Abstract
The core 3 structure of the O-glycan, GlcNAcβ1-3GalNAcα1-Ser/Thr, an important precursor in the biosynthesis of mucin-type glycoproteins, is synthesized by β1,3-N-acetylglucosaminyltransferase 6 (β3Gn-T6; core 3 synthase). We generated an anti-β3Gn-T6 mAb (G8-144 mAb) and performed immunohistochemical analyses. In normal stomach and colon, β3Gn-T6 was strongly expressed in the Golgi region of epithelia. In contrast, its expression was markedly down-regulated in gastric and colorectal carcinomas. Tissue specimens from a familial adenomatous polyposis patient showed a clear correlation between the down-regulation of β3Gn-T6 expression and the degree of dysplasia/neoplasia. In vitro, the level of β3Gn-T6 transcript was increased according to the differentiation of Caco-2 cells. These results suggested that the expression of β3Gn-T6 is closely regulated during differentiation and dedifferentiation. β3Gn-T6 would be a useful marker for distinguishing between benign adenomas and premalignant lesions. HT1080 FP-10 cells stably transfected with the β3Gn-T6 gene showed a decrease in the core 1 structure, Galβ1,3GalNAcα1-Ser/Thr, probably due to competition between the core 1 synthase and core 3 synthase. The migration activity of the transfectants was markedly lower than that of mock transfectants in vitro, and lung metastasis after i.v. injection of the transfectants into nude mice was significantly suppressed. These findings indicated that the core structures of O-glycans are profoundly involved in the metastatic capacity of cancer cells.
Keywords: glycosyltransferase, stomach, familial adenomatous polyposis, immunohistochemical analysis
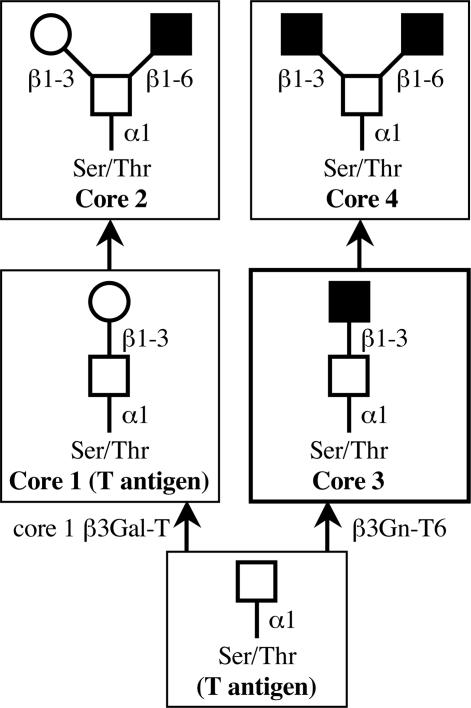
Carbohydrate chains are dramatically altered in cancer cells compared with normal cells. For example, sialyl Lewis a [NeuAcα2-3Galβ1-3(Fucα1–4)GlcNAc, sLea] and sialyl Lewis x [NeuAcα2-3Galβ1-4(Fucα1–3)GlcNAc, sLex] epitopes, well known cancer-associated antigens, increased in cancerous tissues (1–4). The changes occur not only at the terminus of carbohydrate chains on mucins, but also in the core structures of O-glycans. The mucins constitute a heterogeneous group of glycoproteins having O-glycans. Some of them are membrane-associated glycoproteins in epithelial cells and others are secreted from these cells (5, 6). The mucins have been suggested to play important roles in invasion by microbial pathogens, cell adhesion, inflammation, and cancer metastasis (7–15). Mucin-type glycoproteins have clusters of a multitude of O-glycans whose synthesis is mainly initiated by diverse UDP-GalNAc: polypeptide GalNAc-transferases (pp-GalNAc-Ts) (16, 17). After the transfer of a GalNAc residue to a Ser and/or Thr residue by pp-GalNAc-T, sugar residues are sequentially added to the GalNAc on the peptide to form O-glycans. O-glycans can be classified into several different groups according to their core structures (Fig. 1). Core 1 is the major constituent of O-glycans in many cells (18, 19). Core 3 is restricted in its occurrence to mucins from specialized tissues such as the colon (20). Cores 2 and 4 are formed by addition of GlcNAc to GalNAc on cores 1 and 3, respectively. Each core structure is differentially expressed in conjunction with the differentiation and malignant transformation of various cells and tissues (21–25). In normal colonic tissue, the major core structure among O-glycans is the core 3 structure (13, 26–28). However, it has been reported that expression of the core 3 structure is down-regulated and the expression of core 1 and 2 structures is up-regulated in colon cancer tissues and colon cancer cell lines (15, 29). Moreover, the activity of core 3 synthase was reduced to undetectable levels in these tissues and cell lines (30, 31). These results suggested that down-regulation of the expression of the core 3 synthase may cause the loss of the core 3 structure in cancerous tissues and cancer cell lines. Many enzymes involved in synthesizing the core structure of O-glycan may scramble for a GalNAc residue on mucin as a substrate. The core 1 and 3 synthases transfer sugar residues, Gal and GlcNAc, respectively, to the GalNAc residue with a β1,3-linkage. They may compete with GalNAc-peptides as acceptor substrates in the cells.
Fig. 1.
Pathways for the biosynthesis of core 1–4 structures of mucin type O-glycans. GalNAc, Gal, and GlcNAc are indicated with open squares, open circles, and filled squares, respectively. Cancer-associated antigens are given in parentheses.
Recently, we cloned and characterized the gene encoding UDP-GlcNAc:GalNAc-peptide β1,3-N-acetylglucosaminyltransferase (β3Gn-T6) (20). In this study, we developed a monoclonal antibody, named G8-144, which reacts specifically to β3Gn-T6. With this antibody, we immunohistochemically analyzed the expression of β3Gn-T6 by using 39 sets of normal and cancerous tissues and 33 specimens from a patient with familial adenomatous polyposis (FAP). In addition, we established β3Gn-T6-transfected cells and investigated their lung metastatic ability after an i.v. injection into nude mice. Our results demonstrated that the expression of β3Gn-T6 dramatically decreased in cancerous tissues, and the changes to the core structures of O-glycans give rise to phenotypic changes of cancer cells, such as mobility and metastatic activity.
Materials and Methods
mAb. ABALB/c mouse was immunized three times with purified recombinant β3Gn-T6 protein. mAb was generated according to established procedures and named G8-144. The Ig isotype of G8-144 mAb was determined to be IgG1.
Immunohistochemistry. Formalin-fixed, paraffin-embedded specimens were obtained from Fussa Hospital (Tokyo). Paraffin sections (3–4 μm) were deparaffinized in xylene and rehydrated in ethanol and water. After antigen retrieval (10 min of auto-claving in PBS), the sections were rinsed in PBS and treated with 0.3% H2O2 in methanol for 30 min. Then, the sections were washed in PBS and incubated with the primary antibody (G8–144 mAb; diluted 5,000-fold with DAKO Antibody Diluent with Background Reducing Components) overnight at 4°C. The sections were then washed in PBS and incubated with Envision+ peroxidase-linked anti-mouse Ig (DAKO) for 30 min at room temperature. They were washed in PBS, and color was developed in 3,3′-diaminobenzidine (Wako, Osaka) with H2O2. They were washed in running water, counterstained in hematoxylin, dehydrated in ethanol and xylene, and mounted.
Cell Culture. Human colonic adenocarcinoma Caco-2 cells were obtained from American Type Cell Collection. Caco-2 cells were cultured and induced to differentiate according to ref. 32. Cells were collected 0, 7, 14, 21, 28 and 35 days after confluence. They were harvested, washed with PBS and divided into two aliquots. One was homogenized in 0.25 M sucrose in 10 mM Tris·HCl, pH 7.2. The state of differentiation was confirmed by measuring dipeptidyl peptidase IV (DPP-IV) as described (33). The other aliquot was used of the quantitative analysis for the β3Gn-T6 transcript.
Quantitative Analysis of the β3Gn-T6 Transcript in Tissues and Cells. Quantification of the β3Gn-T6 transcript was performed by using the real-time PCR method described in detail in ref. 20. The relative amount of β3Gn-T6 transcript was normalized to the amount of total RNA.
Construction of a Vector Plasmid for Stable Expression of β3Gn-T6 in Cultured Mammalian Cells. The fragment of the ORF encoding β3Gn-T6 was amplified by PCR using the Marathon-Ready cDNA of human stomach tissue (Clontech) as a template. The forward and reverse primer sequences were flanked with attB1 and attB2 sequences, respectively, to create the recombination sites. The forward primer was 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCTTTTCCCTGCCGCAGG-3′, and the reverse primer was 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTGGCCT CAGGAGACCCGGTG-3′. The amplified fragment was inserted between the attP1 and attP2 sites of the pDONR 201 entry vector, and then the insert was transferred into a pDEST12.2 mammalian expression vector by using the GATEWAY system (Invitrogen). The plasmids were named pDEST12.2-β3Gn-T6.
Cell Culture and Transfection. HT1080 FP-10 cells, a highly metastatic variant of human fibrosarcoma cells, were used (34). HT1080 FP-10 cells were transfected with pDEST12.2-β3Gn-T6 expression plasmid DNA using Lipofectamine 2000 reagent (Invitrogen). The cells were selected in the presence of geneticin (1.0 mg/ml; Invitrogen) for 3 weeks. The cells were removed with trypsin, and cloned by limiting dilution in 96-well plates with continued geneticin selection. Five clonal cell lines stably expressing β3Gn-T6 (HT1080 FP-10/β3Gn-T6-1 to 5) and three clonal mock control cell lines (HT1080 FP-10/mock-1 to 3) were established.
Flow Cytometric Analysis. For flow cytometry, the cells pretreated with Neuraminidase from Arthrobacter ureafaciens (Nacalai Tesque, Kyoto), which releases α2-3-, α2-6-, and α2-8-linked NeuAc residues from oligosaccharides, were incubated with FITC-conjugated peanut agglutinin (PNA) lectin (Vector Laboratories).
In Vitro Migration Assay. The in vitro migration assay was performed according to Albini's method (35). Sterile chambers with an 8-μm porous polycarbonate membrane-coated upper surface witha2 μg/0.48 cm2 filter of laminin (TaKaRa) or 1 μg/0.48 cm2 filter of extracellular matrix (ECM; Collaborative Research, MA) were placed in 24-well tissue culture plates. Transfected HT1080 FP-10 cells (8 × 104 cells per well) were seeded on the membrane and incubated for 18 h. The cells that penetrated the pores of the membrane were counted.
Experimental Lung Metastasis Assay. Seven-week-old female BALB/cA nu/nu mice were obtained from Clea Japan Inc. (Tokyo) and used for this experiment. A total of 2 × 106 cells (HT1080 FP-10, HT1080 FP-10/mock-1 or HT1080 FP-10/β3Gn-T6-1) were suspended in 0.5 ml of MEM and injected i.v. into the tail vein of mice. As a control, 0.5 ml of MEM was injected i.v. into the tail vein. Forty-eight days later, the mice were killed and the weight of the lungs was measured (see Table 1). The lungs were then immersed in Bouin's solution for the visualization of metastatic nodules.
Table 1. Cell lines used, mice showing lung metastasis, and mean lung weight.
| Cells | Lung metastases* | Mean lung weight (range),† g |
|---|---|---|
| HT 1080 FP-10/mock-1 | 8/8 | 0.38 (0.19-0.53) |
| HT1080 FP-10/β3Gn-T6-1 | 0/8 | 0.15 (0.14-0.16) |
| None | 0/8 | 0.15 (0.14-0.16) |
Number of mice showing metastasis/number of mice used.
Cells (2 × 106) were injected i.v. into nude mice, and weight of lungs was measured on day 48.
Further Details. For further details, please see Supporting Text and Figs. 6 and 7, which are published as supporting information on the PNAS web site.
Results
Down-Regulation of β3Gn-T6 Expression in Gastric and Colorectal Carcinomas. We measured the amount of β3Gn-T6 transcript in normal and cancerous tissues from gastric and colorectal cancer patients by using real-time PCR. The levels of β3Gn-T6 transcript were significantly lower in both gastric and colorectal cancer samples than in corresponding normal samples (data not shown). Therefore, we developed an anti-β3Gn-T6 monoclonal antibody, named G8-144, to investigate the expression profile of β3Gn-T6 in gastrointestinal tissues. Western blot analysis using recombinant β3Gn-T2 to T6 was indicated that G8–144 mAb recognized β3Gn-T6 but not any of the other β3Gn-Ts (data not shown).
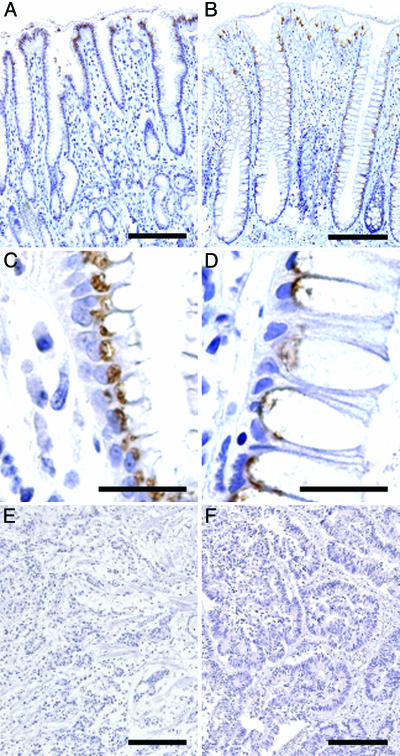
The transcript for β3Gn-T6 is expressed only in stomach and colon as reported in our previous study (20). In this study, normal and cancerous samples of human stomach and colon obtained from 14 gastric and 25 colorectal cancer patients were stained with G8-144 mAb. Representative staining profiles of β3Gn-T6 are shown in Fig. 2. In normal stomach, β3Gn-T6 was detected in the Golgi region, i.e., an apical perinuclear region, of foveolar epithelia, but not in other epithelia (Fig. 2 A and C). In normal colon, β3Gn-T6 was detected in the Golgi region of epithelial cells (Fig. 2 B and D). In contrast to normal tissues, the expression of β3Gn-T6 in carcinomatous lesions of the stomach and colon, β3Gn-T6 was undetectable in all cancerous samples (Fig. 2 E and F, respectively).
Fig. 2.
Expression of β3Gn-T6 in normal gastric tissue (A and C), normal colonic tissue (B and D), and gastric (E) and colorectal (F) carcinoma. Immunostaining was performed with G8-144 mAb. (Scale bars, 100 μmin A, B, E, and F and 10 μmin C and D.)
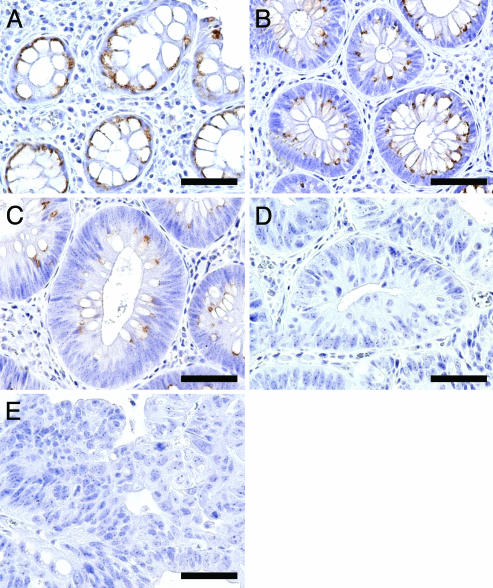
Gradual Down-Regulation of β3Gn-T6 Expression During Progression to Cancer in FAP. To determine further the expression profile of β3Gn-T6 during progression to cancer, an immunohistochemical analysis on 33 specimens from an FAP patient was performed. FAP is a hereditary disease caused by mutations in the APC gene and a colon cancer predisposition syndrome in which many precancerous colonic polyps develop and finally become cancerous (36, 37). Many lesions, which are classified into various atypical stages including benign polyps and adenocarcinomas, were retrieved from one patient, and the expression levels of the respective enzyme could be compared at the different stages without a difference of genetic background. FAP specimens were classified into five categories according to the Vienna classification of gastrointestinal epithelial neoplasia [C1, negative for neoplasia; C2, indefinite for neoplasia/dysplasia; C3, noninvasive low-grade neoplasia (low-grade adenoma/dysplasia); C4, noninvasive high-grade neoplasia (high-grade adenoma/dysplasia, noninvasive carcinoma, and suspicion of invasive carcinoma), and C5, invasive neoplasia (intramucosal carcinoma, submucosal carcinoma, or beyond); see ref. 38].
The results showed that β3Gn-T6 gradually disappeared according to the grade of neoplasia. β3Gn-T6 was abundant in C1 specimens (Fig. 3A), and gradually decreased in C2 and C3 specimens (Fig. 3 B and C, respectively); however, it was undetectable in C4 and C5 specimens (Fig. 3 D and E, respectively).
Fig. 3.
Expression of β3Gn-T6 in FAP lesions. (A) C1. (B) C2. (C) C3. (D) C4. (E) C5. Immunostaining was performed with G8-144 mAb. (Scale bar, 30 μm.)
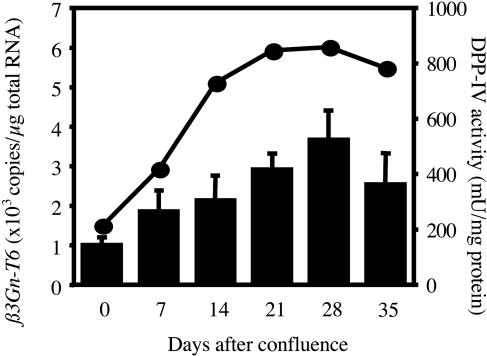
Expression of β3Gn-T6 Transcript During Differentiation. To evaluate whether the expression of β3Gn-T6 is up-regulated during the differentiation, Caco-2 cells were used. Caco-2 cells are derived from human colon adenocarcinoma, and are known to differentiate into enterocyte-like cells spontaneously or in response to sodium butyrate after reaching confluence (39). DPP-IV activity, which is a well established marker for the differentiation of Caco-2 cells, and the amount of β3Gn-T6 transcript were measured at each stage of the differentiation (Fig. 4). Only a few copies of the β3Gn-T6 transcript were detected in undifferentiated Caco-2 cells (0 day in Fig. 4). However, the level of β3Gn-T6 transcript gradually increased in parallel with the DPP-VI activity during the differentiation of Caco-2 cells.
Fig. 4.
Expression of β3Gn-T6 in undifferentiated and differentiated Caco-2 cells. Correlation between the extent of differentiation as indicated by DPP-IV activity (circles) and the expression level of the β3Gn-T6 transcript (bars). One unit of DPP-IV activity is defined as the activity that hydrolyzes 1 μmol of substrate per min at 37°C. Proteins were measured by using a protein assay reagent (Bio-Rad) and BSA as a standard. Standard curves for β3Gn-T6 were generated by serial dilution of plasmid DNA. The relative amount of β3Gn-T6 transcript was normalized to the amount of total RNA. Data were obtained from triplicate experiments and are indicated as the mean ± SD.
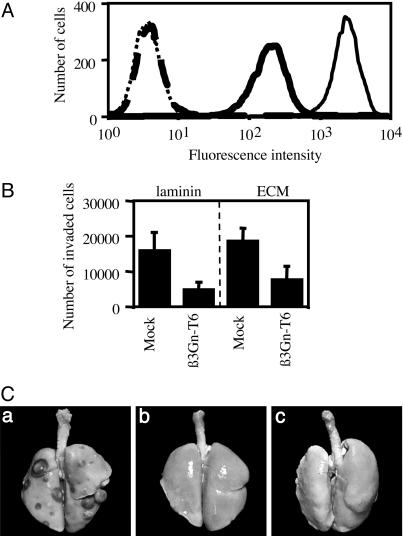
Suppression of T (Core 1) Antigen on the Surface of the Cells Transfected with the β3Gn-T6 Gene (HT1080 FP-10/β3Gn-T6 Cells). The quantitative analysis of the β3Gn-T6 transcript and the immunohistochemical analysis demonstrated that the expression of β3Gn-T6 was dramatically reduced or almost disappeared in gastrointestinal cancer cells. These results suggested that the level of β3Gn-T6 expression might be involved in the phenotypic change of cancer cells, such as metastatic activity. Caco-2 cells are not suitable for a study of metastatic activity. HT1080 FP-10 cells were used for the study of phenotypic change because of their strong metastatic activity (34). Three mock-transfectants (HT1080 FP-10/mock) and five transfectants stably expressing the β3Gn-T6 gene (HT1080 FP-10/β3Gn-T6) were examined for expression of the T (core 1) antigen. For the detection of T antigen, FITC-conjugated PNA lectin was used. The cells were pretreated with Neuraminidase, because PNA lectin does not recognize sialylated T antigen. The results of a representative clone of each transfectant are shown in Fig. 5A. The expression level of the β3Gn-T6 transcript and the flow cytometry profile were nearly the same among the five HT1080 FP-10/β3Gn-T6 cells. The HT1080 FP-10/β3Gn-T6 cells had markedly less of the core 1 structure than the HT1080 FP-10/mock cells. Considering the pathways of synthesis of O-glycans (Fig. 1), the synthesis of the core 1 structure directed by core 1 β3Gal-T possibly interfered with the synthesis of the core 3 structure directed by core 3 β3Gn-T (β3Gn-T6). We could not determine the amount of the core 3 structure in the cells because no tool is available for detecting it.
Fig. 5.
Alteration of the characteristics of HT1080 FP-10 cells transfected with the β3Gn-T6 gene. (A) The expression of T antigen on the surface of HT1080 FP-10/mock and β3Gn-T6 cells was analyzed by flow cytometry. The cells stained with FITC-PNA lectin are indicated by a solid line; unstained cells are indicated by a dotted line. HT1080 FP-10/mock and β3Gn-T6 cells are indicated with a thin and thick line, respectively. (B) Ability of HT1080 FP-10 variant cells to migration in vitro. HT1080 FP-10/mock and β3Gn-T6 cells were seeded onto membranes coated with laminin (Left) or ECM (Right). The cells that penetrated the membrane were counted. Data were obtained from quadruplicate experiments, and the average cell numbers of three HT1080 FP-10/mock or five HT1080 FP-10/β3Gn-T6 cell lines are indicated as the mean ± SD. (C) In vivo metastatic activity of HT1080 FP-10/mock-1 or β3Gn-T6-1 cells. Gross appearance of lungs from nude mice injected with HT1080 FP-10/mock cells (a), HT1080 FP-10/β3Gn-T6 cells (b) and MEM alone (c) is shown.
Reduction of in Vitro Migration Ability and Lung Metastatic Activity of HT1080 FP-10/β3Gn-T6 Cells. The migration activities of the three HT1080 FP-10/mock and five HT1080 FP-10/β3Gn-T6 cells were investigated in vitro. As shown in Fig. 5B, the β3Gn-T6 transfectants obviously migrated less than the mock transfectants on both matrices. Reduced migration rate in the transfectant cells was observed even without matrices (data not shown). We did not observe any difference in the proliferation rates between mock and β3Gn-T6 transfectants (data not shown).
Other two human colonic adenocarcinorma cell lines, HCT-15 and HT29LMM, were transfected with the β3Gn-T6 expression vector. Flow cytometric analysis with PNA lectin showed that the β3Gn-T6 transfected cells had markedly less of the core 1 structure than the mock cells (data not shown). Moreover, in vitro migration assay using these two cell lines showed that the β3Gn-T6 transfectants obviously migrated less than the mock transfectants (data not shown). These results confirmed that the β3Gn-T6 transfectants features, i.e., the decrease of core 1 structure and the reduction of migration ability, are general phenomena.
We then compared the metastatic activity of HT1080 FP-10/β3Gn-T6 cells with that of HT1080 FP-10/mock cells in vivo. One of the clones from each transfectant, HT1080 FP-10/mock-1 and HT1080 FP-10/β3Gn-T6-1, respectively, was selected for this experiment. All eight mice injected with HT1080 FP-10/mock-1 developed lung metastases; however, none of the mice injected with HT1080 FP-10/β3Gn-T6-1 or MEM medium alone showed signs of metastasis even on examination by gross and stereoscopic microscopy after fixation in Bouin's solution (Fig. 5C and Table 1). Typical images of each lung from nude mice are presented in Fig. 5C. The average lung mass of the mice injected with the mock transfectants was nearly equal to that of mice injected with HT1080 FP-10 cells. These results demonstrated that an increase of β3Gn-T6 expression significantly suppressed the lung metastatic activity of HT1080 FP-10 cells in vivo.
Discussion
Core 3 synthase is an important enzyme in the synthesis of mucin-type O-glycans in digestive organs. The core 3 structure was first identified in O-glycans derived from stomach and colon, where the core 3 synthase activity was also very strong (28, 31). Previously, we have cloned and characterized a sixth member of the β3Gn-Ts, named β3Gn-T6, that was identified to be the core 3 synthase (20).
On immunohistochemical analysis, β3Gn-T6 was detected in the Golgi region of both normal gastric and colorectal epithelial cells. It is of interest that, in the stomach, it was restricted to the foveolar epithelia. This finding indicated that mucins produced by foveolar epithelia cells contain the core 3 structure, but mucins derived from subglandular epithelia do not. In contrast to normal tissues, β3Gn-T6 disappeared in both gastric and colorectal cancer.
In FAP samples, the expression level of β3Gn-T6 was well correlated with the histological grade of atypia. β3Gn-T6 was abundantly expressed in C1, then gradually decreased from C2 to C3. β3Gn-T6 disappeared in regions C4 and C5. The regions histologically classified into C2 and C3 are still benign, but C4 lesions are premalignant or carcinomatous. When C3 and C4 show a typical histology, it is not difficult to distinguish them histologically. However, it is sometimes difficult to differentiate between C3 and C4 by histological examination. As described above, C3 and C4 in FAP were clearly distinguishable after the G8-144 mAb staining. Thus, the quantitative measurement of β3Gn-T6 by immunohistochemistry or Western blotting would be a very useful and supportive tool for determining the grade of malignancy, for example, for distinguishing C3 from C4.
The question arises whether cancer cells are derived from β3Gn-T6-expressing cells or β3Gn-T6 nonexpressing cells. Generally, it is difficult to determine the origin cell of a cancer cell. However, we consider that the colon cancer cells are derived from β3Gn-T6-expressing cells, and became β3Gn-T6 nonexpressing cells for the following two reasons. (i) The theory that colon cancer is originated from benign adenoma in accordance with an adenoma–carcinoma sequence is well established. As seen in Fig. 3, the adenoma cells showed sequential phenotypic change during the adenoma–carcinoma sequence, i.e., from adenoma with low-grade atypia to that with high-grade atypia, and finally to cancer. Adenoma with low-grade atypia was occupied almost entirely by goblet cells that expressed β3Gn-T6 the same as normal cells. As the grade of atypia increased, the mucus-containing vacuoles became smaller. This observation apparently showed a gradual decline in the ability to produce mucin and express β3Gn-T6 in accordance with the adenoma–carcinoma sequence. This also reflects the dedifferentiation of goblet cells or of immature cells in the process of differentiating into goblet cells. As the dedifferentiation progressed, the ability to produce mucin was reduced until the mucus vacuoles disappeared. In parallel with the reduction, the expression of β3Gn-T6 gradually decreased during the adenoma–carcinoma sequence, and finally disappeared in premalignant lesions. This finding is consistent with the result of the Caco-2 cell experiment. Caco-2 cells did not express core 3 synthase until they had differentiated. (ii) Many cell lines derived from colorectal and stomach cancers including mucin-producing cancer cells such as KATOIII cells were examined for the expression of core 3 synthase. There was very little or no expression of core 3 synthase in the cancer cell lines examined. This observation strongly indicated that the disappearance of core 3 synthase in cancer cells is closely related to carcinogenesis, and not to the mucin-producing function.
Sialyl Lewis x (sLex) and sialyl Lewis a (sLea) epitopes are well investigated cancer-associated carbohydrate antigens (1–4). They are often expressed in cancer cells derived from gastrointestinal, pancreatic, and lung tissues, and used for the clinical diagnosis of cancer as tumor markers. The sLex and sLea structures are the terminal epitopes of O-glycans carried mainly by core 1 and 2 structures. Therefore, the disappearance of the core 3 synthase in malignant cells may lead to an increase in the core 1 and 2 structures, eventually resulting in the sLex and sLea epitopes.
We performed immunohistochemical analysis using PNA lectin and G8-144 mAb on normal and primary cancerous colorectal tissues and liver metastatic deposits. The expression profiles of β3Gn-T6 were consistent with those of described above. That is, β3Gn-T6 was detected in normal colon regions, but undetectable even in primary colorectal tumors and liver metastatic specimens (data not shown). In contrast to the β3Gn-T6 expression, the core 1 structure detected by PNA was essentially absent in normal region, but detected in primary cancerous region and liver metastatic deposits (data not shown). These results were consistent with many previous studies (40, 41). It was proposed that the appearance of core 1 structures in colorectal cancers is due to incomplete glycosylation (42, 43). Our results suggest another possibility: that the core 1 structure appears due to the disappearance of its competitor, core 3 synthase (β3Gn-T6), in cancerous tissues.
Immunohistochemical analysis with anti-β3Gn-T6 mAb showed that there are no differences between primary tumors and metastatic deposits for the expression of β3Gn-T6. There are many factors involved in the mechanisms of cancer metastasis, and many recent studies indicated that aberrant glycosylation promotes some key events involved in metastasis (summarized in ref. 44). We do not regard that the cancer metastasis is regulated by only β3Gn-T6. However, we demonstrated that all three cell lines transfected with β3Gn-T6 showed significant reduction of migration ability. This finding suggested that disappearance of β3Gn-T6 in colonic cells is one of important factors determining the cancer metastatic ability.
Stable expression of the β3Gn-T6 gene in the cancer cells significantly altered the characteristics of the cells, with regards to motility and metastatic activity. The significant decrease in the ability to migrate in vitro and the lung metastatic activity of HT1080 FP-10/β3Gn-T6 cells (Fig. 5 B and C) strongly indicated that alterations of the O-glycan structure are deeply involved in such activities of cancer cells. This finding demonstrates that the enzyme synthesizing the core structure of O-glycan is significantly involved in cancer metastasis.
In this study, we demonstrated the following points. (i) The measurement of β3Gn-T6 using G8–144 mAb would be useful in deciding the dedifferentiation level of malignant cells derived from gastrointestinal tissues. (ii) O-glycan structures are dramatically regulated in the core structures through the expression of β3Gn-T6. (iii) The structural changes to O-glycan directed by β3Gn-T6 are profoundly concerned with cell motility and metastatic activity.
It will be interesting to identify the glycoproteins involved in the migration and metastatic activities of cancer cells by using this remodeling system of O-glycans.
Supplementary Material
Acknowledgments
We thank Dr. Yuzuru Ikehara, Aich Cancer Center Institute, for critical discussion and suggestions. This work was performed as a part of the R & D Project of Industrial Science and Technology Frontier Program (Research and Development for Establishment and Utilization of a Technical Infrastructure for Japanese Industry) supported by New Energy and Industrial Technology Development Organization (NEDO).
Author contributions: T.I. and H.N. designed research; T.I., R.K., T. Kubota, A.T., and T.H. performed research; T.O. and K.M. contributed new reagents/analytic tools; T.I., T. Kudo, and T. Kawamoto analyzed data; and T.I. and H.N. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: β3Gn-T, β1,3-N-acetylglucosaminyltransferase; FAP, familial adenomatous polyposis; DPP-IV, dipeptidyl peptidase; PNA, peanut agglutinin.
References
- 1.Itzkowitz, S. H., Yuan, M., Fukushi, Y., Palekar, A., Phelps, P. C., Shamsuddin, A. M., Trump, B. F., Hakomori, S. & Kim, Y. S. (1986) Cancer Res. 46, 2627–2632. [PubMed] [Google Scholar]
- 2.Itzkowitz, S. H., Yuan, M., Fukushi, Y., Lee, H., Shi, Z. R., Zurawski, V., Jr., Hakomori, S. & Kim, Y. S. (1988) Cancer Res. 48, 3834–3842. [PubMed] [Google Scholar]
- 3.Gong, E., Hirohashi, S., Shimosato, Y., Watanabe, M., Ino, Y., Teshima, S. & Kodaira, S. (1985) J. Natl. Cancer Inst. 75, 447–454. [PubMed] [Google Scholar]
- 4.Sakamoto, S., Watanabe, T., Tokumaru, T., Takagi, H., Nakazato, H. & Lloyd, K. O. (1989) Cancer Res. 49, 745–752. [PubMed] [Google Scholar]
- 5.Williams, S. J., McGuckin, M. A., Gotley, D. C., Eyre, H. J., Sutherland, G. R. & Antalis, T. M. (1999) Cancer Res. 59, 4083–4089. [PubMed] [Google Scholar]
- 6.Porchet, N., Buisine, M. P., Desseyn, J. L., Moniaux, N., Nollet, S., Degand, P., Pigny, P., Van Seuningen, I., Laine, A. & Aubert, J. P. (1999) J. Soc. Biol. 193, 85–99. [PubMed] [Google Scholar]
- 7.Lillehoj, E. P., Hyun, S. W., Kim, B. T., Zhang, X. G., Lee, D. I., Rowland, S. & Kim, K. C. (2001) Am. J. Physiol. 280, L181–L187. [DOI] [PubMed] [Google Scholar]
- 8.Prakobphol, A., Thomsson, K. A., Hansson, G. C., Rosen, S. D., Singer, M. S., Phillips, N. J., Medzihradszky, K. F., Burlingame, A. L., Leffler, H. & Fisher, S. J. (1998) Biochemistry 37, 4916–4927. [DOI] [PubMed] [Google Scholar]
- 9.Prakobphol, A., Tangemann, K., Rosen, S. D., Hoover, C. I., Leffler, H. & Fisher, S. J. (1999) Biochemistry 38, 6817–6825. [DOI] [PubMed] [Google Scholar]
- 10.Phillips, T. E., Stanley, C. M. & Wilson, J. (1993) Prostaglandins Leukot. Essent. Fatty Acids 48, 423–428. [DOI] [PubMed] [Google Scholar]
- 11.Sreejayan, N., Wittig, B. M., von Stillfried, N., Hennicke, M. S., Meyer, G., Stieber, P., Lamerz, R. & von Ritter, C. (2001) Am. J. Physiol. 280, G1043–G1048. [DOI] [PubMed] [Google Scholar]
- 12.Mack, D. R., Gaginella, T. S. & Sherman, P. M. (1992) Gastroenterology 102, 1199–1211. [PubMed] [Google Scholar]
- 13.Capon, C., Maes, E., Michalski, J. C., Leffler, H. & Kim, Y. S. (2001) Biochem. J. 358, 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bresalier, R. S., Ho, S. B., Schoeppner, H. L., Kim, Y. S., Sleisenger, M. H., Brodt, P. & Byrd, J. C. (1996) Gastroenterology 110, 1354–1367. [DOI] [PubMed] [Google Scholar]
- 15.Kim, Y. S. (1998) Keio J. Med. 47, 10–18. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, Y., Iwasaki, H., Wang, H., Kudo, T., Kalka, T. B., Hennet, T., Kubota, T., Cheng, L., Inaba, N., Gotoh, M., et al. (2003) J. Biol. Chem. 278, 573–584. [DOI] [PubMed] [Google Scholar]
- 17.Schwientek, T., Bennett, E. P., Flores, C., Thacker, J., Hollmann, M., Reis, C. A., Behrens, J., Mandel, U., Keck, B., Schafer, M. A., et al. (2002) J. Biol. Chem. 277, 22623–22638. [DOI] [PubMed] [Google Scholar]
- 18.Kudo, T., Iwai, T., Kubota, T., Iwasaki, H., Takayma, Y., Hiruma, T., Inaba, N., Zhang, Y., Gotoh, M., Togayachi, A. & Narimatsu, H. (2002) J. Biol. Chem. 277, 47724–44731. [DOI] [PubMed] [Google Scholar]
- 19.Ju, T., Brewer, K., D`Souza, A., Cummings, R. D. & Canfield, W. M. (2002) J. Biol. Chem. 277, 178–186. [DOI] [PubMed] [Google Scholar]
- 20.Iwai, T., Inaba, N., Naundorf, A., Zhang, Y., Gotoh, M., Iwasaki, H., Kudo, T., Togayachi, A., Ishizuka, Y., Nakanishi, H. & Narimatsu, H. (2002) J. Biol. Chem. 277, 12802–12809. [DOI] [PubMed] [Google Scholar]
- 21.Piller, F., Piller, V., Fox, R. I. & Fukuda, M. (1988) J. Biol. Chem. 263, 15146–15150. [PubMed] [Google Scholar]
- 22.Yang, J. M., Byrd, J. C., Siddiki, B. B., Chung, Y. S., Okuno, M., Sowa, M., Kim, Y. S., Matta, K. L. & Brockhausen, I. (1994) Glycobiology 4, 873–884. [DOI] [PubMed] [Google Scholar]
- 23.Yousefi, S., Higgins, E., Daoling, Z., Pollex-Kruger, A., Hindsgaul, O. & Dennis, J. W. (1991) J. Biol. Chem. 266, 1772–1782. [PubMed] [Google Scholar]
- 24.Fukuda, M. (1996) Cancer Res. 56, 2237–2244. [PubMed] [Google Scholar]
- 25.Brockhausen, I., Yang, J. M., Burchell, J., Whitehouse, C. & Taylor-Papadimitriou, J. (1995) Eur. J. Biochem. 233, 607–617. [DOI] [PubMed] [Google Scholar]
- 26.Podolsky, D. K. (1985) J. Biol. Chem. 260, 15510–15515. [PubMed] [Google Scholar]
- 27.Podolsky, D. K. (1985) J. Biol. Chem. 260, 8262–8271. [PubMed] [Google Scholar]
- 28.Brockhausen, I., Matta, K. L., Orr, J. & Schachter, H. (1985) Biochemistry 24, 1866–1874. [DOI] [PubMed] [Google Scholar]
- 29.Brockhausen, I. (1999) Biochim. Biophys. Acta 1473, 67–95. [DOI] [PubMed] [Google Scholar]
- 30.Vavasseur, F., Dole, K., Yang, J., Matta, K. L., Myerscough, N., Corfield, A., Paraskeva, C. & Brockhausen, I. (1994) Eur. J. Biochem. 222, 415–424. [DOI] [PubMed] [Google Scholar]
- 31.Vavasseur, F., Yang, J. M., Dole, K., Paulsen, H. & Brockhausen, I. (1995) Glycobiology 5, 351–357. [DOI] [PubMed] [Google Scholar]
- 32.Amano, J. & Oshima, M. (1999) J. Biol. Chem. 274, 21209–21216. [DOI] [PubMed] [Google Scholar]
- 33.Nagatsu, T., Hino, M., Fuyamada, H., Hayakawa, T. & Sakakibara, S. (1976) Anal. Biochem. 74, 466–476. [DOI] [PubMed] [Google Scholar]
- 34.Okada, T., Li, J., Kodaka, M. & Okuno, H. (1998) Clin. Exp. Metastasis 16, 267–274. [DOI] [PubMed] [Google Scholar]
- 35.Albini, A., Iwamoto, Y., Kleinman, H. K., Martin, G. R., Aaronson, S. A., Kozlowski, J. M. & McEwan, R. N. (1987) Cancer Res. 47, 3239–3245. [PubMed] [Google Scholar]
- 36.Bodmer, W. F., Bailey, C. J., Bodmer, J., Bussey, H. J., Ellis, A., Gorman, P., Lucibello, F. C., Murday, V. A., Rider, S. H., Scambler, P., et al. (1987) Nature 328, 614–616. [DOI] [PubMed] [Google Scholar]
- 37.Hampton, G., Leuteritz, G., Ludecke, H. J., Senger, G., Trautmann, U., Thomas, H., Solomon, E., Bodmer, W. F., Horsthemke, B., Claussen, U., et al. (1991) Genomics 11, 247–251. [DOI] [PubMed] [Google Scholar]
- 38.Schlemper, R. J., Riddell, R. H., Kato, Y., Borchard, F., Cooper, H. S., Dawsey, S. M., Dixon, M. F., Fenoglio-Preiser, C. M., Flejou, J. F., Geboes, K., et al. (2000) Gut 47, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siavoshian, S., Blottiere, H. M., Le Foll, E., Kaeffer, B., Cherbut, C. & Galmiche, J. P. (1997) Cell Biol. Int. 21, 281–287. [DOI] [PubMed] [Google Scholar]
- 40.Boland, C. R., Montgomery, C. K. & Kim, Y. S. (1982) Proc. Natl. Acad. Sci. USA 79, 2051–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan, M., Itzkowitz, S. H., Boland, C. R., Kim, Y. D., Tomita, J. T., Palekar, A., Bennington, J. L., Trump, B. F. & Kim, Y. S. (1986) Cancer Res. 46, 4841–4847. [PubMed] [Google Scholar]
- 42.Hakomori, S. (1985) Cancer Res. 45, 2405–2414. [PubMed] [Google Scholar]
- 43.Hakomori, S. & Kannagi, R. (1983) J. Natl. Cancer Inst. 71, 231–251. [PubMed] [Google Scholar]
- 44.Hakomori, S. (2002) Proc. Natl. Acad. Sci. USA 99, 10231–10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.