity as well as the mRNA levels of both CTT1 and CTA1 was significantly decreased in MET18/met18Δ mutant compared with the WT strain (Figures 2(b) and 2(c)), resulting in an increased level of intracellular H2O2 in mutant cells (Figure 2(d)). These results indicate that the cellular redox balance is abolished in MET18-deficient cells due to the decreased antioxidants (catalase) and increased ROS (H2O2) levels.
Since MET18 is a component of the CIA machinery and plays an important role in the maturation of Fe/S proteins [5, 6], one possible explanation for the downregulation of catalase is that the Fe/S cluster transfer required for catalase maturation is inhibited in MET18-deficient yeast, resulting in H2O2 accumulation and an increase in the oxidant burden of the cells. In addition, MET18 could be responsible for or has a role to play in Fe/S cluster transfer or epigenetic modifications in yeasts, affecting their metabolism [7, 13, 23]. Indeed, Wang et al. [13] showed that MET18 can regulate active DNA methylation and methylation pathways in Arabidopsis. Buzas et al. [23] showed that AtDRE2, a member of the CIA, had epigenetic functions that are independent of the CIA. Duan et al. [7] showed that MET18-dependent transfer of the iron cluster was required for ROS1-mediated active DNA methylation. Nevertheless, additional studies will be necessary to specify the role of MET18 in oxidative stress of the yeasts.
H2O2, at high concentrations, primarily contributes to oxidative damage [20, 21]. Dismutation of O2− by SOD is a major source of H2O2 [24]. However, in our study, the activity of SOD in MET18/met18Δ mutants did not appear to be different from that in WT strain (Figure 2(a)), suggesting that other enzymes or factors than SOD are responsible for the increased H2O2 level in MET18-deficient cells. Catalases may downregulate the intracellular level of H2O2 by catalyzing the decomposition of H2O2 into water and oxygen [25]. Accordingly, the increased H2O2 level and enhanced response to oxidative stress have been observed in yeast cells lacking CTA1 and CTT1 [24, 25], which is consistent with our findings. It seems that, compared with WT strains, MET18/met18Δ mutants are more sensitive to the oxidative stress caused by H2O2 and CHP due to the decreased catalase activity and the subsequent H2O2 accumulation.
There are conflicting findings in regard to the role of catalases in yeast response to H2O2. Some studies show that the activity of CTT1 is essential to protect yeast cells against H2O2 challenge, whereas the activity of CTA1 is dispensable [25]. On the other hand, some studies indicate that both CTT1 and CTA1 mutants are sensitive to H2O2, suggesting that CTT1 and CTA1 are equally important for antioxidative response of yeast to H2O2 stress [26]. Our study reveals that the overexpression of either CTT1 or CTA1 in MET18/met18Δ mutants can effectively scavenge the accumulated intracellular H2O2 and thus enhances the cellular resistance to CHP and H2O2 (Figure 3).
Catalase activity is also closely associated with lifespan regulation in various species. It has been reported that overexpression of human catalase targeted to mitochondria can extend mouse lifespan by eliminating mitochondrial H2O2 [2]. Consistently, reduced mitochondrial ROS as well as increased antioxidant enzyme activities of SOD, catalase, and glutathione peroxidase 1 has been observed in long-lived white-footed mice compared with common laboratory mice [27]. The role of catalase in the yeast replicative aging may be complicated by genetic background (haploid or diploid) and nutrient availability (glucose or ethanol) [28]. Several lines of evidence show that catalases and excess H2O2 exert positive and negative effects on RLS in yeast, respectively [29]. Our study found that loss of one copy of MET18 leads to the reduction in yeast RLS which can be rescued by CTT1, but not by CTA1 (Figure 4), indicating that inactivity of CTT1 is a driving force of senescence in MET18/met18Δ mutants. These findings are in agreement with the previous study that CTT1, but not CTA1, is required for the maintenance of RLS in yeast cultured in glucose-containing media [30]. MET18 deficiency leads to shortened RLS or aging in yeast, at least in part, through inhibition of catalases. It is known that CTT1 and CTA1 are not submitted to the same regulatory mechanisms in response to nutrient availability and oxidative stress [30]. This could explain, at least in part, why CTT1 and CTA1 did not have the same effect on RLS in the present study, but additional studies are necessary to address this issue. Nevertheless, Rona et al. [2] showed that CTT1 overexpression increased the lifespan of calorie-restricted S. cerevisiae deficient in Sod1 through increased peroxide degradation catalyzed by catalase. Similarly, Zhao et al. [31] showed that CTT1 overexpression increased the replicative lifespan of S. cerevisiae sensitive to MMS and deficient in KSP1. On the other hand, Ohtsuka et al. [32] showed that CTT1 was required for the H2O2-resistant phenotype, but that CTT1 overexpression did not increase the chronological lifespan of the yeast. These previous studies provide some clues about the role of catalases as modulators of replicative lifespan in S. cerevisiae.
5. Conclusions
In conclusion, our study demonstrates for the first time that MET18 deficiency in yeast impairs oxidative stress tolerance and shortens lifespan, at least partly, through the inhibition of catalase activity and accumulation of intracellular H2O2. Further investigation is needed to examine the role of MET18 in other Fe/S transfer-associated processes such as epigenetic modifications in eukaryotes.
Acknowledgments
This work was financially supported by grants from The National Natural Science Foundation of China (31101051 and 81170327), The Natural Science Foundation of Guangdong Province (9252402301000002), and The Natural Science on the Surface of Guangdong Medical University (M2014013). The authors thank Matt Kaeberlein and Brian K. Kennedy for technical assistance.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this article.
References
- 1.Zorov D. B., Juhaszova M., Sollott S. J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiological Reviews. 2014;94(3):909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rona G., Herdeiro R., Mathias C. J., Torres F. A., Pereira M. D., Eleutherio E. CTT1 overexpression increases life span of calorie-restricted Saccharomyces cerevisiae deficient in Sod1. Biogerontology. 2015;16(3):343–351. doi: 10.1007/s10522-015-9550-7. [DOI] [PubMed] [Google Scholar]
- 3.Krumova K., Cosa G. Singlet Oxygen: Applications in Biosciences and Nanosciences. Vol. 1. London, UK: Royal Society of Chemistry; 2016. Chapter 1: overview of reactive oxygen species; pp. 1–21. (Comprehensive Series in Photochemical & Photobiological Sciences). [DOI] [Google Scholar]
- 4.Poprac P., Jomova K., Simunkova M., Kollar V., Rhodes C. J., Valko M. Targeting free radicals in oxidative stress-related human diseases. Trends in Pharmacological Sciences. 2017;38(7):592–607. doi: 10.1016/j.tips.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Stehling O., Vashisht A. A., Mascarenhas J., et al. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science. 2012;337(6091):195–199. doi: 10.1126/science.1219723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gari K., Ortiz A. M. L., Borel V., Flynn H., Skehel J. M., Boulton S. J. MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science. 2012;337(6091):243–245. doi: 10.1126/science.1219664. [DOI] [PubMed] [Google Scholar]
- 7.Duan C.-G., Wang X., Tang K., et al. MET18 connects the cytosolic iron-sulfur cluster assembly pathway to active DNA demethylation in arabidopsis. PLoS Genetics. 2015;11(10) doi: 10.1371/journal.pgen.1005559.e1005559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paul V. D., Lill R. Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochimica et Biophysica Acta. 2015;1853(6):1528–1539. doi: 10.1016/j.bbamcr.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 9.Sharma A. K., Pallesen L. J., Spang R. J., Walden W. E. Cytosolic iron-sulfur cluster assembly (CIA) system: factors, mechanism, and relevance to cellular iron regulation. Journal of Biological Chemistry. 2010;285(35):26745–26751. doi: 10.1074/jbc.R110.122218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stehling O., Lill R. The role of mitochondria in cellular iron-sulfur protein biogenesis: mechanisms, connected processes, and diseases. Cold Spring Harbor Perspectives in Biology. 2013;5(8):p. a011312. doi: 10.1101/cshperspect.a011312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paul V. D., Mühlenhoff U., Stümpfig M., et al. The deca-GX3 proteins Yae1-Lto1 function as adaptors recruiting the ABC protein Rli1 for iron-sulfur cluster insertion. eLife. 2015;4(2015) doi: 10.7554/eLife.08231.e08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seki M., Takeda Y., Iwai K., Tanaka K. IOP1 protein is an external component of the human cytosolic iron-sulfur cluster assembly (CIA) machinery and functions in the MMS19 protein-dependent CIA pathway. Journal of Biological Chemistry. 2013;288(23):16680–16689. doi: 10.1074/jbc.M112.416602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X., Li Q., Yuan W., et al. The cytosolic Fe-S cluster assembly component MET18 is required for the full enzymatic activity of ROS1 in active DNA demethylation. Scientific Reports. 2016;6 doi: 10.1038/srep26443.26443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao W., Zheng H.-Z., Niu Y.-J., et al. CIA2 deficiency results in impaired oxidative stress response and enhanced intracellular basal UPR activity in Saccharomyces cerevisiae. FEMS microbiology letters. 2015;362(6) doi: 10.1093/femsle/fnv013. [DOI] [PubMed] [Google Scholar]
- 15.Steffen K. K., Kennedy B. K., Kaeberlein M. Measuring replicative life span in the budding yeast. Journal of Visualized Experiments. 2009;(28) doi: 10.3791/1209.e1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao W., Fang B. X., Niu Y. J., et al. Nar1 deficiency results in shortened lifespan and sensitivity to paraquat that is rescued by increased expression of mitochondrial superoxide dismutase. Mechanisms of Ageing and Development. 2014;138(1):53–58. doi: 10.1016/j.mad.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Enya B., Young T., Kirchmaier A. Effects of H4K16 and gene mutation on sensitivity to DNA damaging agents and silencing. FASEB. 2017;31(1):754–755. [Google Scholar]
- 18.Hu P., Tirelli N. Scavenging ROS: superoxide dismutase/catalase mimetics by the use of an oxidation-sensitive nanocarrier/enzyme conjugate. Bioconjugate Chemistry. 2012;23(3):438–449. doi: 10.1021/bc200449k. [DOI] [PubMed] [Google Scholar]
- 19.Patlevič P., Vašková J., Švorc P., Vaško L., Švorc P. Reactive oxygen species and antioxidant defense in human gastrointestinal diseases. Integrative Medicine Research. 2016;5(4):250–258. doi: 10.1016/j.imr.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schleit J., Johnson S. C., Bennett C. F., et al. Molecular mechanisms underlying genotype-dependent responses to dietary restriction. Aging Cell. 2013;12(6):1050–1061. doi: 10.1111/acel.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan S.-X., Teo M., Lam Y. T., Dawes I. W., Perrone G. G. Cu, Zn superoxide dismutase and NADP(H)homeostasis are required for tolerance of endoplasmic reticulum stress in Saccharomyces cerevisiae. Molecular Biology of the Cell. 2009;20(5):1493–1508. doi: 10.1091/mbc.E08-07-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birben E., Sahiner U. M., Sackesen C., Erzurum S., Kalayci O. Oxidative stress and antioxidant defense. World Allergy Organization Journal. 2012;5(1):9–19. doi: 10.1097/WOX.0b013e3182439613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buzas D. M., Nakamura M., Kinoshita T. Epigenetic role for the conserved Fe-S cluster biogenesis protein AtDRE2 in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(37):13565–13570. doi: 10.1073/pnas.1404058111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mesquita A., Weinberger M., Silva A., et al. Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(34):15123–15128. doi: 10.1073/pnas.1004432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martins D., English A. M. Catalase activity is stimulated by H2O2 in rich culture medium and is required for H2O2 resistance and adaptation in yeast. Redox Biology. 2014;2(1):308–313. doi: 10.1016/j.redox.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrisic L., Collinson E. J., Tehlivets O., et al. Transcriptional and antioxidative responses to endogenous polyunsaturated fatty acid accumulation in yeast. Molecular and Cellular Biochemistry. 2015;399(1-2):27–37. doi: 10.1007/s11010-014-2229-6. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y., Pulliam D. A., Liu Y., et al. Reduced mitochondrial ROS, enhanced antioxidant defense, and distinct age-related changes in oxidative damage in muscles of long-lived Peromyscus leucopus. American Journal of Physiology—Regulatory Integrative and Comparative Physiology. 2013;304(5):R343–R355. doi: 10.1152/ajpregu.00139.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawalek A., Lefevre S., Veenhuis M., Van der Klei I. Peroxisomal catalase deficiency modulates yeast lifespan depending on growth conditions. Aging. 2013;5(1):67–83. doi: 10.18632/aging.100519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maoz N., Gabay O., Ben-Asher H. W., Cohen H. Y. The yeast forkhead HCM1 controls life span independent of calorie restriction. Journals of Gerontology—Series A Biological Sciences and Medical Sciences. 2015;70(4):444–453. doi: 10.1093/gerona/glu059. [DOI] [PubMed] [Google Scholar]
- 30.Van Zandycke S. M., Sohier P. J., Smart K. A. The impact of catalase expression on the replicative lifespan of Saccharomyces cerevisiae. Mechanisms of Ageing and Development. 2002;123(4):365–373. doi: 10.1016/S0047-6374(01)00382-7. [DOI] [PubMed] [Google Scholar]
- 31.Zhao W., Zheng H., Zhou T., et al. CTT1 overexpression increases the replicative lifespan of MMS-sensitive Saccharomyces cerevisiae deficient in KSP1. Mechanisms of Ageing and Development. 2017;164:27–36. doi: 10.1016/j.mad.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 32.Ohtsuka H., Azuma K., Kubota S., et al. Chronological lifespan extension by Ecl1 family proteins depends on Prr1 response regulator in fission yeast. Genes to Cells. 2012;17(1):39–52. doi: 10.1111/j.1365-2443.2011.01571.x. [DOI] [PubMed] [Google Scholar]