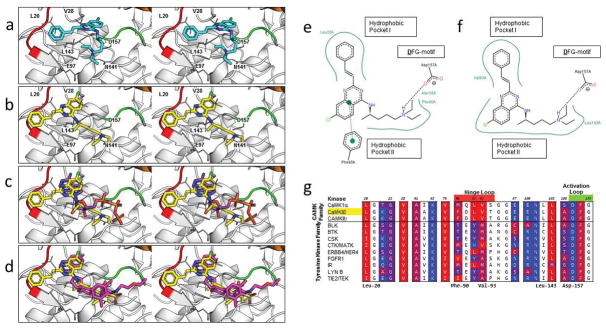
Figure 4.
Characterization of the binding mode of 1 to CaMKIIδ. (a) Stereoview of the binding mode of 1 from the original virtual screening against CaMKIIδ in the R configuration. (b) Stereoview of the predicted binding mode of 1 after analysis of the compound series against CaMKIIδ, CaMKIIγ, and MELK. (c) Stereoview of ATP (PDB ID: 1ATP, 2.20 Å) is superimposed on the predicted binding mode of 1 (yellow). (d) Stereoview of an experimentally crystallized inhibitor featuring a quinazoline ring structure in a similar conformation (PDB ID: 3QKM, 2.20 Å, magenta) as the predicted binding mode of 1 (yellow). In (a–d), a crystal structure of CaMKIIδ (PDB ID: 2WEL, 1.90 Å) is shown as cartoon (gray) with the hinge loop (red), DFG-motif (green), and αC helix (orange) highlighted. The protein is shown with the glycine-rich loop removed (Residues Glu-19 to Arg-29) to expose the ATP binding pocket. Key residues between 1 and the protein kinase are shown in stick format. Two-dimensional representation of the (e) virtual screening and (f) Glide-predicted binding modes of 1 against CaMKIIδ, showing the key interactions between the atoms of the ligand and the ATP binding pocket of the receptor. Generated by PoseView (http://poseview.zbh.uni-hamburg.de/poseview). (g) A multiple sequence alignment of the top candidates of 6 was generated in Cluster Omega and the residues of CaMKIIδ within 4 Å of the original virtual screening binding mode of 1 were identified (in color). These residues were annotated based on the hydrophobicity of the residue, ranging from red (most hydrophobic) to blue (most hydrophilic). Residues in the hinge loop and activation loop are highlighted red and green, respectively, on the residue numbers.