Abstract
Many functional RNAs have an evolutionarily conserved secondary structure. Conservation of RNA base pairing induces pairwise covariations in sequence alignments. We developed a computational method that quantitatively tests whether covariation analysis supports the presence of a conserved RNA secondary structure: R-scape (RNA Structural Covariation Above Phylogenetic Expectation). R-scape analysis finds no statistically significant support for the proposed secondary structures of the long noncoding RNAs HOTAIR, SRA, and Xist.
Pairwise covariations in RNA alignments provide a means of deducing evolutionarily conserved RNA secondary structures.1–5 In turn, a conserved secondary structure provides positive evidence for a noncoding RNA function. The first manual covariation analyses of small numbers of aligned RNA sequences used rules of thumb to infer conserved structures from a few compensatory base pair substitutions2;6. A “compensatory base pair substitution” means observing two substitutions at a pair of positions of an RNA sequence alignment that preserve Watson-Crick or G:U base pairing. As the number of aligned sequences grows, apparent compensatory base pair substitutions may be observed by chance. The potential for seeing chance compensatory substitutions is exacerbated by phylogenetic correlations: two independent single residue substitutions that fortuitously look compensatory can propagate into several descendants, and appear like several compensatory base pair substitutions (Fig. 1).
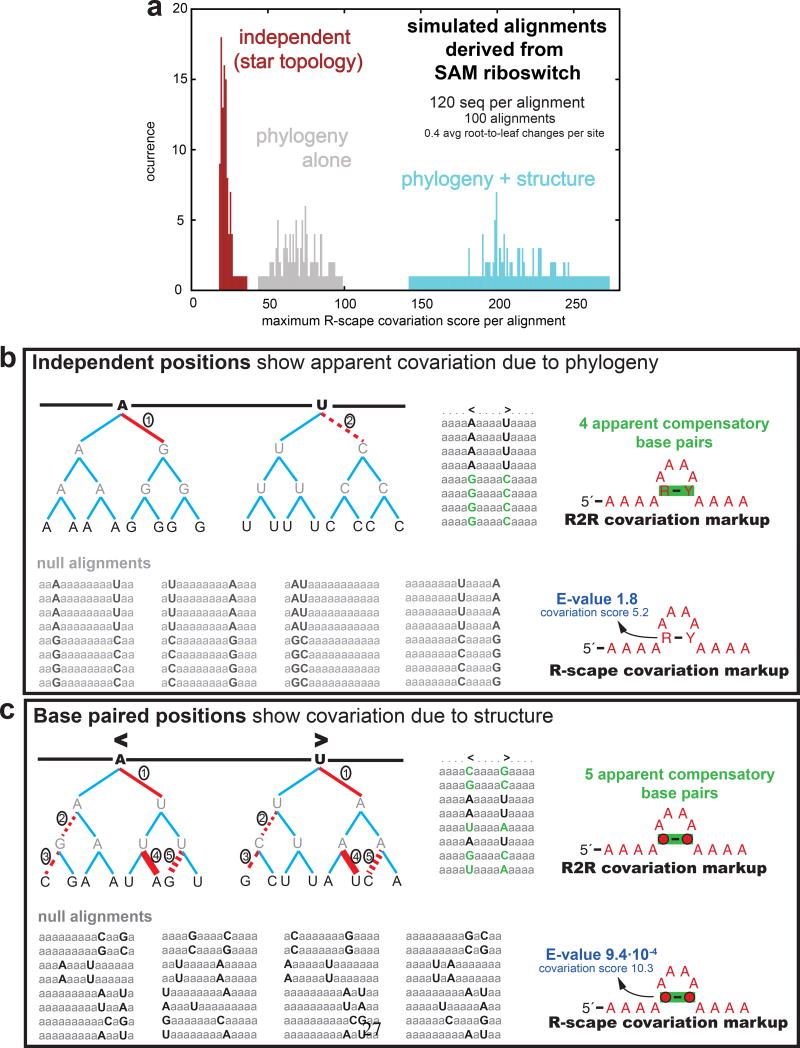
Figure 1. Independent substitutions on a tree can create confounding covariations.
(a) Illustrative example showing a histogram of maximum covariance score per alignment, over 100 synthetic alignments simulated under three evolutionary models: no phylogeny/no structure constraint (red); phylogeny alone (grey); or phylogeny plus structure constraint (cyan). (b) Toy alignment (top left) with two independent substitutions (marked 1,2) on the same branch, resulting in an apparent pairwise covariation annotated by R2R (top right). R-scape simulated null alignments (bottom left) retain this confounding covariation signal, and it is judged insignificant (bottom right). (c) Toy alignment with five compensatory base pair substitutions (marked 1–5) showing a covariation pattern that is destroyed in the R-scape simulated null alignments, and thus judged significant.
Because an RNA can function as an unstructured sequence7, and because some RNAs, particularly some long noncoding RNAs (lncRNAs), may be unannotated coding mRNAs or various sorts of transcriptional noise8, there is a need for computational tools that distinguish whether or not sequence alignment analysis provides statistical support for an evolutionarily conserved RNA secondary structure. There is extensive literature on RNA covariation analysis methods9–12, but these methods have been underutilized, perhaps because no one computational tool has yet adequately combined covariation analysis with statistical significance testing, computational efficiency, and accessibility.
For example, the evidence for structure conservation in the lncRNA HOTAIR13 consisted of using the RNA drawing program R2R14 to annotate an alignment of 33 sequences, using a proposed consensus structure based on chemical and enzymatic probing experiments. Examination of this HOTAIR alignment shows that in most cases only a single compensatory base pair substitution supports each proposed covarying base pair, while many substitutions disrupt the proposed pair. R2R was intended for visualization of known RNA structures, not to quantitate evidence for structure conservation14. It annotates a consensus base pair as covarying if any compensatory base pair substitution is observed, even just one, and it does not consider substitutions that conflict with the proposed structure.
We have developed an accessible tool, R-scape (RNA Structural Covariation Above Phylogenetic Expectation), that analyzes a multiple RNA sequence alignment and quantitates the statistical support for evolutionary conservation of an RNA structure. A pairwise covariation statistic is calculated for each alignment column pair, and statistically significant covariation is interpreted as evidence for a conserved RNA base pair. We compared several covariation statistics on a test set of annotated consensus structures for 104 RNA sequence alignments from Rfam15 (Methods), counting a false negative when an annotated base pair has a covariation statistic below threshold, and a false positive when an unstructured pair scores above threshold. The G-test statistic16 was more robust than other statistics tested, including mutual information (Methods; Supplementary Fig. 1). A background correction17 further improves covariation detection (Supplementary Fig. 1a). R-scape calculates the average product corrected (APC) G-test covariation statistic by default.
The APC G-test statistic can be calculated rapidly even for deep sequence alignments, but it does not explicitly deal with confounding covariation caused by phylogenetic correlation (Fig. 1a)5. Methods that do tend to be computationally expensive9;11;12 (Supplementary Fig. 2). R-scape instead determines the statistical significance of the observed covariation scores, by simulating alignments under a null hypothesis in which phylogenetic relationships are preserved but columns evolve independently. For example, in Fig. 1b, a toy alignment (top) has two independent substitutions in two different columns, in the same tree branch, resulting in four apparent compensatory base pairs in the alignment. In R-scape sampled null alignments (bottom), the same two substitutions are made on the same branch, but with their sequence positions randomized, and the apparent covariation remains. In another toy alignment (Fig. 1c, top), a covarying base pair has five compensatory base pair substitutions in five different branches. In R-scape sampled null alignments (Fig. 1c, bottom), after randomizing the sequence position of each substitution on each branch, the five compensatory pair substitutions become ten uncorrelated substitutions, destroying the correlation seen in the alignment.
For a given alignment, R-scape produces many simulated null alignments (default 20) and calculates an APC G-test statistic for each alignment column pair, thus collecting an expected null distribution conditioned on the input alignment’s characteristics including its length in columns, sequence number, pairwise identity, base composition, substitution types, and phylogenetic correlation (Methods). This empirical null distribution of the covariation statistic estimates the probability of obtaining a false positive on null data (the p-value) at any threshold. From a p-value, we calculate an expectation value (E-value) by multiplying by total number of column pairs evaluated. An E-value E(x) is the number of column pairs expected to give a covariation score of at least x when they are evolving independently, under no RNA structure constraint. A significant E-value is ≪ 1.
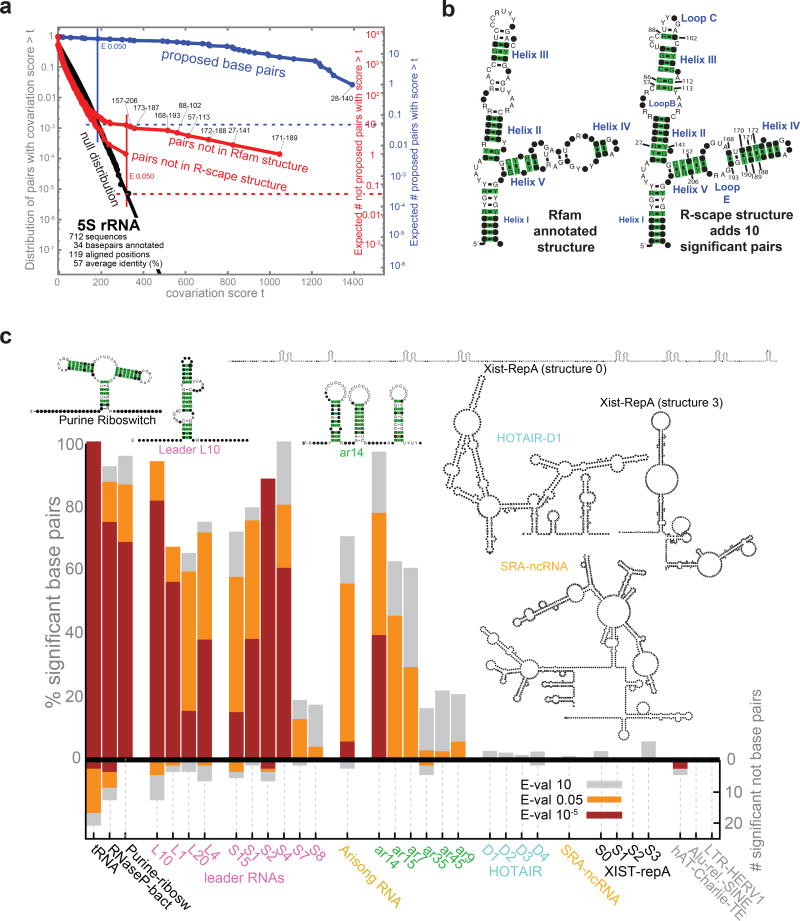
Fig. 2a shows an example of a known structural RNA, 5S ribosomal RNA. R-scape analysis of the Rfam 5S alignment shows significant covariation support for 22/34 base pairs in the annotated consensus structure. There are also eight significant pairs not in the Rfam-annotated consensus structure, which provides an example of how R-scape can not just support but improve a structural annotation (Fig. 2b). Using an optional feature that predicts a new consensus secondary structure that includes the maximum number of significantly covarying pairs, R-scape proposes a modified 5S rRNA consensus structure in which 32/38 base pairs are significant. The R-scape structure is in agreement with the accepted 5S rRNA consensus structure18, suggesting there are some errors in the curated Rfam structure. Three other examples are shown in Supplementary Fig. 3, showing R-scape support for consensus structures of two small noncoding RNAs and a cis-regulatory mRNA structure.
Figure 2. Covariation analysis of known or proposed RNA secondary structures.
(a) 5S rRNA as an example of an known structural RNA with significant covariation support. The plot (left) shows the expected null distribution (fit: black lines; data: black circles), compared to covariances observed for pairs in (blue) or not in (red) the annotated structure. (b) Covariation support for the Rfam annotated 5S rRNA structure, versus an alternative structure proposed by R-scape to include all significantly covarying pairs. Significant pairs (at E< 0.05) are highlighted in green. Coordinates are alignment column positions. Specific nucleotides are shown when their weighted frequency in the column exceeds 50%; black dots represent more variable positions. (c) On the positive y-axis, plot shows percentage of base pairs supported by covariation at three thresholds (red, E< 10–5; orange, E < 0.05; grey, E < 10). Negative y-axis shows the number of additional significantly covarying pairs not in the proposed structure. Transfer RNA (tRNA), RNase P RNA, and the purine riboswitch (leftmost) are examples of functional structural RNAs with strong statistical support; three human DNA repeat elements (rightmost) are negative controls with no known RNA secondary structure constraint.
Fig. 2c summarizes the covariation evidence for several RNA structures proposed in the literature. For alignments of typical, known structural RNAs (transfer RNA, bacterial RNAse P RNA, purine riboswitch), 70–100% of the annotated base pairs are supported by R-scape with E-values < 10–5. For a recent study of ten γ-proteobacterial mRNA leader structures that autoregulate ribosomal protein synthesis19, eight proposed structures have many significant covarying pairs, and only two show weaker support (one, the S7 leader, overlaps the ribosomal protein L5 coding region and therefore has restricted variation). For a recent screen identifying six small RNAs in α-proteobacteria20, three (αr14, α15, and αr7) have good covariation support.
For the proposed HOTAIR lncRNA structure, using the same alignment used by Somarowthu et al.13, no significantly covarying base pairs are found for any of the four proposed domain structures. This result differs from the previous HOTAIR analysis13 because R-scape accounts for the fact that the observed sequence variation is more frequently inconsistent than consistent with the proposed structure. Details of the analysis of proposed helices H7 and H10 are in Supplementary Fig. 4.
For the proposed ncSRA lncRNA structure21, R-scape does not find any significantly covarying pairs. As with HOTAIR, the picture is of sequence variation, not covariation. Details of the analysis of putative helices H3 and H4, and putative helices H9, H20, and H21, are provided in Supplementary Fig. 5 and Supplementary Fig. 6.
For the Xist lncRNA, consensus structures have been proposed for the repeat A (RepA) region, with compensatory base changes cited as support22, but R-scape shows no significant covariation support for any base pair. The alignment in the published analysis has only 10 sequences22, which limits power in identifying covariation support.
A different RepA secondary structure has been proposed, and said to have covariation support for four base pairs in an alignment of 13 sequences23. Applying the same criteria used by Fang et al.23 systematically to all column pairs shows 541 pairs with equivalent support (Supplementary Fig. 7), 538 of which are inconsistent with the proposed structure. This highlights another source of confounding signal. Independent G→A and U→C substitutions in conserved G+A and U+C columns (454/541 pairs in this RepA alignment) create an appearance of covariation support because of G:U wobble base pairing. R-scape null alignments reproduce this sequence variation pattern, so R-scape finds no statistically significant covariation support.
Failure to identify significant covariation support for an evolutionarily conserved RNA secondary structure does not necessarily mean that a structure is not present. Deeper alignments or a more powerful statistical analysis might reveal a more subtle conserved secondary structure for any of these RNAs. The published Xist RepA alignments have few sequences (10–13), for example, and the pattern of sequence conservation is consistent with a large number of possible base pairs, too many to deduce any single conserved structure; here especially, deeper alignments may help. In contrast, the published HOTAIR and ncSRA alignments are already relatively deep (30–60 sequences) and their pattern of sequence variation is inconsistent with their proposed structures. Here it may be more likely that any functions of these RNAs may depend more on their linear sequences than on conserved secondary structure7;24. There are many other lncRNAs, and lncRNA function and structure remain controversial and difficult to study. Tools like R-scape will be useful for quantitative analysis of the covariation evidence supporting proposed structures of lncRNAs, or indeed of any RNA.
Methods
R-scape: RNA Structural Covariation Above Phylogenetic Expectation
An R-scape web server is at eddylab.org/R-scape. The current version of the R-scape source code is freely downloadable from eddylab.org, and an archived tarball of the version used in this paper (version 0.2.2) is included as part of the online Supplementary Information. The source code for the web server is freely available at github.com/EddyRivasLab/R-scape-web.
The input to the R-scape program is a multiple RNA sequence alignment, typically with a consensus structure, in Stockholm format. The output is a list of pairs of alignment columns that significantly covary, ranked by their E-value. The source code distribution includes examples of input and output files, and documentation. The R-scape web server is a wrapper around the R-scape program.
If a consensus RNA structure is provided for the input alignment, R-scape evaluates the covariation support for that consensus structure. Optionally, R-scape can calculate an independent consensus structure, which is the maximum likelihood secondary structure constrained to use all significantly covarying pairs (for a given E-value cutoff). This constrained folding method uses the probabilistic “basic grammar” model introduced with the RNA folding method Tornado25. This algorithm cannot predict pseudoknots, but the presence of nested significant pairs not in the structure but compatible with it (depicted in orange in the web application) are a good indication of their possible presence.
By default, R-scape (v0.2.2) uses two external programs: FastTree26 (v2.1.8) to build a phylogenetic tree from the given alignment by approximate maximum likelihood, and a modified version of R2R14 (v1.0.4) to draw the consensus structure annotated with the covarying base pairs with E-values smaller than a given E-value cutoff.
Different covariation statistics
We tested the following statistics for measuring pairwise covariations, out of many different statistics that have been suggested. Given two alignment columns i, j:
where a, b are (non-gap) residues; N is the total number of aligned sequences; is the observed count of a : b pairs in columns i, j (only counting when both a,b are residues); Nij. is the total number of residue pairs in columns i, j (only counting when both a,b are residues); is the observed frequency of pair a : b in columns is the expected frequency of pair a : b assuming i, j are independent, where are the marginal frequencies of a residues in column i (averaged to all other positions) is the number of pairs involving at least one gap symbol. The definition of Bi,j used in the RAF and RAFS statistics is complicated and not shown here; their definition can be found elsewhere10.
We also tested two background corrections that can be applied to any of the above covariation statistics17. Let COV(i, j) be a covariation statistic; then:
where is an average covariation for an individual column i, and is the average covariation overall.
By default R-scape uses the APC G-test statistic, as this was the most robust statistic in our benchmark tests (Supplementary Fig. 1; also, see below). Compared to the related and more commonly used Mutual Information (MI) statistic, the G-test (based on observed counts) is different from MI (based on frequencies) for alignments with gaps. A column pair with many gaps could have similar MI to another column pair with no gaps, but the G-test score of the former will have smaller magnitude (fewer number of effective sequences) than the G-test score for the latter. This difference makes G-test a more robust statistic than MI on alignments with gaps (Supplementary Fig. 1b).
Comparing the G-test to the RAFS statistic, while RAFS has better sensitivity (as has been reported before10), RAFS is more prone to report covariations among non base pairs, especially for E-values larger than 10–4 (worse positive predictive value), and it is also more affected by alignment gaps. This could either be because RAFS is less specific (more false positives), or because it is more sensitive to true (tertiary structure) non-Watson-Crick interactions. We tested this by simulating alignments in which we preserve the base paired columns, but replaced all unpaired columns using the null model of phylogenetically dependent but position-independent changes. Results for these partially simulated alignments are similar to those of the original alignments in Supplementary Fig. 1b (data not shown), suggesting that RAFS is less specific. The RAFS statistic is also slower to calculate, scaling with the square of the number of sequences in the alignment.
R-scape filters the input alignment before collecting observed counts such that columns with greater than 50% gap symbols are ignored. After this filtering, relative sequence weights are calculated using the GSC algorithm32. These steps help compensate for the fact that simple correlation statistics do not take the phylogenetic relationship of the sequences into account. Some covariation statistics have been developed that directly account for phylogeny9;11, but at a cost of increased computational and model complexity.
The covariation statistics can either be applied as 16-class tests (taking the summation over a : b over the 16 possible base pairs) or as 2-class tests where pairs are separated into two groups according to whether they correspond to canonical Watson-Crick pairs (including G:U) or not33. A two-class test is better on a small number of sequences (see Supplementary Fig. 1) and short alignments (not shown), because with fewer classes, it is less susceptible to statistical fluctuations in small numbers of counts. The RAFS statistic is intrinsically of the 2-class type. A two-class test looks specifically at whether covariation is consistent with Watson-Crick/G:U base pairing; a 16-class test detects any pairwise correlation, including non-Watson-Crick pairing seen in RNA tertiary contacts. For alignments with more than 8 sequences, we use the 16-class covariation as it performs similarly to the 2-class covariations for larger numbers of sequences, while also allowing identification of possible non-Watson-Crick covariations (Supplementary Fig. 1c).
The same approach could be applied to other covariation statistics. A maximum entropy approach called Direct Coupling Analysis (DCA), first introduced for protein sequence covariation analysis34, has been applied to RNA35;36. DCA produces pairwise pseudoenergies based on pairwise correlation statistics observed in a multiple alignment. Strong pseudoenergies are thought to reflect direct structural interactions. Currently, applications of DCA methods to structure prediction are simply taking N top ranking pseudoenergies for an arbitrarily chosen N, without using a measure of statistical significance. The general approach used in R-scape could provide a means for assigning significance to DCA scores, allowing better discrimination of signal and noise, and allowing more meaningful comparison to other covariation statistics.
Benchmarking to choose the default covariation statistic
We evaluated the different covariation statistics on a test set of 104 alignments taken from seed alignments in the Rfam sequence family database15, manually chosen to give wide representation of known structural RNAs with well studied, more reliable consensus secondary RNA structures, and to have at least 40 sequences in the alignment. The test set includes: 2 tmRNA families, vault RNA, 6S RNA, U7 small nuclear RNA, 9 rRNA families (including 5.8S, 3 small subunit, and 3 large subunit rRNA families); 3 signal recognition particle RNAs; selenocysteine tRNA; 14 riboswitch RNAs; 1 leader peptide; 23 other cis-regulatory RNAs; 9 spliceosomal RNAs; 5 ribozymes; 13 sRNAs; 8 group-II introns; 5 miRNAs; 3 C/D-box snoRNAs; and 5 other RNA genes. The percentage identity of the alignments ranges from 41% to 80% (defined as the average pairwise % identity over all aligned sequence pairs, with pairwise identity calculated as the ratio of identical positions divided by the minimum length of the two sequences). The number of sequences varies from 44 (Glycine riboswitch) to 956 (glnA, a bacterial regulatory sRNA).
In the analysis presented in Supplementary Fig. 1, we aggregate all alignments together. The total number of consensus base pairs is 7,483, and the total number of alignment columns is 50,769. When columns with more than 50% gaps are removed, the average percentage identity of the alignments (ranging from 42% to 79%) remains similar to that of the original alignments. The number of base pairs remains unchanged, but the number of analyzed columns is reduced to 28,526. A complete list with summary statistics, and the alignments themselves are provided in the online Supplementary Information.
We tested the different covariation statistics on the 104 test alignments and their trusted consensus secondary structures, measuring the fraction of base pairs detected (and the fraction of detected pairs that are base pairs) at different E-value significance thresholds. The results, leading to the choice of the APC G-test statistic as the default, were summarized above.
The ability to detect significantly covarying base pairs depends on many factors, including the quality of the alignment, and the number and diversity of the sequences in it. We tested the average effect of varying sequence number in the test alignments (Supplementary Fig. 1d). Typically, about 60% of base pairs are detected as significant at E < 0.05 when an input alignment contains 40 sequences, depending on other details of the alignment such as percent identity. More sequences have diminishing benefit. Fewer sequences compromises detection; with only 10 sequences in the alignment, few base pairs are detected. Supplementary Fig. 1e shows a scatter plot of % detected base pairs (at an E < 0.05 threshold) versus average % identity in the alignment, showing that there is substantial variation from alignment to alignment, some of which is accounted for by sequence diversity (unsurprisingly). These results emphasize that the failure to detect significant covariation for an individual base pair does not necessarily mean that the base pair is not present in a conserved structure.
Significance calculations on simulated null data
In R-scape, we calculate the significance of covariation scores by simulating phylogenetically related sequences under a null hypothesis of independently evolving columns. Given an alignment, we estimate a tree by approximate maximum- likelihood using the FastTree method (version 2.1.7 SSE3)26, root the tree by midpoint rooting, and assign substitutions to branches by maximum parsimony (using the Fitch algorithm37). We then simulate an alignment of the same depth (in sequences) and width (in columns), starting from a parsimoniously inferred root sequence. For each ancestral node, we introduce the same set of single nucleotide substitutions observed on each original descendant branch, while randomizing their positions. For example, if we are introducing an A→G substitution on a branch, we choose a random A in the ancestral sequence and substitute it to a G in the descendant. The result is a sampled null alignment which has exactly the same base composition as the input alignment, exactly the same set of single nucleotide substitutions, and similar pairwise percentage identities as the original sequences, while any correlated pairwise substitution has been scrambled.
E-value estimation
R-scape uses simulated null alignments (above) to estimate the expected number of false positives (E-value) as a function of covariation score. Because simulated null alignments are generated by a resampling strategy on the input alignment, false positive estimation takes into account the characteristics of the input alignment, including base composition, number of sequences, average pairwise identity, and phylogeny. By default, 20 synthetic null alignments are generated and scored, and the tail of the resulting survival distribution for covariation scores is fitted to a truncated gamma distribution by maximum likelihood38 to estimate P(score > x), the probability that one tested column pair would give a covariation score better than x.
The expected number of false positives is then E = NP(score > x), where N is the number of column pairs tested. R-scape calculates two different E-values. One assumes that we are testing the support for a given secondary structure, in which case N is the number of proposed base pairs in that structure (and N scales with the length of the alignment L). The second assumes that we are testing for any other column pair that shows statistically significant covariation, in which case N is the total number of possible pairs in the alignment, less those in the proposed structure (and N scales with L2).
Because E-values are based on stochastic simulations, there is some run-to-run variability. R-scape E-values are typically reproducible to an accuracy of about 2-fold in different runs of the program.
R-scape uses the default E-value of 0.05 to define a significant covarying pair, which means that it would be expected to detect about 5 false positives overall in 100 different analyzed alignments.
Computational efficiency
R-scape is fast and memory-efficient. RNA sequence alignments smaller than ∼1,000 columns take under 10 secs each and under 50 MB of memory (Supplementary Fig. 2). As an example of a large RNA, the Rfam bacterial small subunit ribosomal RNA seed alignment15 of length 1980 columns and 99 sequences takes 49 secs and 800 MB of memory. The 956 sequence alignment of the glnA glutamine riboswitch RNA (274 alignment columns) takes 9 seconds and 16 MB of memory. Empirically, R-scape computation time scales approximately NL1.2 for sequence number N and alignment length L (Supplementary Fig. 2). Thus R-scape can be run systematically on any RNA alignment(s).
Simulations of sequences related by phylogeny alone, or by phylogeny and RNA structure
Fig. 1a uses simulated data to illustrate the effect on covariation statistics for sequences related by phylogeny alone, versus sequences related by phylogeny and with structurally constrained positions. From an alignment of a known structural RNA, we produce synthetic alignments guided by the consensus structure, alignment, and phylogeny of the given RNA according to one of the three scenarios: (1) simulated sequences are evolved following the phylogeny and the structure of the RNA; (2) simulated sequences follow the phylogeny but positions are independent from each other; and (3) sequences and positions are independent from each other (alignment is not structural, and follows a star topology).
As an example, Fig. 1a starts with the Rfam seed alignment of the SAM riboswitch. From a sub-alignment of a random set of 120 sequences from the original Rfam SAM riboswitch alignment (433 sequences total), a phylogenetic tree was calculated using FastTree26. Starting from a random SAM riboswitch sequence as the root, three different evolutionary models are applied. The structural/phylogenetic model generates sequences along the branches of the SAM riboswitch tree, with base pairs in the ancestral sequence substituted under a 16×16 base pair substitution process. The phylogenetic model generates sequences along the branches of the SAM riboswitch tree, but each ancestral position evolves independently. The independent model evolves extant sequences directly from the root, using the overall distance from root to leaf of the SAM riboswitch tree. In Fig. 1a, for each simulated alignment, we collect the maximum R-scape covariation score from all possible pairs. We generated 100 synthetic alignments for each of the three methods.
The structural/phylogenetic model depends on a 16×16 rate matrix (a base pair evolving to another base pair) calculated from the base paired positions in a collection of SSU and LSU ribosomal RNA alignments, and a 4×4 rate matrix calculated from the unpaired positions of the same alignments. The phylogenetic and independent models use a 4×4 rate matrix created from the same alignment but using all aligned columns. The rate matrices were obtained as the logarithm of the conditional probability (substitution) matrices obtained for the rRNA alignments, normalized to one substitution per site. Insertions and deletions were created using the AIF evolutionary model39 parameterized with these rate matrices. Individual insertions (possibly consisting of several residues) are assumed to be independent from each other, and once created they do not evolve.
The code to produce these simulations (R-scape-sim) from a Stockholm alignment of a structural RNA as input is provided as part of the R-scape source code package, including the rate matrices used.
Comparison to other methods
We identified two other methods (MICA and CoMap, from package CoMap v1.5.1) that calculate covariations and provide an estimation of statistical significance12. MICA (mutual information coevolution analysis) implements the Mutual Information statistic. We used MICA using a background corrected MIp covariation statistic, and z-scores to estimate significance. CoMap (cosubstitution mapping) uses a phylogenetic tree to calculate its covariation statistic. We used parametric bootstrapping to estimate significance for CoMap scores. (Those are the default settings provided with the CoMap v1.5.1 package.)
In Supplementary Fig. 2, we show that R-scape is at least as fast as the phylogenetic-free method MICA, while at the same time it performs favorably when compared with the fully phylogenetic method CoMap. The program CoMap was also run using a phylogenetic tree created with PhyML40 with similar results (not shown).
Provenance of structural alignments
The Arisong ciliate ncRNA alignment for 69 sequences was provided by S. Jung, as an updated version of the alignment given in the original manuscript41. The six α-proteobacteria ncRNA alignments (αr7, αr9, αr14, αr15, αr35, and αr45) were provided by C. del Val, as updated versions of the alignments given in the original manuscript42. The alignments for the 10 γ-proteobacteria ribosomal protein mRNA leader regions were obtained from the supplemental material of Fu et al. 19. The alignments for the four HOTAIR domains D1-D4 and their proposed secondary structures were provided by S. Somarowthu13.
We were unable to obtain alignments for the SRA ncRNA described in Novikova et al.21 from the authors. Instead a close approximation was produced by reproducing the proposed secondary structure of the human ncSRA by hand from Supplementary Fig. 1 of the above manuscript, and imposing it as a consensus structure in the Multiz100way alignment of the ncSRA region obtained from the UCSC human genome browser (http://genome.ucsc.edu). This alignment includes 76 mammalian species.
The alignment for the Xist repA region described in Maenner et al.22 was obtained from their Supplemental Fig. 5. Four alternative secondary structures (named here S0 to S3) were presented22. The consensus structures for the mouse sequence were reproduced by hand from Figs. 2, 3,4,5. The alignment includes 10 vertebrate sequences, with average length of 438 nucleotides, and average percentage identity of 77%.
The alignment for the Xist RepA region described in Fang et al.23 was provided by W. Moss. The alignment includes 13 vertebrate sequences, with average length of 423 nucleotides, and average percentage identity of 75%. In the alignment, we imposed by hand the proposed mouse secondary structure obtained by targeted structure-seq23.
Rfam alignments where obtained from Rfam v12.0 seed alignments15. The human repeat alignments were obtained from Dfam v2.0 seed alignments 43.
All the alignments used in this analysis are provided in Stockholm format as part of the online Supplementary Information. Details of the properties of the alignments are provided in Supplementary Fig. 8.
Supplementary Material
Acknowledgments
We thank S.E.R. Egnor for suggesting the name R-scape, and the Centro de Ciencias de Benasque Pedro Pascual in Spain, where part of this manuscript was drafted.
Footnotes
Supplementary information is linked to the online version of the paper.
Authors contribution E.R. and S.R.E. designed the method and wrote the manuscript. E.R. wrote the code, and designed and carried out the experiments. J. C. wrote the R-scape web application.
Author information An R-scape web server is at eddylab.org/R-scape and source code can be downloaded from a link on that page. The authors declare that they have no competing financial interests. Correspondence and requests for materials should be addressed to E.R. (elenarivas@fas.harvard.edu).
References
- 1.Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, Zamir A. Structure of a ribonucleic acid. Science. 1965;14:1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- 2.Noller HF, Kop J, Wheaton V, Brosius J, Gutell RR, Kopylov AM, Dohme F, Herr W, Stahl DA, Gupta R, Woese CR. Secondary structure model for 23S ribosomal RNA. Nucl. Acids Res. 1981;9:6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pace NR, Smith DK, Olsen GJ, James BD. Phylogenetic comparative analysis and the secondary structure of Ribonuclease P RNA – a review. Gene. 1989;82:65–75. doi: 10.1016/0378-1119(89)90031-0. [DOI] [PubMed] [Google Scholar]
- 4.Williams KP, Bartel DP. Phylogenetic analysis of tmRNA secondary structure. RNA. 1996;2:1306–1310. [PMC free article] [PubMed] [Google Scholar]
- 5.Michel F, Costa M, Massire C, Westhof E. Modeling RNA tertiary structure from patterns of sequence variation. Meth. Enzymol. 2000;317:491–510. doi: 10.1016/s0076-6879(00)17031-4. [DOI] [PubMed] [Google Scholar]
- 6.Gutell RR, Power A, Hertz GZ, Putz EJ, Stormo GD. Identifying constraints on the higher-order structure of RNA: Continued development and application of comparative sequence analysis methods. Nucl. Acids Res. 1992;20:5785–5795. doi: 10.1093/nar/20.21.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidovich C, Cech TR. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA. 2015;21:2007–2022. doi: 10.1261/rna.053918.115. no. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji Z, Song R, Regev A, Struhl K. Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife. 2016;4:e08890. doi: 10.7554/eLife.08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akmaev VR, Kelley ST, Stormo GD. Phylogenetically enhanced statistical tools for RNA structure prediction. Bioinformatics. 2000;16:501–512. doi: 10.1093/bioinformatics/16.6.501. [DOI] [PubMed] [Google Scholar]
- 10.Lindgreen S, PP G, Krogh A. Measuring covariation in RNA alignments: physical realism improves information measures. Bioinformatics. 2006;22:2988–2995. doi: 10.1093/bioinformatics/btl514. [DOI] [PubMed] [Google Scholar]
- 11.Yeang C-H, Darot JFJ, Noller HF, Haussler D. Detecting the coevolution of biosequences -an example of RNA interaction prediction. Mol. Biol. Evol. 2007;24:2119–2131. doi: 10.1093/molbev/msm142. [DOI] [PubMed] [Google Scholar]
- 12.Dutheil JY. Detecting coevolving positions in a molecule: why and how to account for phylogeny. Brief. Bioinf. 2012;13:228–243. doi: 10.1093/bib/bbr048. [DOI] [PubMed] [Google Scholar]
- 13.Somarowthu S, Legiewicz M, Chillón I, Marcia M, Liu F, Pyle AM. HOTAIR forms an intricate and modular secondary structure. Mol. Cell. 2015;58:353–361. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinberg Z, Breaker RR. R2R – software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011;12:3. doi: 10.1186/1471-2105-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, Floden EW, Gardner PP, Jones TA, Tate J, Finn RD. Rfam 12.0: updates to the RNA families database. Nucl. Acids Res. 2015;43:D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woolf B. The log likelihood ratio test (the G-test) Annals of Human Genetics. 1957;21:397–409. doi: 10.1111/j.1469-1809.1972.tb00293.x. [DOI] [PubMed] [Google Scholar]
- 17.Dunn SD, Wahl LM, Gloor GB. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact predictions. Bioinformatics. 2007;24:333–340. doi: 10.1093/bioinformatics/btm604. [DOI] [PubMed] [Google Scholar]
- 18.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 5S ribosomal RNA database. Nucl. Acids Res. 2002;30:176–178. doi: 10.1093/nar/30.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Y, Deiorio-Haggar K, Anthony J, Meyer MM. Most RNAs regulating ribosomal protein biosynthesis in Escherichia coli are narrowly distributed to Gammaproteobacteria. Nucl. Acids Res. 2013;41:3491–3503. doi: 10.1093/nar/gkt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.del Val C, Rivas E, Torres-Quesada O, Toro N, Jiménez-Zurdo JI. Identification of differentially expressed small non-coding RNAs in the legume endosymbiont Sinorhizobium meliloti by comparative genomics. Mol. Microbiol. 2007;66:1080–1091. doi: 10.1111/j.1365-2958.2007.05978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucl. Acids Res. 2012;40:5034–5051. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianfrani S, Dorsselaer AV, Clerc P, Avner P, Visvikis A, Branlant C. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLOS Biol. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang R, Moss WN, Rutenberg-Schoenberg M, Simon MD. Probing Xist RNA structure in cells using targeted structure-seq. PLOS Genet. 2015;11:e1005668. doi: 10.1371/journal.pgen.1005668. no. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivas E, Lang R, Eddy SR. A range of complex probabilistic models for RNA secondary structure prediction that include the nearest neighbor model and more. RNA. 2012;18:193–212. doi: 10.1261/rna.030049.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price MN, Dehal PS, Arkin AP. FastTree 2 - approximately maximum-likelihood trees for large alignments. PLOS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon C. A note on the concept of entropy. Bell System Tech. J. 1948;27:379–423. [Google Scholar]
- 28.Gutell RR, Larsen N, Woese CR. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin L, Gloor G, Dunn S, Wahl L. Using information theory to search for co-evolving residues in proteins. Bioinformatics. 2005;21:4116–4124. doi: 10.1093/bioinformatics/bti671. [DOI] [PubMed] [Google Scholar]
- 30.Fodor AA, Aldrich RW. Influence of conservation on calculations of amino acid co- variance in multiple sequence alignments. Proteins: Structure, Function, and Bioinformatics. 2004;56:211–221. doi: 10.1002/prot.20098. no. 2. [DOI] [PubMed] [Google Scholar]
- 31.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 32.Gerstein M, Sonnhammer ELL, Chothia C. Volume changes in protein evolution. J. Mol. Biol. 1994;235:1067–1078. doi: 10.1016/0022-2836(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 33.Gorodkin J, Staerfeldt H, Lund O, Brunak S. MatrixPlot: visualizing sequence constraints. Bioinformatics. 1999;15:769–770. doi: 10.1093/bioinformatics/15.9.769. [DOI] [PubMed] [Google Scholar]
- 34.Weigt M, White RA, Szurmant H, Hoch JA, Hwa T. Identification of direct residue contacts in protein-protein interaction by message passing. Proc. Natl. Acad. Sci. USA. 2009;106:67–72. doi: 10.1073/pnas.0805923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Leonardis E, Lutz B, Ratz S, Cocco S, Monasson R, Schug A, Weigt M. Direct-Coupling Analysis of nucleotide coevolution facilitates RNA secondary and tertiary structure prediction. Nucl. Acids Res. 2015;43:10444–10455. doi: 10.1093/nar/gkv932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinreb C, Riesselman AJ, Ingraham JB, Gross T, Sander C, Marks DS. 3D RNA and functional interactions from evolutionary couplings. Cell. 2016;165:963–975. doi: 10.1016/j.cell.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 1971:406–416. [Google Scholar]
- 38.Goebel B, Dawy Z, Hagenauer J, Mueller JC. An approximation to the distribution of finite sample size mutual information estimates; IEEE International Conference on Communications, 2005. ICC 2005; 2005. pp. 1102–1106. IEEE. [Google Scholar]
- 39.Rivas E, Eddy SR. Parameterizing sequence alignment with an explicit evolutionary model. BMC Bioinformatics. 2015;16:406. doi: 10.1186/s12859-015-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 41.Jung S, Swart EC, Minx PJ, Magrini V, Mardis ER, Landweber LF, Eddy SR. Exploiting Oxytricha trifallax nanochromosomes to screen for noncoding RNA genes. Nucl. Acids Res. 2011;39:7529–7547. doi: 10.1093/nar/gkr501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.del Val C, Romero-Zaliz R, Torres-Quesada O, Peregrina A, Toro N, Jiménez- Zurdo JI. A survey of sRNA families in α-proteobacteria. RNA Biol. 2012;9:119–129. doi: 10.4161/rna.18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wheeler TJ, Clements J, Eddy SR, Hubley R, Jones TA, Jurka J, Smit AFA, Finn RD. Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucl. Acids Res. 2013;41:D70–D82. doi: 10.1093/nar/gks1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.