Abstract
The enzyme YkvM from Bacillus subtilis was identified previously along with three other enzymes (YkvJKL) in a bioinformatics search for enzymes involved in the biosynthesis of queuosine, a 7-deazaguanine modified nucleoside found in tRNAGUN of Bacteria and Eukarya. Genetic analysis of ykvJKLM mutants in Acinetobacter confirmed that each was essential for queuosine biosynthesis, and the genes were renamed queCDEF. QueF exhibits significant homology to the type I GTP cyclohydrolases characterized by FolE. Given that GTP is the precursor to queuosine and that a cyclohydrolase-like reaction was postulated as the initial step in queuosine biosynthesis, QueF was proposed to be the putative cyclohydrolase-like enzyme responsible for this reaction. We have cloned the queF genes from B. subtilis and Escherichia coli and characterized the recombinant enzymes. Contrary to the predictions based on sequence analysis, we discovered that the enzymes, in fact, catalyze a mechanistically unrelated reaction, the NADPH-dependent reduction of 7-cyano-7-deazaguanineto7-aminomethyl-7-deazaguanine, a late step in the biosynthesis of queuosine. We report here in vitro and in vivo studies that demonstrate this catalytic activity, as well as preliminary biochemical and bioinformatics analysis that provide insight into the structure of this family of enzymes.
Keywords: tRNA, modified base
The avalanche of new protein structures that have been reported over the last decade (see summary at www.rcsb.org/pdb/holdings.html) has made it clear that the number of scaffolds that are used to produce all of the proteins in a cell is surprisingly limited, with ≈80% of the proteins using one of the 400 structural folds identified to date (1, 2). Specific functions evolve by duplication, recombination, and divergence of this core repertoire (3). Analysis of the functions of the different members of a protein structural family reveal that, in general, catalytic mechanisms and chemistries are conserved in a given family whereas substrate specificity changes (4). Much rarer are the cases in which the reactions catalyzed differ among members of the family (3); the best characterized examples being the TIM barrel superfamilies (5) and the enolase superfamily (6). Understanding the molecular paths that lead to the evolution of one function from another in a given superfamily is one of the next challenges of structural biology, impacting not only our understanding of how proteins evolve, but also the task of correctly annotating the genes identified by whole-genome sequencing (7, 8).
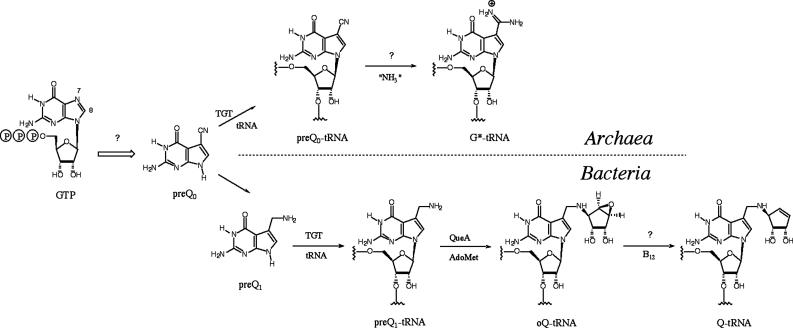
We recently used comparative genomic techniques (9) to discover four previously uncharacterized bacterial genes families (queCDEF) involved in the biosynthesis of the modified nucleoside queuosine (10). Three of these families (queCDE) have homologs in Archaea and are therefore implicated in the biosynthesis of the related modified nucleoside archaeosine (Fig. 1). Both nucleosides share an unusual 7-deazaguanosine core structure but diverge in their phylogenetic distribution, location in the tRNA, and presumed function: queuosine is found in the wobble position of tRNAGUN in Eukarya and Bacteria (11) and is thought to be involved in translational modulation (12), whereas archaeosine is located at position 15 in the majority of archaeal tRNAs (13), where it is thought to function in structural stabilization of the tRNA (13). Pioneering work in the 1970s showed that queuosine is derived from GTP or a related metabolite (14), and 7-cyano-7-deazaguanine (preQ0) and 7-aminomethyl-7-deazaguanine (preQ1) are pathway intermediates (15, 16) (Fig. 1). A key observation in these studies was that C-8 of the guanylate residue is lost in the conversion to queuosine (14), a process reminiscent of the biosynthesis of the antibiotics toyocamycin (17, 18) and tubercidin (19) and the pterins and folic acid (20). In these latter cases, the loss of C-8 occurs through the action of GTP cyclohydrolase I, which converts GTP to dihydroneopterin triphosphate. The structural similarities between 7-deazaguanine and the pterin core, and the common loss of C-8 in the biosynthesis, led to the proposal that a cyclohydrolase-like reaction was the first step in the biosynthesis of the 7-deazaguanine modified nucleosides (21).
Fig. 1.
The biosynthetic pathways to queuosine and archaeosine.
Sequence analysis revealed that QueF is homologous to the GTP cyclohydrolase I (FolE) family, and given the essential role of QueF in queuosine biosynthesis, we hypothesized that QueF was the cyclohydrolase-like enzyme responsible for the first step of queuosine biosynthesis (10). Notably, whereas Archaea lack a QueF homolog, the recently discovered GTP cyclohydrolase III enzyme (22) has been proposed to be involved in the biosynthesis of archaeosine, consistent with an orthologous displacement of the queF gene in Archaea, and providing a putative catalytic activity for QueF.
The results reported here show that, contrary to our initial hypothesis, the members of the QueF family are not cyclohydrolases, but are instead NADPH-dependent oxidoreductases involved in a late step in the queuosine biosynthetic pathway. The sequence homology between the QueF and the FolE families has allowed us to develop a preliminary understanding of the putative structure of the active-site and the structural differences responsible for the divergent substrate specificities and chemistries of these two enzyme families and to predict important structural differences within the QueF family.
Materials and Methods
Cloning of folE, ribA, and yqcD from Escherichia coli and ykvM from Bacillus subtilis.The folE, ribA, and queF (yqcD) genes from E. coli K12, and queF (ykvM) from B. subtilis JH642 (trpC2 pheA1) were cloned into the pET-30Xa vector by using ligation-independent cloning as described by Novagen. PCR amplification was done by using 50 units/ml PfuUltra DNA polymerase, the supplied buffer, 200 μM dNTPs, 20 ng of E. coli or B. subtilis genomic DNA, and 250 ng of each primer. The primers used for each gene were the following: sense (folE), 5′-GGTATTGAGGGTCGCATGCCAGCACTCAGTAAG-3′; antisense (folE), 5′-AGAGGAGAGTTAGAGCCTCAGTTGTGATGACGC-3′; sense (ribA), 5′-GGTATTGAGGGTCGCATGCAGCTTAAACGTGTG-3′; antisense (ribA) 5′-AGAGGAGAGTTAGAGCCTTATTTGTTCAGCAAAT-3′, sense (yqcD) 5′-GGTATTGAGGGTCGCATGTCTTCTTATGCA-3′; antisense (yqcD), 5′-AGAGGAGAGTTAGAGCCTTATTGCCGAACCAGTC-3′; sense (ykvM), 5′-GGTATTGAGGGTCGCATGACGACAAGAAAA-3′; antisense (ykvM) 5′-AGAGGAGAGTTAGAGCCTTAACGATTATCAAT-3′.
The PCR program included an initial hold at 95°C for 45 sec, followed by 30 cycles of 95°C for 45 sec, 60°C for 45 sec, and 72°C for 2 min. The amplified PCR products were gel purified with 1% agarose before annealing and transformation. The integrity of the resulting constructs was confirmed by sequencing.
Expression and Purification of His-6 Fusion Proteins. Recombinant His-6 fusion proteins from pET30 constructs were overproduced in E. coli BL21(DE3) cells and purified by Ni2+-agarose affinity chromatography by using standard protocols. All enzymes were >90% pure based on SDS/PAGE analysis and were stored in 100 mM Tris·HCl (pH 8.0)/300 mM (FolE) or 50 mM (all others) KCl/30% glycerol at –80°C.
Factor Xa cleavage of His-6-YqcD and His-6-YkvM was carried out according to the manufacturer's instructions, and the wild-type proteins were purified by binding the cleaved leader peptides and unreacted fusion protein to the Ni2+-agarose affinity resin. The purified proteins were subsequently dialyzed into the above storage buffers and stored at –80°C.
PreQ0 Reduction Assays. PreQ0 was synthesized as described in ref. 23. Routine assays for the reduction of preQ0 to preQ1 were performed in 100 mM Hepes (pH 7.5)/1mMDTT/50–100 mM KCl/1–100 μM preQ0/2–150 μM NADPH/noted concentration of E. coli or B. subtilis QueF at 30°C. The progress of the reaction was monitored by following the loss of absorbance at 334, 340, and 365 nm (24). Alternatively, the reaction was monitored by reverse-phase HPLC (Bondclone 10 C18, 300 × 3.9 mm) by using a series of linear gradients from 20 mM ammonium acetate (pH 6.0) to 75% methanol in 20 mM ammonium acetate (pH 6.0). Data represent a minimum of three replicates with a relative standard error of <10%. Reactions were initiated by the addition of enzyme unless otherwise noted.
Isolation and NMR Spectroscopy of QueF Reaction Product PreQ1. PreQ1 was produced in a reaction containing 50 mM phosphate (pH 7.25), 100 mM KCl, 0.5 mM preQ0, 0.5 mM NADPH, and 200 μg/ml E. coli His-6-QueF. After reacting at 30°C for 3 h, the protein was removed by ultrafiltration, and the sample was purified by reverse-phase HPLC as described above. A proton spectrum was acquired in deuterium oxide at 400.14 MHz with a spectral width of 5,618 Hz, and methanol was the internal standard: 1H-NMR (400.14 MHz, 2H2O) δ 4.18 (2H, s, C10-H2); 6.86 (1H, s, C8-H). A heteronuclear single quantum coherence experiment was performed in deuterium oxide, and methanol was the internal standard to obtain the chemical shifts of the carbons with attached protons: 13C-NMR (100.62 MHz, 2H2O) δ 36 (C10); 118.5 (C8).
Synthesis and Purification of PreQo-Nucleoside. Minihelix RNA was in vitro transcribed and modified by the insertion of preQ0 essentially as described in ref. 25. The protein was subsequently removed by heat precipitation, and the RNA was precipitated with isopropanol. The RNA was then digested and dephosphorylated (26), and preQ0-nucleoside was purified by reverse-phase HPLC (Supelcosil LC-18S, 5 μm, 250 × 4.6 mm) by using a series of linear gradients from 25 mM ammonium acetate (pH 6.0) to 50% acetonitrile in 25 mM ammonium acetate (pH 6.0).
tRNA Analysis. Bulk tRNA was prepared from Acinetobacter ADP1 (27) and ΔykvM::SacB KmR (10), then hydrolyzed and analyzed by HPLC as described in refs. 26 and 28.
Results and Discussion
QueF Is Not a GTP Cyclohydrolase. The QueF family was described as a putative GTP cyclohydrolase-like enzyme (10) based on its sequence homology to GTP cyclohydrolase I (FolE) (for example, see Fig. 5) and, therefore, was hypothesized to catalyze the initial step in queuosine biosynthesis (10). To test this hypothesis, the queF genes of B. subtilis (ykvM) and E. coli (yqcD) were cloned through PCR into pET30 expression vectors. The genes from both E. coli and B. subtilis were cloned because the encoded enzymes form two subclasses, with the E. coli enzyme ≈60% larger than that from B. subtilis and possessing notable sequence differences (vide infra), and we sought to confirm the activity of both and investigate any structural and functional consequences of these differences. The E. coli genes encoding GTP cyclohydrolase I (folE) and GTP cyclohydrolase II (ribA) were cloned in parallel so that the enzymes would be available to serve as positive controls for cyclohydrolase activity assays. All genes were cloned into pET30 vectors containing an in-frame His-6 cassette and a Factor Xa site to allow for affinity purification of the recombinant proteins and subsequent Factor Xa cleavage at the starting Met to generate wild-type proteins.
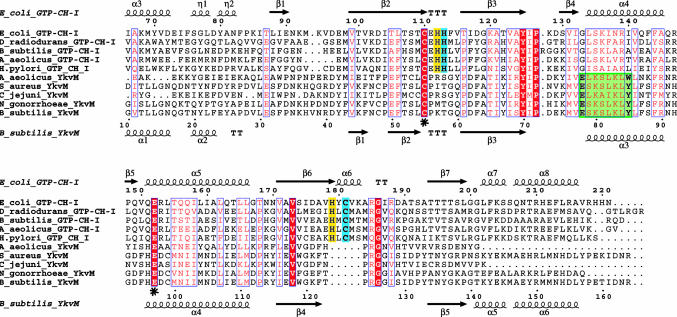
Fig. 5.
Alignment of unimodular FolE (GTP cyclohydrolase I) and YkvM sequences. For clarity and space, only sequences from select organisms are shown from among 60 sequences in the original alignment, and the N-termini have been truncated. Sequence numbers of every 10th residue are shown for E. coli FolE and B. subtilis YkvM. Secondary structure elements and nomenclature as defined by the crystal structure of E. coli FolE and by the 3D homology model of B. subtilis YkvM are shown at Upper and Lower, respectively. The conserved Cys and Glu found in the substrate binding pocket of both protein families are indicated by asterisks. The QueF motif, specific for the QueF family, is highlighted in green. The zinc binding His and Cys residues found in FolE and not in QueF are highlighted in blue. Other catalytic residues in FolE not found in QueF are highlighted in yellow. The absence of the zinc-binding and catalytic residues of FolE is the best identifier of QueF sequences in genome databases.
Possible GTP cyclohydrolase-like activity was investigated by using three different assays [radiochemical-based release of [14C]formic acid (29), fluorescence (30), and HPLC analysis of reactions (31)] and a large screen of assay conditions [including those for cyclohydrolase I, II, and III activity (22, 32)]. Using FolE and RibA as positive controls, we were unable to detect any cyclohydrolase-like enzymatic activity for the QueF enzyme from either B. subtilis or E. coli (data not shown). To maximize the sensitivity of the HPLC assay [U-ribosyl-14C]GTP was synthesized (25) to facilitate the detection of new products that might not be visible by UV-visible detection in HPLC assays. As above, no enzymatic activity was observed for the QueF enzymes. The related metabolites GMP, GDP, and guanosine were also investigated as potential substrates of the QueF enzymes, but no products were observed when monitoring the reactions by HPLC.
QueF Is an NADPH-Dependent Nitrile Oxidoreductase. The failure to observe cyclohydrolase-like activity with E. coli and B. subtilis QueF prompted us to consider alternative roles this gene family might play in queuosine biosynthesis. We had initially reconciled that the lack of a QueF homolog in Archaea to the intervention of the recently discovered type III cyclohydrolase (22) in the biosynthesis of archaeosine. However, the lack of a QueF homolog in Archaea is also consistent with QueF possessing an activity occurring after the formation of preQ0, because preQ0 is the last common intermediate in the pathways to queuosine and archaeosine (Fig. 1). Thus, potential enzymatic activities for QueF included the conversion of preQ0 to preQ1 or the conversion of epoxyqueuosine to queuosine. Both transformations involve biologically unprecedented reductions; in the first, a nitrile to an amine, and in the second, the conversion of an epoxide to an alkene. Because evidence suggests that the second conversion depends on vitamin B12 (33) and no putative B12-binding motif was present in any of the QueF sequences, we first probed the ability of the QueF enzymes to catalyze the conversion of preQ0 to preQ1.
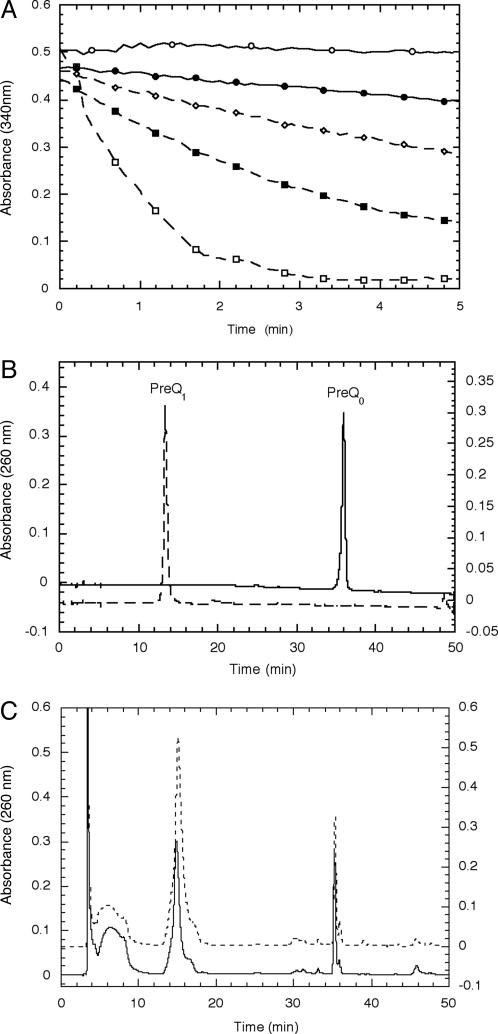
PreQ0 oxidoreductase activity was tested with a number of redox cofactors, and activity was observed in the presence of NADPH. Using a continuous UV-based assay (Fig. 2A), the rate of NADPH oxidation was shown to depend on enzyme and substrate concentrations, consistent with QueF acting as the catalyst in the redox reaction of preQ0 and NADPH. To confirm that preQ1 was the reduced product as predicted, reaction assays were analyzed by HPLC (Fig. 2 B and C). A new peak appeared at ≈14 min that coeluted with authentic preQ1 prepared synthetically (34) and had an identical UV-visible spectrum. The new peak was isolated and analyzed by proton NMR, which showed a spectrum identical to authentic preQ1.
Fig. 2.
Activity of E. coli QueF (assays with B. subtilis QueF gave qualitatively identical results). (A) UV continuous assays of NADPH consumption. Assays were performed in 100 mM Tris·HCl (pH 7.5)/100 μM preQ0/100 μM NADPH and monitored at 340 nm. The concentrations of enzyme are the following: ○, no enzyme; •, 19 μg/ml; ⋄, 38 μg/ml; ▪, 95 μg/ml; □, 190 μg/ml. (B) HPLC chromatogram of synthetic preQ1 (dashed) and preQ0 (solid). (C) HPLC chromatograms of an E. coli QueF reaction (solid) and the reaction spiked with authentic preQ1 (dashed).
Using the continuous UV assay, we carried out a preliminary characterization of the enzyme activity to determine the kinetic constants. Analysis of velocity data with variable NADPH and constant, saturating preQ0 provided a Km for NADPH of 36 μM, consistent with the Km values for other bacterial NADPH-dependent oxidoreductases (35). The measured kcat of 0.6 min–1, while low, is comparable with the two subsequent enzymes in the pathway (25, 36). Attempts to determine the Km for preQ0 were hampered by the inability to obtain accurate rate data at concentrations <1 μM because of the poor signal to noise at such low conversions of NADPH. However, based on our data we can conclude that the KM < 1 μM. The enzyme TGT (Fig. 1), which inserts the product of QueF (preQ1) into the tRNA, has a Km for preQ1 of 0.39 μM (36).
Having shown that QueF catalyzed the conversion of preQ0 to preQ1 in vitro, we sought to demonstrate that this activity was also exhibited in vivo. A disruption in the queF gene leading to a loss of QueF activity should result in the buildup of preQ0, and because the bacterial TGT is able to incorporate preQ0 into tRNA (albeit with somewhat lower efficiency than it incorporates preQ1; ref. 36), analysis of the tRNA from a ΔqueF bacterial strain should reveal the presence of preQ0 nucleoside.
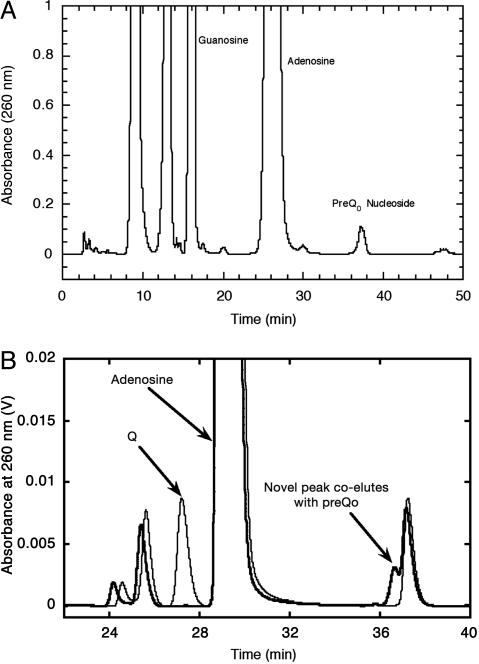
We synthesized authentic preQ0 nucleoside to use as an HPLC standard by carrying out the TGT-mediated exchange of preQ0 for guanine in a synthetic 17-mer RNA corresponding to the anticodon stem-loop of E. coli tRNAAsn (25), followed by enzymatic digestion of the RNA, dephosphorylation of the mononucleotides, and isolation of the nucleoside by HPLC (Fig. 3A). Subsequent analysis of the constituent nucleosides of total tRNA from the YkvM-deficient Acinetobacter ADP1 ΔykvM::sacB-KmR (10) revealed a new peak that coeluted with authentic preQ0-nucleoside (Fig. 3B).
Fig. 3.
HPLC analysis of preQ0 nucleoside. (A) HPLC chromatogram of the constituent nucleosides from digestion of preQ0-containing 17-mer RNA. (B) HPLC chromatograms of the nucleoside components of unfractionated tRNA from wild-type Acinetobacter ADP1 (light line) and Acinetobacter ADP1 ΔykvM::sacB-KmR (heavy line).
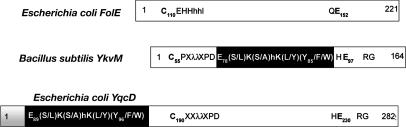
QueF Belongs to the T Fold Structural Superfamily. FolE and QueF are clearly members of the same structural superfamily; the homology score between the two families (detected by Psi-blast; www.ncbi.nlm.nih.gov/blast) is ≈25% sequence identity and 40% similarity in a 100-aa stretch. The FolE family contains two structural subfamilies: homodecameric enzymes of unimodular 26-kDa subunits exemplified by bacterial and mammalian GTP-CH-I (37, 38) and bimodular 50-kDa proteins of two tandem GTP-CH-I-like domains, each containing half the active site residues, and forming lower-order quaternary structures as found in plant GTP-CH-I (39). Similarly, the QueF proteins form two subfamilies, type I proteins exemplified by B. subtilis YkvM and type II proteins exemplified by E. coli YqcD (Fig. 4). The type I proteins are comparable in size with bacterial and mammalian FolE, whereas the type II proteins are larger and are predicted to be comprised of two domains, similar to plant FolE.
Fig. 4.
Primary structure organization of the YqcD and YkvM subfamilies of QueF.
To differentiate the QueF family from the FolE family, because they are both annotated as GTP cyclohydrolase I enzymes in the databases, we generated a clustalw alignment (40) of 30 unimodular FolE sequences and 30 YkvM sequences (Fig. 5). Two major features differentiate the QueF and FolE families. First, the strictly conserved pattern E78(S/L)K(S/A)hK(L/Y)(Y/F/W)85 (residue numbers are those of B. subtilis YkvM, h is hydrophobic amino acid) is characteristic of the QueF family, but is not found in the FolE family, and will be referred to herein as the QueF motif. Two residues, Cys-55 and Glu-97, flank the QueF motif, are strictly conserved in both protein families, and correspond to Cys-110 and Glu-152 in E. coli FolE. Second, four catalytically important residues in FolE (31, 41), His-112, 113, and 179 and Cys-181 (E. coli FolE numbering), are absent in QueF sequences (Fig. 5). Notably, His-113 and Cys-181 provide ligands for Zn2+ (42), indicating the absence of a zinc-binding site in QueF.
The crystal structure of E. coli FolE reveals a homodecamer of two pentameric substructures, each constructed by a cyclic arrangement of the four-stranded β-sheets of the five monomers to form a 20-stranded β-barrel (37). The interfaces between the monomeric subunits each contain a zinc- and a GTP-binding site. The three residues C110, H113, and C181 are involved in zinc binding (41, 42), whereas Glu-152 forms a salt bridge with the C2-NH2 of the guanine moiety of bound GTP. FolE is part of a structural superfamily of functionally distant pterin/purine binding proteins that use a common oligomerization of the characteristic tunneling-fold, comprised of an antiparallel β-sheet and two helices, to form a β2nαn barrel (43). Two barrels join in a head-to-head fashion to form a tunnel-like center. Other members of the FolE structural superfamily are 6-pyruvol tetrahydropterin synthase (n = 3) (44), urate oxidase (n = 4) (45), and dihydroneopterin adolase (n = 4) (46), which all similarly bind planar substrates of purine/pterin at the interface of monomers and use a positionally conserved Glu/Gln to anchor the substrate, although their chemistries and catalytic mechanisms are unrelated. The homology of QueF and FolE families clearly suggests that QueF belongs to the tunneling-fold structural superfamily.
The C-terminal domain of the bimodular E. coli QueF (YqcD) (Fig. 4) contains the region of clear homology to the bacterial and mammalian GTP-CH-I subfamily. The N-terminal domain has often been annotated as a membrane-spanning domain, but transmembrane prediction programs (47) run on YqcD do not detect any transmembrane segments. Instead, the QueF motif can be easily detected in this domain, whereas the flanking and invariant cysteine and glutamate residues (Cys-190 and Glu-230 in E. coli YqcD residue numbers) are only present in the C-terminal domain. The splitting of active-site residues between the two domains of YqcD is very similar to that seen in bimodular FolE, in which neither domain contains the full set of active site residues nor is active when expressed separately (39). Further, the pattern of active-site splitting is the same in both proteins, with a similarly located conserved central sequence motif split from two flanking sequences, which are ≈40 residues apart. The splitting of the YqcD active site suggests that a gene duplication occurred, with each domain retaining some of the residues of the putative active site. As in bimodular FolE, such a duplication event and redistribution of active-site residues could allow the YqcD proteins to evolve a simpler quaternary structure than the YkvM proteins.
To test this hypothesis, we determined the native quaternary structures of B. subtilis and E. coli QueF (YkvM and YqcD, respectively) by gel filtration chromatography (data not shown). The elution volume of YkvM (His-6-tagged and wild-type) is consistent with a molecular weight corresponding to a dodecamer (12.2 and 11.9 subunits, respectively), whereas YqcD (His-6-tagged and wild-type) eluted with a volume consistent with the molecular weight of a dimer (1.8 and 1.9 subunits, respectively). Notably, unimodular FolE exists as a homodecamer, and bimodular FolE as a dimer.
The discovery of oxidoreductase activity within the FolE scaffold is an intriguing example of structural and functional evolution, particularly in light of the need to bind a second organic substrate, the cofactor NADPH. The specificity of the QueF motif to the QueF family suggests that these residues might be involved in NADPH binding. Additionally, the binding of a modified base to QueF, instead of the nucleotide to FolE, in principle leaves vacant in QueF the binding site occupied by the ribosyl portion of GTP. This putative “empty” ribosyl pocket might also contribute to NADPH binding. The veracity of our predictions will be tested formally when the crystal structures of the QueF enzymes are solved.
Conclusions
The biochemical and genetic data clearly establish that QueF is not a GTP cyclohydrolase as suggested by sequence homology but a previously uncharacterized class of oxidoreductase that carries out the unprecedented reduction of a nitrile group (preQ0) to a primary amine (preQ1). Currently, four types of enzymes are known to be involved in the metabolism of nitriles (48): nitrilase, which catalyzes the hydrolysis of nitriles to the corresponding acids and ammonia; nitrile hydratase, which catalyzes the partial hydrolysis of nitriles to form amides; oxygenase, which oxidizes α to the cyano group to form cyanohydrins; and hydroxynitrile lyase, which cleave a wide range of cyanohydrins into aldehydes or ketones and hydrogen cyanide. The QueF family represents a fifth class of enzymes responsible for nitrile metabolism, the four-electron reduction to form primary amines. This enzyme family provides a compelling example of a protein scaffold being recruited for a significantly different function and underscores the problem of relying on genome annotations and blast scores to predict function, particularly in the case of enzyme superfamilies (49). In addition to the fundamental interest generated by the discovery of a previously uncharacterized enzyme activity, the discovery of biological nitrile reduction may have important applications to industrial biocatalysis, because nitrile-containing compounds and the amines derived from them are ubiquitous intermediates in the pharmaceutical, specialty, and commodity chemical industries.
Acknowledgments
We thank Dr. Michiko Nakano (Oregon Health and Science University, Beaverton) for the B. subtilis strain, Integrated Genomics (Chicago) for ergo access, Ross Overbeek (The Fellowship for Integration of Genomes, Argonne, IL) for seed access, and Paul Schimmel for his unwavering support. This work was supported by National Science Foundation Grants MCB-9733746 (to D.I.-R.) and MCB-0128901 (to V.d.C.-L.).
Author contributions: V.d.C.-L. and D.I.-R. designed research; S.G.V.L., J.S.R., M.A.S., V.d.C.-L., and B.L. performed research; S.G.V.L., J.S.R., M.A.S., V.d.C.-L., and D.I.-R. analyzed data; and M.A.S., V.d.C.-L., and D.I.-R. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FolE, GTP cyclohydrolase I; preQ0, 7-cyano-7-deazaguanine; preQ1, 7-aminomethyl-7-deazaguanine.
References
- 1.Coulson, A. F. & Moult, J. (2002) Proteins 46, 61–71. [DOI] [PubMed] [Google Scholar]
- 2.Babbitt, P. C. & Gerlt, J. A. (1997) J. Biol. Chem. 272, 30591–30594. [DOI] [PubMed] [Google Scholar]
- 3.Chothia, C., Gough, J., Vogel, C. & Teichmann, S. A. (2003) Science 300, 1701–1703. [DOI] [PubMed] [Google Scholar]
- 4.Teichmann, S. A., Rison, S. C., Thornton, J. M., Riley, M., Gough, J. & Chothia, C. (2001) J. Mol. Biol. 311, 693–708. [DOI] [PubMed] [Google Scholar]
- 5.Copley, R. R. & Bork, P. (2000) J. Mol. Biol. 303, 627–641. [DOI] [PubMed] [Google Scholar]
- 6.Meng, E. C., Polacco, B. J. & Babbitt, P. C. (2004) Proteins 55, 962–976. [DOI] [PubMed] [Google Scholar]
- 7.Babbitt, P. C. (2003) Curr. Opin. Chem. Biol. 7, 230–237. [DOI] [PubMed] [Google Scholar]
- 8.Kinch, L. N. & Grishin, N. V. (2002) Curr. Opin. Struct. Biol. 12, 400–408. [DOI] [PubMed] [Google Scholar]
- 9.Osterman, A. & Overbeek, R. (2003) Curr. Opin. Chem. Biol. 7, 238–251. [DOI] [PubMed] [Google Scholar]
- 10.Reader, J. S., Metzgar, D., Schimmel, P. & de Crecy-Lagard, V. (2004) J. Biol. Chem. 279, 6280–6285. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama, S., Miyazawa, T., Iitaka, Y., Yamaizumi, Z., Kasai, H. & Nishimura, S. (1979) Nature 282, 107–109. [DOI] [PubMed] [Google Scholar]
- 12.Kersten, H. & Kersten, W. (1990) in Chromatography and Modification of Nucleosides Part B, eds. Gehrke, C. W. & Kuo, K. C. T. (Elsevier, Amsterdam), pp. B69–B108.
- 13.Gregson, J. M., Crain, P. F., Edmonds, C. G., Gupta, R., Hashizume, T., Phillipson, D. W. & McCloskey, J. A. (1993) J. Biol. Chem. 268, 10076–10086. [PubMed] [Google Scholar]
- 14.Kuchino, Y., Kasai, H., Nihei, K. & Nishimura, S. (1976) Nucleic Acids Res. 3, 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noguchi, S., Yamaizumi, Z., Ohgi, T., Goto, T., Nishimura, Y., Hirota, Y. & Nishimura, S. (1978) Nucleic Acids Res. 5, 4215–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okada, N., Noguchi, S., Nishimura, S., Ohgi, T., Goto, T., Crain, P. F. & McCloskey, J. A. (1978) Nucleic Acids Res. 5, 2289–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uematsu, T. & Suhadolnik, R. J. (1970) Biochemistry 9, 1260–1266. [DOI] [PubMed] [Google Scholar]
- 18.Suhadolnik, R. J. & Uematsu, T. (1970) J. Biol. Chem. 245, 4365–4371. [PubMed] [Google Scholar]
- 19.Smulson, M. E. & Suhadolnik, R. J. (1967) J. Biol. Chem. 242, 2872–2876. [PubMed] [Google Scholar]
- 20.Krumdieck, C. L., Shaw, E. & Baugh, C. M. (1966) J. Biol. Chem. 241, 383–387. [PubMed] [Google Scholar]
- 21.Iwata-Reuyl, D. (2003) Bioorg. Chem. 31, 24–43. [DOI] [PubMed] [Google Scholar]
- 22.Graham, D. E., Xu, H. & White, R. H. (2002) Biochemistry 41, 15074–15084. [DOI] [PubMed] [Google Scholar]
- 23.Migawa, M. T., Hinkley, J. M., Hoops, G. C. & Townsend, L. B. (1996) Synth. Commun. 26, 3317–3322. [Google Scholar]
- 24.Ziegenhorn, J., Senn, M. & Bucher, T. (1976) Clin. Chem. 22, 151–160. [PubMed] [Google Scholar]
- 25.Van Lanen, S. G., Kinzie, S. D., Matthieu, S., Link, T., Culp, J. & Iwata-Reuyl, D. (2003) J. Biol. Chem. 278, 10491–10499. [DOI] [PubMed] [Google Scholar]
- 26.Crain, P. F. (1990) Methods Enzymol. 193, 782–790. [DOI] [PubMed] [Google Scholar]
- 27.Metzgar, D., Bacher, J. M., Pezo, V., Reader, J., Doring, V., Schimmel, P., Marliere, P. & Crecy-Lagard, V. d. (2004) Nucleic Acids Res. 32, 5780–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pomerantz, S. C. & McCloskey, J. A. (1990) Methods Enzymol. 193, 796–824. [DOI] [PubMed] [Google Scholar]
- 29.Yim, J. J. & Brown, G. M. (1976) J. Biol. Chem. 251, 5087–5094. [PubMed] [Google Scholar]
- 30.Hatakeyama, K. & Yoneyama, T. (1998) Methods Mol. Biol. 100, 265–272. [DOI] [PubMed] [Google Scholar]
- 31.Nar, H., Huber, R., Auerbach, G., Fischer, M., Hosl, C., Ritz, H., Bracher, A., Meining, W., Eberhardt, S. & Bacher, A. (1995) Proc. Natl. Acad. Sci. USA 92, 12120–12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foor, F. & Brown, G. M. (1975) J. Biol. Chem. 250, 3545–3551. [PubMed] [Google Scholar]
- 33.Frey, B., McCloskey, J. A., Kersten, W. & Kersten, H. (1988) J. Bacteriol. 170, 2078–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akimoto, H., Imaniya, E., Hitaka, T., Nomura, H. & Nishimura, S. (1988) J. Chem. Soc. Perkin Trans. 1, 1637–1644.
- 35.Smith, E. L., Austen, B. M., Blumenthal, K. M. & Nye, J. F. (1975) in The Enzymes, ed. Boyer, P. D. (Academic, New York), Vol. XI, pp. 293–367. [Google Scholar]
- 36.Hoops, G. C., Townsend, L. B. & Garcia, G. A. (1995) Biochemistry 34, 15381–15387. [DOI] [PubMed] [Google Scholar]
- 37.Nar, H., Huber, R., Meining, W., Schmid, C., Weinkauf, S. & Bacher, A. (1995) Structure 3, 459–466. [DOI] [PubMed] [Google Scholar]
- 38.Maier, J., Witter, K., Gutlich, M., Ziegler, I., Werner, T. & Ninnemann, H. (1995) Biochem. Biophys. Res. Commun. 212, 705–711. [DOI] [PubMed] [Google Scholar]
- 39.Basset, G., Quinlivan, E. P., Ziemak, M. J., Diaz De La Garza, R., Fischer, M., Schiffmann, S., Bacher, A., Gregory, J. F., 3rd, & Hanson, A. D. (2002) Proc. Natl. Acad. Sci. USA 99, 12489–12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. G. & Thompson, J. D. (2003) Nucleic Acids Res. 31, 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rebelo, J., Auerbach, G., Bader, G., Bracher, A., Nar, H., Hosl, C., Schramek, N., Kaiser, J., Bacher, A., Huber, R. & Fischer, M. (2003) J. Mol. Biol. 326, 503–516. [DOI] [PubMed] [Google Scholar]
- 42.Auerbach, G., Herrmann, A., Bracher, A., Bader, G., Gutlich, M., Fischer, M., Neukamm, M., Garrido-Franco, M., Richardson, J., Nar, H., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 13567–13572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colloc'h, N., Poupon, A. & Mornon, J. P. (2000) Proteins 39, 142–154. [DOI] [PubMed] [Google Scholar]
- 44.Nar, H., Huber, R., Heizmann, C. W., Thony, B. & Burgisser, D. (1994) EMBO J. 13, 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colloc'h, N., el Hajji, M., Bachet, B., L'Hermite, G., Schiltz, M., Prange, T., Castro, B. & Mornon, J. P. (1997) Nat. Struct. Biol. 4, 947–952. [DOI] [PubMed] [Google Scholar]
- 46.Hennig, M., D'Arcy, A., Hampele, I. C., Page, M. G., Oefner, C. & Dale, G. E. (1998) Nat. Struct. Biol. 5, 357–362. [DOI] [PubMed] [Google Scholar]
- 47.Hofmann, K. & Stoffel, W. (1992) Comput. Appl. Biosci. 8, 331–337. [DOI] [PubMed] [Google Scholar]
- 48.Banerjee, A., Sharma, R. & Banerjee, U. C. (2002) Appl. Microbiol. Biotechnol. 60, 33–44. [DOI] [PubMed] [Google Scholar]
- 49.Gerlt, J. A. & Babbitt, P. C. (2000) Genome Biol. 1, 1–10.11178226 [Google Scholar]