Abstract
The HEN1 RNA 2’-O-methyltransferase plays important roles in biogenesis of small non-coding RNAs in plants and proved a valuable tool for selective transfer of functional groups from cofactor analogues onto miRNA and siRNA duplexes in vitro. Herein, we demonstrate versatile HEN1-mediated methylation and alkylation of small RNA strands in heteroduplexes with a range of complementary synthetic DNA oligonucleotides carrying user-defined moieties such as internal or 3’-terminal extensions or chemical reporter groups. The observed DNA-guided covalent functionalization of RNA broadens our understanding of the substrate specificity of HEN1 and paves the ways for developing novel chemo-enzymatic tools with potential applications in miRNomics, synthetic biology and nanomedicine.
Keywords: HEN1, methyltransferase, labeling, non-coding RNA, microRNA, siRNA, synthetic AdoMet analogue
Table of Contents
A new method for addressable chemo-enzymatic labeling of miRNAs and siRNAs exploits an unexpected activity of the HEN1 2′-O-methyltransferase to transfer functional groups to the 3′ end of the RNA strand in DNA/RNA heteroduplexes. The observed high tolerance of HEN1 to structural variation in the addressing DNA probe permits a variety of user-defined single- or dual-reporter applications, where one tag can be synthetically incorporated in the DNA probe and the second is covalently attached by HEN1 to the target RNA.
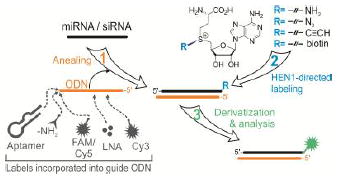
Small non-coding RNAs (ncRNAs) such as microRNAs (miRNAs) and small interfering RNAs (siRNAs) post-transcriptionally control gene expression in eukaryotic organisms.[1] They maintain genome stability and integrity, and govern a wide range of human physiological and pathological processes. Unique expression patterns of small RNAs for different types of tissues, stem cells, disease stages or therapy responses uncover a great potential for diagnosis, prognosis and targeted development of the novel therapies for human diseases.[2] Moreover, a high stability of ncRNA biomarkers in biological fluids provides diverse opportunities for development of non-invasive monitoring methods.[3] Although there is a range of general enzymatic and non-enzymatic techniques for RNA modifications,[4-6] the exploration of non-coding RNomes depends on reliable tools for selective covalent labeling of small RNAs, which is difficult to achieve using current methods.
The existing toolbox for targeted covalent labeling of RNA includes methyltransferase-directed 5’-cap functionalization in mRNAs,[7,8] internal programmable sequence-specific modification of RNAs using complex C/D box RNPs[9] systems or modification of a specific position in a particular tRNA.[10] However, none of these options is amenable for addressable targeting of small RNAs. Recently, the HEN1 small RNAs methyltransferase from Arabidopsis thaliana[11-13] has been exploited for selective covalent labeling of miRNA/miRNA* duplexes for their enrichment and analysis.[14] Naturally, HEN1 uses the cofactor S-adenosyl-L-methionine (AdoMet) for the 2′-O-methylation of the 3′-terminal extending nucleotides in small RNA duplexes, but is inert towards single RNA strands (Figure S1). The above strategy, methyltransferase-directed Transfer of Activated Groups (mTAG), combined the cognate specificity of the HEN1 methyltransferase and a synthetic AdoMet analogue carrying a sulfonium-bound extended side chain replacing the methyl group. However, practical implementation of this approach turn out to be limited due to the short life-times of mature microRNA duplexes in mammalian cells, which contrasts with a vast abundance of individual RNA strands.[15,16] Given that DNA/RNA hybrids are known to adopt a standard A-helix geometry characteristic of RNA duplexes,[17,18] we assumed that non-methylatable oligonucleotide probes could be used for hybridization with endogenous RNA strands to produce duplexes for selective modification with HEN1. Thus, we examined several types of model antisense oligonucleotides that bring certain practical advantages such as improved hybridization properties or higher durability in cellular environments.
In accord with these expectations, we found that HEN1 was able to perform efficient 3’-terminal methylation of the RNA strand in a series of model miRNA/DNA heteroduplexes (Figure S2A and Scheme S1). No HEN1-dependent modification was detectable in the DNA strands, which lack the 2’-OH group but retain the adjacent 3’-OH group in the 3’-terminal nucleotide, in DNA/DNA or RNA/DNA duplexes (Figure S2B and S9A, respectively), indicating a high regiospecificity of the reaction. Remarkably, the rates of methyl-group transfer onto miR173 or miR173* strand under single turnover conditions in miR173/DNA173-R2:D2 and miR173*/DNA173-R2:D2 heteroduplexes were similar or even greater than those of the natural miR173/miR173* substrates (Figure S2C and Table S1). Typically, the methyl group transfer to a particular RNA strand in a RNA/RNA duplex shows two kinetic phases: a faster phase corresponding to direct binding and modification of the target strand and a slower phase accounting for binding and release of the complementary strand followed by rebinding and methylation of the target strand.[11] In contrast, the methylation of miR173 and miR173* strands in RNA/DNA heteroduplexes followed single-exponential kinetics. This may suggest that HEN1 poorly interacts with a DNA strand and thus effectively binds the RNA/DNA duplex in a single orientation positioning the 3’-terminal nucleotide of the RNA strand in the catalytic center.
Crystallographic evidence[19] suggests that, in contrast to the target strand, which is closely engulfed by the active site and the so-called 5’-capping site, the 3’-terminus of the non-target strand is exposed on the outer surface of the protein suggesting its tolerance to structural modifications or extensions (Figure S3A). Indeed, we found no obvious effect on the methylation efficiency of the RNA strand even with DNA probes that contained 3’-terminal fluorophores (FAM or Cy5) or internal (Cy3) as well as locked nucleic acid (LNA) derivatives (Figure S3B). We then examined the modification of three miRNA-derived (miR173, miR-26a and let-7a) strands in RNA/DNA heteroduplexes bearing 0-7 nucleotide 3’-overhangs on the DNA strand and found that all of the examined substrates could be faithfully methylated by HEN1 under standard experimental conditions (Figure S4 and Figure S5, top row). Moreover, efficient methyltransferase activity in vitro was observed with blunt-ended RNA/DNA as well as RNA/RNA duplexes that contained no 3’-overhangs on either end (Figure S6). Slight variations of the single-turnover methylation rates observed with the blunt-ended and 3’-overhang RNA/RNA duplexes (Table S1) argue against a previously suggested requirement of dinucleotide 3’-overhangs in double-stranded substrates for the HEN1 activity in vitro.[13,20] The broader substrate specificity of the HEN1 methyltransferase observed in this study suggests that the well-established preference for ncRNA duplexes with 3’-dinucleotide overhangs in vivo may derive from interactions with partner proteins such as HYL1 and/or DCL1 rather than HEN1 itself.[21]
We next determined whether these RNA/DNA heteroduplexes can be selectively modified by HEN1 using synthetic AdoMet cofactor analogues carrying extended side-chains with functional amino (2-3), azide (4), alkyne (5) or biotin groups (6-7) (Figure 1) as previously reported for RNA/RNA duplexes.[14] Indeed, we found that the extended side-chains were efficiently transferred to the RNA strand in a range of RNA/DNA duplexes (Figure 1 and S7, S8); the modification levels with cofactors 2-6 were in the range of 70 98%. ESI-MS analysis (Figure S9 and Table S2) of the nucleoside composition in HEN1-modified RNA/DNA hybrids confirmed the transfer of a full-length side chain on the ribose moiety of a 3’-terminal nucleotide. The nature of the 3’-terminal ribonucleotide had a minor influence on the HEN1-directed alkylation efficiency (C, 96–99%; G, 92–96%; A, 76–92%; U, 62%) (Figure S10) suggesting that the presented method is suitable for labeling a diverse variety of types and isoforms of small ncRNAs. Moreover, the methyltransferase tolerated 3’-terminal and internal DNA modifications such as covalently attached amino groups, FAM or Cy3 fluorophores as well as an oligonucleotide containing several internal locked nucleic acid (LNA) nucleotides (Figure 1).
Figure 1. Selective oligonucleotide-directed 3’-terminal 2’-O-labeling of short RNA strands using HEN1 methyltransferase.
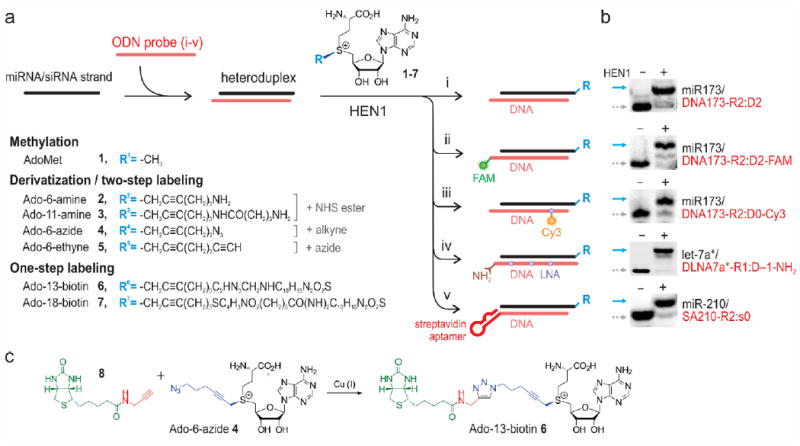
(a) RNA of interest is annealed to a complementary oligodeoxynucleotide (i), optionally containing 3’-terminal (ii) or internal (iii) affinity or fluorescence reporters, multiple modifications (iv) or a 3’-terminal DNA aptamer (v) followed by HEN1 methyltransferase-catalyzed transfer of extended side chains containing functional or reporter groups from synthetic cofactor analogues 1-7 for two-step (compounds 1-5) or one-step (compounds 6-7) 2’-O-labeling at their 3’-terminal nucleotides. (b) Gel electrophoretic analysis of representative examples of 3’-terminal modification of miRNAs with the Ado-11-amine cofactor 3 in heteroduplex structures (i-v) depicted on left; solid arrows point at bands corresponding to covalently modified RNA strands, dotted arrows – to input RNAs. (c) Synthesis of Ado-13-biotin 6 from Ado-6-azide 4.
To demonstrate that a synthetic DNA probe can selectively address the HEN1-mediated labeling to a predefined miRNA strand, we conducted the alkylation reaction in a mixture of three distinct miRNAs. The obtained data clearly showed that only the miRNA strand that was complementary to the added DNA probe was efficiently modified (Figure S11A). We further explored the selectivity of the method in multicomponent RNA mixtures. As shown in Figure S11B, the addition of up to a 100-fold excess of E. coli total RNA did not alter the modification efficiency of the spiked miR173 or miR-210 strands. Moreover, both a standard and a 3’-FAM-modifed DNA strand efficiently directed the derivatization of miR173 in the presence of a 10-fold excess of total RNA from U2OS osteosarcoma cells (Figure S11C).
In the second step, a range of commercially available reporter groups such as fluorophores or biotin can be conjugated to the covalently derivatized RNA strands to afford its selective visualization or purification (Figure S12B-D and Figure S13). Alternatively, a one-step biotin labeling of miRNA can be achieved using appropriate extended cofactor analogues (Figure S12A). As the modification yields of some miRNAs using the previously described Ado-18-biotin cofactor[14] proved rather low, we went on to prepare a new enantiomerically pure analogue, Ado-13-biotin 6, from the above mentioned azide cofactor 4 (Figure 1C). A high yield conjugation of 3-biotinoylamidoprop-1-yne was achieved by copper-assisted alkyne-azide cycloaddition (click) reaction under mildly acidic conditions. The new cofactor exhibited a significantly higher (87% versus 19%) trans-alkylation activity towards the least “favorable” miR-210/DNA210-R2:D2 heteroduplex (Figure S14). Akin to the AdoMet-dependent methylations described above, HEN1-directed transalkylations showed no obligatory requirement for the 3’-overhangs of the target RNA strand (Figure S8). Moreover, in many cases we found that shifting the DNA sequence to form blunt-ended heteroduplexes with target RNAs led to a higher efficiency of RNA modification. An even higher tolerance has been observed towards variations of the 3’-overhangs of the DNA strand (Figure S5, bottom row). Ultimately, we examined if HEN1 could tolerate a large functional ligand at the 3’-terminus of the guide oligonucleotide which could be suitable for the development of novel biotechnological or therapeutic applications. As a proof of principle, DNA oligonucleotides complementary to miR173 or miR-210 were extended at their 3’-termini to include a 29-mer streptavidin-specific DNA aptamer St-2-1.[22] Using these substrates we observed that HEN1 was able to both methylate and alkylate the miRNA strands in duplexes regardless of their 3’-overhang structure (Figure S15). Insertion of a tetranucleotide spacer between the aptamer and the complementary DNA sequences further improved the modification efficiency. The alkylated miRNA/aptamer was compatible with purification using streptavidin-coated magnetic beads. Importantly, we found that the double-stranded miR-210/SA210-R2:s0 structure interacted much more efficiently with the magnetic beads than SA210-R2:s0 alone (Figure S16) suggesting that hybridization with the target RNA enhanced the accessibility of the aptamer by eliminating fold-back interactions in the DNA strand.
In real biological systems, the DNA-directed HEN1-labeling of target RNA is prone to producing detectable background signal due to labeling of native miRNA/siRNA duplexes. This shortcoming could be resolved by using two-reporter detection whereby one reporter group (fluorophore, quencher or affinity tag) is synthetically incorporated in the targeting DNA probe and the other reporter is selectively deposited in the mTAG reaction (Figure S17). As an example, we tested whether our aptamer-based approach is suitable for selective click labeling and subsequent affinity extraction of desired ncRNA from a complex RNA pool. For this, we prepared four miR-26a, miR173, miR-210 and let-7a mixtures in which only one of the miRNAs was 5’-P33-labeled and thus visible on PAGE. After annealing with the miR-210-specific DNA aptamer, the samples were incubated with HEN1 and Ado-6-azide cofactor followed by labeling with a Cy5.5-alkyne dye. Only miR-210 was selectively labeled (Figure S18) and effectively enriched using streptavidin beads (Figure 2). The fluorescent band of the magnetic beads-bound (Bd) fraction co-localized precisely with the position of the P33-miR-210 band confirming that the Cy5.5 fluorophore was selectively attached to miR-210 (Figure 2 and Figure S18, bottom panels).
Figure 2. Aptamer-addressed selective labeling and extraction of miRNA.
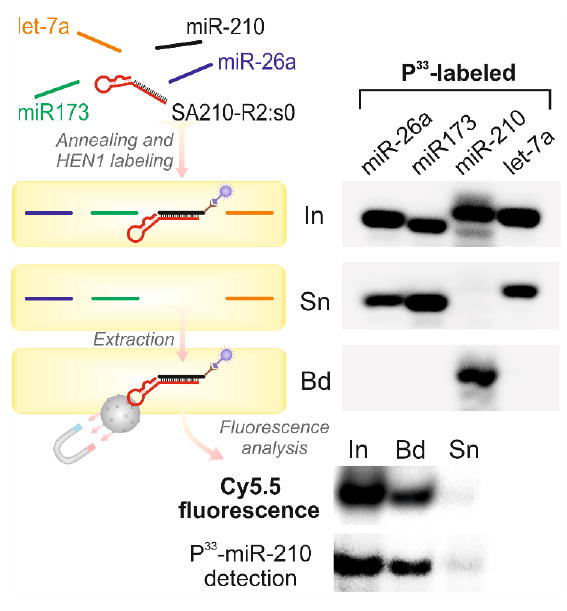
Four mixtures of miR-26a, miR173, miR-210 and let-7a miRNAs each containing differentP33-labeled miRNA were annealed to a streptavidin-specific aptamer SA210-R2:j0 complementary to miR-210. Samples were incubated with HEN1 and Ado-6-azide 4 cofactor, click-labeled with Cy5.5 alkyne fluorophore and immobilized on streptavidin-coated magnetic beads. P33-miRNAs were analyzed in input (In), supernatant (Sn) and magnetic beads (Bd) fractions after separation on a denaturing polyacrylamide gel. Cy5.5 fluorescence was detected.
A proof of principle experiment of a two-fluorophore assay utilizing Förster resonance energy transfer (FRET) in solution was carried out using miRNA/DNA heteroduplex labeled with cyanine dyes Cy3 (synthetic DNA probe) and Cy5 (3’-end labeled miRNA) (Figure 3, top panel). A clear FRET signal, which is characterized by a decrease of yellow emission of Cy3 and an increase of farred emission from Cy5, can only be detected when both the donor and acceptor chromophores are present within the miRNA/DNA heteroduplex (Figure 3, bellow panel). Similarly, the applicability of dual-fluorophore labeling was validated in a conventional polyacrylamide gel electrophoresis format (Figure S19). A strong signal was observed corresponding to FRET from the DNA-borne Cy3 to the Cy5.5 fluorophore that was attached to the 3’-terminus of the microRNA strand via HEN1-directed covalent alkynylation followed by a Cu(I)-assisted click reaction, but not in heteroduplexes carrying either one fluorophore (Figure S19b). Thus, our results demonstrate the first selective dual-reporter FRET-based detection of miRNA, providing a solid basis for the adaptation of this approach for analysis of desired microRNAs in biological samples.
Figure 3. A dual-reporter Förster resonance energy transfer (FRET) assay for selective microRNA detection in solution.
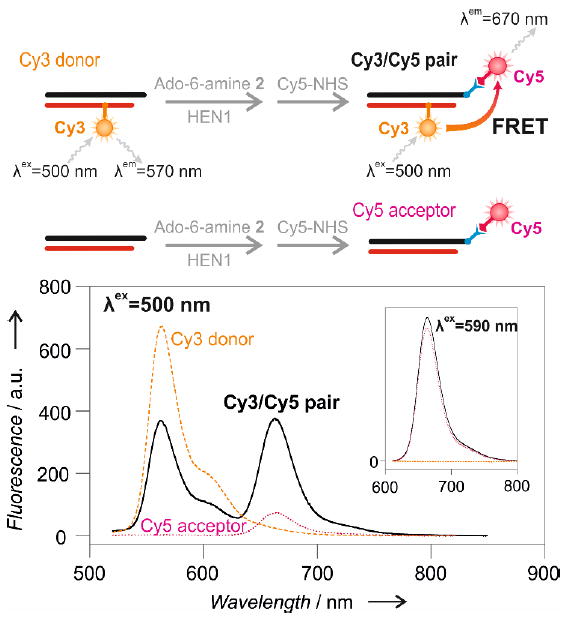
Top, schematic outline of FRET from a Cy3 reporter incorporated into the DNA strand of the miR173/DNA173-R2:D0-Cy3 duplex to a Cy5 dye attached by two-step HEN1-directed labeling to the 3’-end of the miRNA strand. Bottom, emission spectra at λex = 500 nm of the Cy3 donor (yellow), the Cy5 acceptor (magenta) and the Cy3/Cy5 pair (black). Inset, control spectra at λex = 590 nm show similar emission levels from the Cy5 fluorophore in the Cy5 acceptor and Cy3/Cy5 pair samples
Altogether, we demonstrate a novel chemo-enzymatic tool for addressable 3’-terminal covalent functionalization and labeling of individual small RNA strands in vitro. This paves the way to a broad range of potential applications of this approach in miRNomics research, nanomedicine and next generation analytical tools.
Supplementary Material
Acknowledgments
The authors thank Audronė Rukšėnaitė and Georgij Kostiuk for valuable help with MS and FRET analysis. This work was supported by the Research Council of Lithuania (grant number MIP-059/2015 to G.V.) and the NIH (HG007200 to S.K.).
Contributor Information
Aleksandr Osipenko, Institute of Biotechnology, Vilnius University, LT-10257 Vilnius, Lithuania.
Alexandra Plotnikova, Institute of Biotechnology, Vilnius University, LT-10257 Vilnius, Lithuania.
Milda Nainytė, Institute of Biotechnology, Vilnius University, LT-10257 Vilnius, Lithuania; Faculty of Chemistry and Geosciences, Vilnius University, Vilnius LT-03225, Lithuania.
Dr. Viktoras Masevičius, Institute of Biotechnology, Vilnius University, LT-10257 Vilnius, Lithuania Faculty of Chemistry and Geosciences, Vilnius University, Vilnius LT-03225, Lithuania.
Prof. Saulius Klimašauskas, Institute of Biotechnology, Vilnius University, LT-10257 Vilnius, Lithuania
Dr. Giedrius Vilkaitis, Institute of Biotechnology, Vilnius University, LT-10257 Vilnius, Lithuania
References
- 1.Ha M, Kim VN. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 2.Mendell JT, Olson EN. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ortiz-Quintero B. Cell Prolif. 2016;49:281–303. doi: 10.1111/cpr.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillingham D, Shahid R. Curr Opin Chem Biol. 2015;25:110–114. doi: 10.1016/j.cbpa.2014.12.025. [DOI] [PubMed] [Google Scholar]
- 5.Schulz D, Rentmeister A. ChemBioChem. 2014;15:2342–2347. doi: 10.1002/cbic.201402240. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Sousa R, Wang Y-X. BioEssays. 2016;38:192–200. doi: 10.1002/bies.201500119. [DOI] [PubMed] [Google Scholar]
- 7.Muttach F, Rentmeister A. Angew Chem Int Ed Engl. 2016;55:1917–1920. doi: 10.1002/anie.201507577. [DOI] [PubMed] [Google Scholar]
- 8.Muttach F, Mäsing F, Studer A, Rentmeister A. Chem Weinh Bergstr Ger. 2017 doi: 10.1002/chem.201605663. [DOI] [PubMed] [Google Scholar]
- 9.Tomkuviene M, Clouet-d’Orval B, Cerniauskas I, Weinhold E, Klimasauskas S. Nucleic Acids Res. 2012;40:6765–6773. doi: 10.1093/nar/gks381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motorin Y, Burhenne J, Teimer R, Koynov K, Willnow S, Weinhold E, Helm M. Nucleic Acids Res. 2011;39:1943–1952. doi: 10.1093/nar/gkq825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plotnikova A, Baranauskė S, Osipenko A, Klimašauskas S, Vilkaitis G. Biochem J. 2013;453:281–290. doi: 10.1042/BJ20121699. [DOI] [PubMed] [Google Scholar]
- 12.Vilkaitis G, Plotnikova A, Klimasauskas S. RNA. 2010;16:1935–1942. doi: 10.1261/rna.2281410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plotnikova A, Osipenko A, Masevičius V, Vilkaitis G, Klimašauskas S. J Am Chem Soc. 2014;136:13550–13553. doi: 10.1021/ja507390s. [DOI] [PubMed] [Google Scholar]
- 15.Guo Y, Liu J, Elfenbein SJ, Ma Y, Zhong M, Qiu C, Ding Y, Lu J. Nucleic Acids Res. 2015;43:2326–2341. doi: 10.1093/nar/gkv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marzi MJ, Ghini F, Cerruti B, de Pretis S, Bonetti P, Giacomelli C, Gorski MM, Kress T, Pelizzola M, Muller H, et al. Genome Res. 2016;26:554–565. doi: 10.1101/gr.198788.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong Y, Sundaralingam M. Structure. 1998;6:1493–1501. doi: 10.1016/s0969-2126(98)00148-8. [DOI] [PubMed] [Google Scholar]
- 18.Anosova I, Kowal EA, Dunn MR, Chaput JC, Van Horn WD, Egli M. Nucleic Acids Res. 2016;44:1007–1021. doi: 10.1093/nar/gkv1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Ji L, Huang Q, Vassylyev DG, Chen X, Ma J-B. Nature. 2009;461:823–827. doi: 10.1038/nature08433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z, Ebright YW, Yu B, Chen X. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baranauskė S, Mickutė M, Plotnikova A, Finke A, Venclovas Č, Klimašauskas S, Vilkaitis G. Nucleic Acids Res. 2015;43:2802–2812. doi: 10.1093/nar/gkv102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bing T, Yang X, Mei H, Cao Z, Shangguan D. Bioorg Med Chem. 2010;18:1798–1805. doi: 10.1016/j.bmc.2010.01.054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


