Abstract
Neuron-glial interactions are crucial for growth, guidance and ensheathment of axons across species. In the Drosophila CNS midline, neuron-glial interactions underlie ensheathment of commissural axons by midline glial (MG) cells in a manner similar to mammalian oligodendrocytes. Although there has been some advance in the study of neuron-glial interactions and ensheathment of axons in the CNS midline, key aspects of axonal ensheathment are still not fully understood. One of the limitations has been the unavailability of MG membrane markers that could highlight the glial processes wrapping the axons. Previous studies have identified two key molecular players from the neuronal and glial cell types in the CNS midline. These are the neuronal transmembrane protein Neurexin IV (Nrx IV) and the membrane-anchored MG protein Wrapper, both of which interact in trans to mediate neuron-glial interactions and ensheathment of commissural axons. In the current study, we attempt to further our understanding of MG biology and try to overcome some of the technical difficulties posed by the lack of a robust MG driver that will specifically allow expression or knockdown of genes in MG. We report the generation of BAC transgenic flies of wrapper-GAL4 and demonstrate how these flies could be used as a genetic tool to understand MG biology. We have utilized the GAL4/UAS system to drive GFP-reporter lines (membrane-bound mCD8-GFP; microtubule-associated tau-GFP) and nuclear lacZ using wrapper-GAL4 to highlight the MG cells and/or their processes that surround and perform axonal ensheathment functions in the embryonic midline. We also describe the utility of the wrapper-GAL4 driver line to down-regulate known MG genes specifically in Wrapper-positive cells. Finally, we validate the functionality of the wrapper-GAL4 driver by rescue of wrapper mutant phenotypes and lethality. Together, these studies provide us with a versatile genetic tool to investigate MG functions and will aid in future investigations where genetic screens using wrapper-GAL4 could be designed to identify novel molecular players at the Drosophila midline and unravel key aspects of MG biology.
Keywords: Neurexin IV, Wrapper, wrapper-GAL4, Midline Glia, Axonal Ensheathment, Commissures
Introduction
Drosophila has emerged as a powerful model system for the study of neuron-glial biology due to its genetic, molecular, and behavioral tractability. Although Drosophila has a relatively simple nervous system compared to vertebrates, it exhibits stereotypic glial organization and neuron-glial interactions similar to vertebrates. The establishment of neuron-glial interactions during development is essential for correct functioning of the nervous system. Any disruption in interactions between the neurons and glial cells has structural and physiological consequences that lead to severe pathologies in the nervous system (Bhat, 2003; Edenfeld et al., 2005; Nave, 2010).
One of the key features of neuron-glial interactions is ensheathment of axons. In vertebrates, oligodendrocytes in the CNS and Schwann cells in the PNS help maintain neuronal functions by wrapping axons with sheaths of myelin (Herbert and Monk, 2016; Bhat, 2003). This process of myelination is important for fast saltatory conduction of nerve impulses and in the organization of the axons into distinct molecular domains, namely the Nodes of Ranvier, paranodes, juxtaparanodes and internodes (Thaxton and Bhat 2009; Thaxton et al., 2011; Buttermore et al., 2013). Studies from our laboratory and others have established a key tripartite complex of highly conserved proteins, namely Contactin-associated protein (CASPR), Contactin and Neurofascins (Thaxton and Bhat, 2009; Charles et al., 2002), and their Drosophila counterparts in the PNS: Neurexin IV (Nrx IV), Contactin (Cont) and Neuroglian (Faivre-Sarrailh et al., 2004; Banerjee et al., 2006a, b) to be crucial for neuron-glial interactions and ensheathment of axons.
Although a number of genes have been identified that show specific expression in MG (Crews, 2010), the mechanistic details of how MG interact with midline neurons and their axons are still poorly understood. The Drosophila embryonic midline has been compared to the vertebrate spinal cord (floorplate) as both have a specialized set of cells at the ventral midline comprised of neurons and glial cells. Both cell types exist as a scaffold that ensheath commissural axons and both are important signaling centers (Jacobs, 2000; Lane et al., 2004; Crews, 2010). The Drosophila midline has long been exploited as a model system to dissect axon guidance pathways and has provided fundamental information about the Slit/Roundabout repulsive and Netrin/Frazzled attractive axon guidance cues (Hidalgo, 2003; O'Donnell et al., 2009; Dickson and Zou, 2010; Fulkerson and Estes, 2011; Neuhaus-Follini and Bashaw, 2015). More recent studies in Drosophila embryonic CNS midline have demonstrated the requirement of Nrx IV and Wrapper in neuron-glial interactions and ensheathment of commissural axons (Stork et al., 2009; Wheeler et al., 2009; Slováková and Carmena, 2011; Jacobs 2000; Noordermeer et al., 1998). While Wrapper does not have a direct mammalian ortholog, its structure is very similar to proteins found to interact with Nrx IV/Caspr such as Cont/TAG1 (Pavlou et al., 2002). Both Wrapper and Cont are Ig-superfamily proteins with Ig/FNIII domains and are inserted in the membrane via their GPI anchors.
Despite recent advances in the study of neuron-glial interactions and identification of proteins that mediate these interactions in the CNS midline, there is still a lack of key reagents that could provide insights into how the MG cells wrap the commissural axons and identify deficits in axonal wrapping in mutants. To address these limitations, here we report the generation and characterization of wrapper-GAL4 and its utilization to further our understanding of the molecular basis of axo-glial interactions underlying the ensheathment of axons during CNS development.
Material and Methods
Generation of wrap-GAL4
wrapper-GAL4 was generated using Bacterial Artificial Chromosome (BAC)-based recombination (Carreira-Rosario et al., 2013; Ejsmont et al., 2011). We used BAC CH321-61P08 (Venken et al., 2009; Venken et al., 2006), which contains the complete wrapper locus and the adjacent loci. This BAC was modified by inserting yeast GAL4 sequence immediately after the wrapper start codon followed by a neo cassette and the rest of the BAC sequence. The resultant modified BAC was injected into Drosophila embryos containing acceptor sites to create transgenic flies carrying wrapper-GAL4 integrated into their genome on 2nd and 3rd chromosomes.
Fly stocks
Fly stocks used in this study were: Canton S as the wild type control, nrxIV4304 (Baumgartner et al., 1996), wrapper175, UAS-wrapper (Noordermeer et al., 1998), wrapper-GAL4 (Janelia GAL4 stock) wrapper-GAL4 (this study) and UAS-Cont (Faivre-Sarrailh et al., 2004). All other fly stocks, including the UAS-slitRNAi (stock numbers 31467, 31468) and UAS-wrapperRNAi (stock number 29561), were obtained from Bloomington Stock Center.
Immunohistochemistry, β-Galactosidase staining and confocal imaging
Immunostaining experiments were performed as previously described (Banerjee et al., 2006). Primary antibodies used were: anti-BP102 (DSHB, 1:500), anti-Slit (DSHB, 1:250), anti-Wrapper (Wheeler et al., 2009), anti-Nrx IV (Baumgartner et al., 1996), anti-Cont (Faivre-Sarrailh et al., 2004), anti-GFP (Life Technologies, 1:500) and anti-β-Gal (Promega, 1:500). Fluorescent Alexa Fluor 488, 568 and 647 (Life Technologies, 1:200) were used as secondary antibodies. Confocal images were acquired using Zeiss LSM710 and image editing was done using Adobe Photoshop. β-Galactosidase histochemistry was performed according to Sonnenfeld and Jacobs (1995).
Electron Microscopy
For transmission electron microscopy (TEM), wild type and embryos from specified genotypes were dechorionated and processed for TEM as previously described (Stollewerk et al., 1996; Banerjee et al., 2006), with some modifications for the β-galactosidase assayed embryos. Briefly, embryos were collected and dechorionated in 100% bleach for 2.5 minutes. Embryos were then washed in 0.1M Phosphate buffer, and fixed in 5% glutaraldehyde in 0.1M phosphate buffer, pH 7.0. Embryos were manually devitelinized in PBS and placed at 37oC in a β-galactosidase staining solution (PBS, pH 7.4, 10mM MgCl2, 31 μM K3[Fe(CN)6], 31 μM K4[Fe(CN)6], 0.2% X-gal) until color developed. Embryos were washed with 0.1M Phosphate buffer, and post-fixed for 30 mins in 18.5% glutaraldehyde in 0.1M sodium phosphate buffer, pH 7.0 and then with 2% acrolein/ 4% glutaraldehyde in 0.1 M sodium phosphate buffer for 15 minutes. They were subsequently placed in 4% glutaraldehyde in 1M sodium cacodylate buffer, followed by 1 hr in cacodylate-buffered 1% osmium tetroxide and stained en bloc with 2% uranyl acetate, rinsed, dehydrated in increasing concentrations of cold ethanol, and embedded in PolyBed (Polysciences, Warrington, PA). One-micrometer sections were cut and stained with a combination of uranyl acetate and lead citrate. Electron micrographs were obtained using a Zeiss Leo 910 TEM microscope with a digital camera. Images were processed using Adobe Photoshop software.
Survival assay
Three sets of 100 embryos and third instar larvae of the specified genotypes were analyzed for the survival assay. Briefly, 0-2 hour embryos of all genotypes were collected and aged at 25°C. Survival of embryos to 1st instar larvae were scored after 24-26 hours from egg lay. It was observed that the embryos that did not hatch past 26 hours after egg lay, failed to hatch altogether. For correct identification of mutant and rescue genotypes, non-GFP embryos were sorted at stage 16 of development. For third instar larval to adult survival assay, three sets of 100 larvae of the specified genotypes were scored for eclosion to adulthood.
Quantification of Wrapper fluorescence intensity
Fluorescence intensity measurements for Wrapper was quantified from confocal slices of embryonic stage 16 Z-stack images compressed using maximum projection functions. Regions of interest were selected for each MG from abdominal segments 2-8 (A2-A8) of embryos of designated genotypes and used for assessment and quantification of fluorescence intensity using Image J software. For each embryo, average background fluorescence was subtracted from fluorescence intensity measurements of Wrapper from MG of segments A2-A8. 15 embryos (n=15) from each genotype were assayed for Wrapper levels and subjected to statistical analysis.
Statistical Analysis
All statistical analyses were performed using the Graphpad PRISM software. Statistical significance was determined by one way ANOVA followed by post hoc Tukey's multiple comparison test and Student's t-test. Error bars represent mean + SEM (***p<0.001, **p<0.01, *p<0.05, ns – not significant).
Results and Discussion
Generation of wrapper-GAL4 for Midline Glial Expression
MG are known to ensheath the commissural axons in the CNS midline. However, the process of MG ensheathment is not well characterized partly due to the lack of known MG membrane proteins that are robustly expressed in MG processes, or GAL4 drivers that express specifically in the MG cells. Some of the previously generated midline GAL4 drivers such as sim-GAL4 (Fig. 1A), slit-GAL4 (Fig. 1B) (Awasaki and Lee, 2011; Xiao et al., 1996; Scholz et al., 1997) and wrapper-GAL4 (Fig. 1C) (Janelia GAL4 collection) have expression that is not limited to MG. In addition to MG, these GAL4s are either expressed in midline neurons or non-neuronal tissues. The immunoglobulin (Ig)-superfamily protein, Wrapper, was identified and reported as a MG-specific protein (Noordermeer et al., 1998) that mediates neuron-glial interactions and ensheathment of commissural axons in the CNS midline (Stork et al., 2009; Wheeler et al., 2009). Although Wrapper is a GPI-linked protein, its endogenous expression could not be detected in the MG processes. Because the previously generated wrapper-GAL4 driver (Fig. 1C) did not mimic the endogenous Wrapper expression pattern, we decided to create a Bacterial Artificial Chromosome (BAC) that contained GAL4 sequence inserted into the wrapper locus (Fig 1D; Carreira-Rosario et al., 2013; Ejsmont et al., 2011). The insertion site was immediately following the wrapper start codon (ATG; Fig 1D). No genomic sequences were deleted from the wrapper locus. Since endogenous Wrapper was restricted mostly to the MG soma, we expected our transgenic wrapper-GAL4 driver line to allow expression of genes and reporters specifically in Wrapper-positive cells. The modified BAC was subsequently injected into embryos to generate transgenic wrapper-GAL4 strains. After establishing a wrapper-GAL4 line, the flies were crossed to UAS-mCD8-GFP to express a membrane-bound GFP reporter to confirm GFP expression in the midline (Fig. 1E, E′, green). These embryos were co-stained with BP102 that labels the CNS axonal scaffold (Fig. 1E′, red). The expression of GFP in MG confirmed the successful generation of a wrapper-GAL4 driver with specific expression in the MG cells. To further determine whether the wrapper-Gal4 line that we generated caused any increase in Wrapper levels, we measured the fluorescence intensity of Wrapper in MG of wrapper-Gal4 homozygous embryos. As shown in SFig. 1, no significant differences in Wrapper levels were observed when compared to the wild type controls.
Figure 1. Generation and expression of wrapper-GAL4.
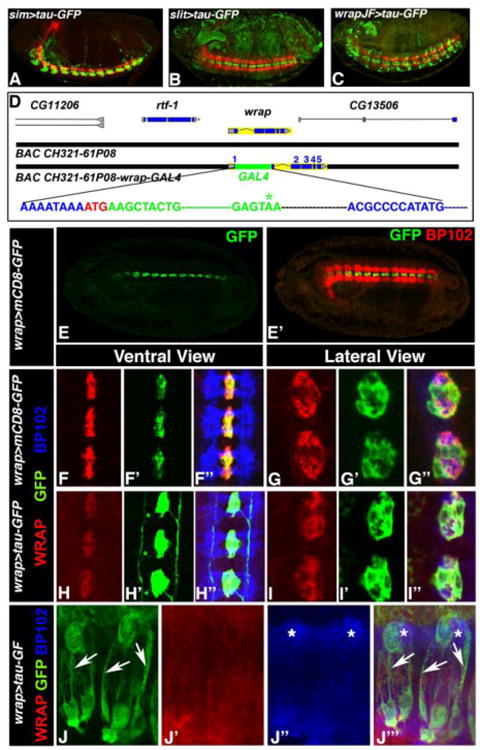
(A-C) Confocal images of Stage 16 embryos of sim-GAL4/UAS-tauGFP (A), slit-GAL4/UAS-tauGFP (B) and wrapper-GAL4/UAS-tauGFP (C, Janelia GAL4 stock abbreviated as JF) show expression of GFP (green) and BP102 (red). (D) Design of the wrapper-GAL4 construct using BAC CH321-61P08. The DNA sequence in blue is from the wrapper gene with its initiator codon ATG shown in red. The sequence in green is the GAL4 sequence and the termination codon is shown in green with asterisk. Further downstream the wrapper sequence continues in blue. No wrapper sequences were deleted but only disrupted with inframe insertion of GAL4. (E) Confocal image of Stage 16 embryo of wrapper-GAL4/UAS-mCD8-GFP in ventral view shows GFP expression in MG (E) and BP102 in CNS axons (E′). (F-F″ and G-G″) Higher magnification confocal images of Stage 16 embryos of wrapper-GAL4/UAS-mCD8-GFP (F-F″, ventral view, and G-G″ lateral view) shows an overlap of Wrapper with GFP (F″ and G″). (H-H″ and I-I″) Confocal images of Stage 16 wrapper-GAL4/UAS-tau-GFP embryos in ventral view (H-H″) and lateral view (I-I′) also display an overlap of Wrapper (H and I) with GFP (H′ and I′) at the MG. GFP expression can also be seen in glial processes (green asterisk, H′). BP102 (blue) was used for labeling CNS axonal scaffold. Embryos in A, B and C show a lateral view and are oriented anterior to the left and dorsal top. Embryo in E shows a sagittal view with anterior to the left. Higher magnification images in F-I″ have anterior to the top. (J-J‴) Confocal images of MG and their ensheathing processes in third instar ventral nerve cord of wrapper-Gal4/UAS-tau-GFP labeled with GFP (J, J‴, arrows), Wrapper (J′, J‴) and BP102 (J″, J‴, asterisks).
We next wanted to analyze in more detail to what extent the wrapper-GAL4 recapitulates the endogenous Wrapper expression in wrapper-GAL4/UAS-mCD8-GFP embryos during CNS midline development (Fig. 1F-F″, G-G″). To this end, we immunostained stages 12-16 wrapper-GAL4/UAS-mCD8-GFP embryos with anti-GFP (green), anti-Wrapper (red) and anti-BP102 (blue) (data shown for stage 16, Fig. 1F-F″, G-G″). Across all stages of embryonic development that we analyzed, GFP showed colocalization with the endogenous Wrapper in both the ventral (Fig. 1F-F″) and lateral views (Fig. 1G-G″). The lateral view allowed us to study the axonal wrapping by MG cells where the commissural axons labeled by anti-BP102 (blue, Fig. 1G″) are seen to be completely surrounded by Wrapper (Fig. 1G) and GFP (Fig. 1G′) expressed in Wrapper-positive MG cells.
Although the membrane-bound mCD8-GFP mimicked the endogenous Wrapper with high fidelity, it did not label the MG processes. Therefore, in order to visualize MG processes, we recombined the UAS-tau-GFP reporter with wrapper-GAL4 to increase the copy number and intensity of the GFP expression. Since tau-GFP binds to the microtubule network, it facilitated the visualization of the glial cytoplasm and processes (Williams et al., 2000) in embryos of wrapper-GAL4,UAS-tau-GFP (Fig. 1H-H″). Embryos were immunostained with anti-GFP (Fig. 1H′, I′, green), anti-Wrapper (Fig. 1H, I, red) and anti-BP102 (Fig. 1H″, I″, blue). While tau-GFP expression overlapped with endogenous Wrapper (Fig. 1H, H′), we observed additional expression of GFP in cellular projections (green asterisks, Fig. 1H′) that emanated from MG cells and otherwise not visible with anti-Wrapper (Fig. 1H). These projections seemed to pass through the commissural axon tracts in stage 13 (data not shown) and ran along the entire length of the longitudinal axons at later developmental time points (Fig. 1H′, SFig.2). It is important to note that the UAS-tauGFP reporter embryos have some leaky GFP expression mostly in the periphery (SFig. 2). Since MG are mostly known for ensheathment of commissural axons, the presence of longitudinal processes may indicate that MG may have additional functions along longitudinal axonal tracts. These functions may include providing trophic support or ensheathment of axons as well as signal transduction interfaces. MG processes were observed outside the midline area and they seemed very transient. At this stage it is not clear whether some processes are eliminated as development continues. This transient MG process formation will be further explored using EM and immuno-EM to precisely define the dynamic nature of the process formation and axonal ensheathment during midline development. Together these data sets demonstrate that the newly generated wrapper-GAL4 better recapitulates expression in Wrapper-positive MG cells than any of the existing MG drivers (shown in Fig. 1A-C) and that it can be used as a tool to specifically study MG cell development and function.
Having examined the wrapper-Gal4 expression in the embryonic stages, we extended our studies to visualize MG in the third instar larval ventral nerve cord (VNC). A lateral view of the third instar VNC of wrapper-Gal4;tau-GFP showed GFP expression in the MG cells and their membrane processes (Fig. 1J, J‴, arrows) ensheathing the commissural axons labeled with BP102 (Fig. 1J″, J‴, asterisks) as reported previously (Jacobs, 2000). Wrapper expression (Fig. 1J′) in the lateral view was detected in the MG membrane at lower intensity than GFP. These data suggest that the newly engineered wrapper-Gal4 could be used to study MG beyond embryonic development.
wrapper-GAL4, a Tool for Analysis of Midline Glial Cell Biology
We next wanted to test the utility and versatility of the wrapper-GAL4 as a genetic tool to study MG biology at the embryonic CNS. We crossed the wrapper-GAL4 flies to the UAS-lacZ reporter line to analyze expression of lacZ, as detected by β-galactosidase activity in Wrapper-positive cells in wrapper-GAL4/UAS-lacZ embryonic midline (Fig. 2A, B). The substrate 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) is converted to an insoluble product which forms blue crystals (Sonnenfeld and Jacobs, 1995). Our goal was to utilize the β-galactosidase-expressing wrapper-GAL4/UAS-lacZ embryos for ultrastructural analysis such that the β-galactosidase generated reaction product crystals enabled the visualization of the MG soma and their cellular processes with exceptional clarity. As a comparison, we first performed transmission electron microscopy (TEM) analysis of wild type embryos (Fig. 2C-E). Cross-sections through the abdominal segments of the wild type embryos reveal the commissures (Com) separated by midline glia (MG) with neuronal cell bodies (asterisks) on the ventral side (Fig. 2C). Higher magnification micrographs from the wild type midline reveal MG cells (MG, Fig. 2D) and commissural axons (Ax, Fig. 2E) surrounded by MG processes (arrowhead, Fig. 2E). TEM of wrapper-GAL4/UAS- lacZ embryos show the accumulation of crystals surrounding the MG nuclei (Fig. 2F,G) as well as the glial processes. The presence of crystals in the TEM of wrapper-GAL4/UAS- lacZ embryos provided an ease and definitive identification of the MG cells and simplified following the anatomical features of glial processes, thus providing a better resolution in the study and understanding of axonal ensheathment in a wild type context. Together the ultrastructural studies demonstrate that wrapper-GAL4 could be utilized for the study of MG cell ultrastructure and processes that traverse and ensheath axons not only across different stages of embryonic development but also could be utilized to study the deficits in the mutants of known and yet to be identified MG genes.
Figure 2. Utility of wrapper-GAL4 in the study of CNS midline.
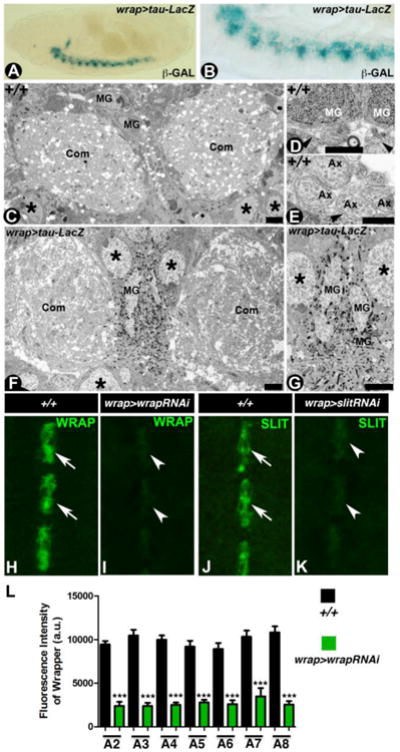
(A, B) β-Gal staining of wrapper-GAL4/UAS-tau-lacZ stage 16 embryo show lacZ expression specifically in the MG at a low magnification (A) and high magnification (B). (C-E) Electron micrographs of embryonic stage 16 wild type midline displayed commissural axon tracts (Com) separated by MG (C). Also visible are the neuronal cell bodies in the CNS (asterisk). Higher magnification images shown with MG and their processes (arrowheads, D) and axons (Ax, E) surrounded by MG processes (arrowhead). (F, G) Electron micrographs of stage 16 wrapper-GAL4/UAS-tau-lacZ reveal MG cells and processes decorated by β-Gal reaction product crystals (F, G). Note the commissural axon tracts crossing the midline are surrounded by MG processes and are highlighted by β-Gal reaction product crystals (G). (H-L) Confocal images of stage 16 embryonic midline of wrapper-GAL4/UAS-wrapper-RNAi (I) and wrapper-GAL4/UAS-slit-RNAi (K) show a reduction in the levels of Wrapper (I, quantified in L) and Slit (K) compared to their wild type counterparts (H and J, respectively). Scale bars: C, F= 5μm; D, E= 0.2 μm; G= 2 μm. Anterior is to the left in A, B and to the top in H-K. a.u.= arbitrary units
To further determine the efficacy of the wrapper-GAL4, we next wanted to test whether wrapper-GAL4 could be effective in knocking down known genes that are MG-specific, such as wrapper and slit (Noordermeer et al., 1998; Kidd et al., 1999; Simpson et al., 2000). Immunohistochemical analyses of wrapper-GAL4/UAS-wrapperRNAi (Fig. 2I) and wrapper-GAL4/UAS-slitRNAi (Fig. 2K) when compared to their respective wild type counterparts (Fig. 2H and 2J, respectively), showed a significant downregulation of the endogenous Wrapper (compare Fig. 2I with 2H; also see Fig. 2L) and Slit (Fig. 2K with 2J) expression levels in the MG cells. It is also important to note that the fluorescence intensity of Wrapper did not show any significant differences between individual segments analyzed within embryos of the same genotype (such as comparing all black bars that represent wild type and all green bars representing the Wrapper knockdown, Fig. 2L) suggesting a uniform knockdown under wrapper-Gal4. Although both Wrapper and Slit showed significant knockdown, we did not come across embryos that exhibited a complete loss of either Wrapper or Slit as is observed in their respective null mutant embryos (data not shown). A more robust knockdown could be achieved by: (1) having more copies of the UAS-RNAi and/or the GAL4, (2) raising flies at a higher temperature and (3) introducing Dicer together with the RNAi. Together, these data sets revealed that wrapper-GAL4 could be used as a genetic tool to address specific aspects of MG cells using loss or gain of function strategies.
Wrapper-GAL4 Expression and Rescue of wrapper Mutant Phenotypes and Lethality
Previous studies have identified Nrx IV and Wrapper as key molecular players in mediating neuron-glial interactions and in the developmental organization of the Drosophila embryonic CNS midline (Wheeler et al., 2009; Stork et al., 2009; Slovakova and Carmena, 2011). wrapper mutant embryos displayed mislocalization of Nrx IV (Wheeler et al., 2009; Stork et al., 2009) in the CNS and significant lethality (Noordermeer et al., 1998). None of the previously published studies, however, reported a rescue of the aforementioned wrapper mutant phenotypes and/or lethality. With the generation of wrapper-GAL4, we were in a position to address some of these gaps remaining from the previous studies by performing functional rescue analysis of wrapper mutant phenotypes.
We first carried out immunofluorescence analysis of Nrx IV, Wrapper and BP102 in stage 16 embryos of wild type (Fig. 3A), wrapper-/- (Fig. 3B), nrx IV-/- (Fig. 3C) and wrapper-/-; wrapper-GAL4/UAS-wrapper (Fig. 3D). Consistent with previous studies, wild type embryos showed a strong asymmetric localization of Nrx IV (arrowheads, Fig. 3A, A‴) in midline neurons that juxtapose Wrapper in MG (Fig. 3A; Banerjee et al., 2010), and low levels of Nrx IV in lateral CNS neurons. nrx IV mutants showed aberrant Wrapper localization (Fig. 3B′, B‴) resulting from malformed MG, and a disrupted axonal scaffold (Fig. 3B″, B‴). wrapper mutants, on the other hand, showed a striking mislocalization of Nrx IV (arrowheads, Fig. 3C, C‴) in the form of a more uniform distribution in neuronal cell bodies and an elevated expression throughout the lateral CNS neurons and axons. wrapper-/-; wrapper-GAL4/UAS-wrapper embryos displayed a complete rescue of Nrx IV localization comparable to wild type embryos (compare Fig. 3D with 3A) and normal distribution of Wrapper in the MG (Fig. 3D′). These results highlight the importance of wrapper-GAL4 in the phenotypic rescue of wrapper mutants.
Figure 3. wrapper-GAL4 and rescue of wrapper-/- phenotypes.
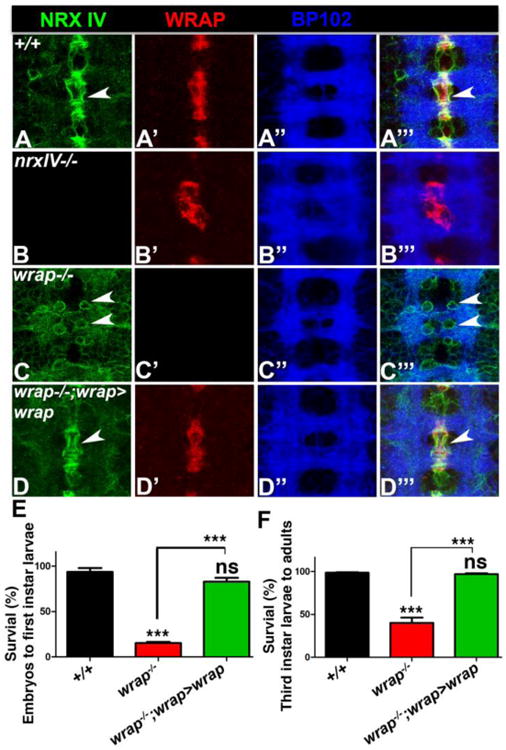
(A-D) Localization of Nrx IV (green), Wrapper (red) and BP102 (blue) in stage 16 embryos of wild type (A), nrx IV-/- (B), wrapper-/- (C) and wrapper-/-;wrapper-GAL4/UAS-wrapper (D). Anterior is to the top in A-D. (E, F) Survival assay of embryonic to 1st instar larvae (E) and 3rd instar larvae to adults (F) in wild type, wrapper-/- and wrapper-/-;wrapper-GAL4/UAS-wrapper.
We next wanted to test if wrapper-GAL4 was capable of rescuing the lethality of wrapper loss-of-function. We conducted a survival assay on wild type, wrapper-/- and wrapper-/-; wrapper-GAL4/UAS-wrapper (Fig. 3E, F) to check for survival of embryos into first instar larvae as well as survival of third instar larvae to adulthood. Our findings suggest that a vast majority of wrapper-/- embryos displayed a failure to hatch into first instar larvae which gets fully rescued in wrapper-/-;wrapper-GAL4/UAS-wrapper embryos (Fig. 3E). Interestingly, wrapper-/- that made it to the third instar larval stage had a higher chance of survival to adulthood (Fig. 3F) than survival of wrapper-/-embryos to first instar larvae (Fig. 3E). While there was still a significantly higher lethality of wrapper-/- larvae to emerge as adults, the larvae from wrapper-/-;wrapper-GAL4/UAS-wrapper were completely rescued to wild type levels (Fig. 3F). These data suggest that wrapper-GAL4 could be used for functional rescue of wrapper mutant phenotypes and potentially that of other MG mutants.
Contactin, a GPI-anchored Ig-superfamily protein, can Substitute for Wrapper in Midline Glial Cells
Having tested the usefulness of wrapper-GAL4 in the rescue of wrapper mutant lethality and aberrant Nrx IV midline localization, we wanted to test whether Drosophila Contactin will be able to rescue wrapper mutants based on the similarities in their domain structure and the fact that there is no direct vertebrate homolog of wrapper. Drosophila contactin and neurexin IV have vertebrate/human homologs Contactin and Caspr1, respectively, and are binding partners that form a major part of the axo-glial junctional machinery (Bhat, 2003; Banerjee et al., 2006b). We reasoned that in the absence of a vertebrate wrapper homolog, proteins that have similar domain structure as Wrapper and that bind to Nrx IV could potentially substitute for Wrapper function in its absence, and thus making Wrapper relevant to our understanding of axon-glia biology in an invertebrate system.
As shown in Fig. 4, Contactin (Cont) shares domain similarity with Wrapper (Fig. 4A) and binds to Nrx IV (Banerjee et al., 2006), and might potentially be able to substitute for Wrapper when expressed in MG cells of wrapper mutants. We immunostained wild type (Fig. 4B), wrapper-/- (Fig. 4C) and wrapper-/-;wrapper-GAL4/UAS-Cont (Fig. 4D, E) embryos with anti-Wrapper (blue), anti-Nrx IV (green) and anti-Cont (red). As expected, in wild type embryos Wrapper localized to MG cells (Fig. 4B) and Nrx IV to midline neurons (Fig. 4B′), while Cont (red arrowhead, Fig. 4B″, B‴) localized to a specific subset of glia in the CNS, called the channel glia (Ito et al., 1995; Giangrande 1996; Hartenstein 2011; Freeman 2015). Interestingly, in wrapper-/- apart from Nrx IV misdistribution (Fig. 4C′, C‴), Cont localization in channel glia seemed unaffected (red arrowhead, Fig. 4C″, C‴) suggesting that Cont does not play a role together with the Wrapper/Nrx IV protein complex in the midline. In wrapper-/-;wrapper-GAL4/UAS-Cont rescue embryos (Fig. 4D-D‴), in the absence of Wrapper, Cont expression was detected in wrapper-GAL4 domain in MG (Fig. 4D″, D‴), together with a remarkable rescue of Nrx IV localization characterized by its typical asymmetry in the midline neurons now juxtaposing the Cont expressing MG cells. The red arrowhead (Fig. 4D″, D‴) points to the presumptive location of the channel glia. A higher magnification of a lateral view of MG clearly revealed the asymmetric pattern of Nrx IV localization (Fig. 4E′, E‴) and Cont in MG (white arrowhead, Fig. 4E″, E‴) and its endogenous channel glia localization (red arrowhead, Fig. 4E″, E‴). These results validate the efficiency of wrapper-GAL4 as a powerful tool for genetic manipulations in the CNS MG cells of Drosophila.
Figure 4. Contactin can substitute for Wrapper Function in MG to restore Nrx IV localization.
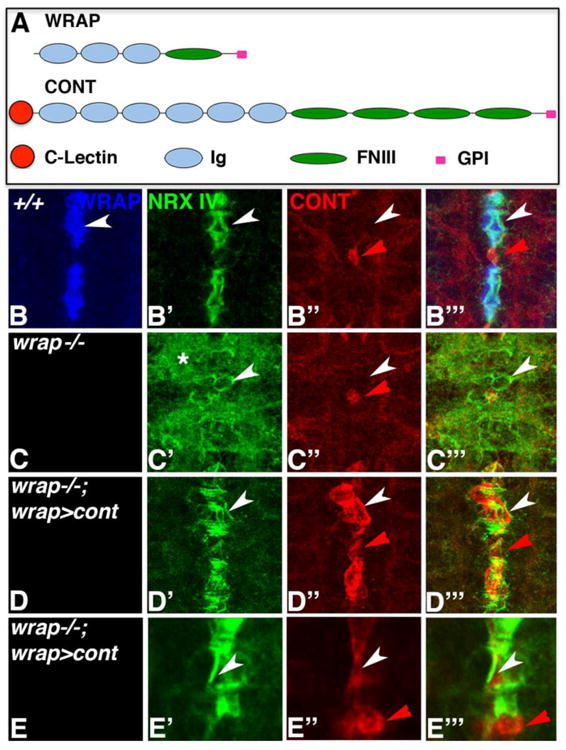
(A) Comparison of domain structure of Drosophila Wrapper (WRAP) and Contactin (CONT). (B-E) Localization of Wrapper (blue), Nrx IV (green) and Cont (red) in wild type (B), wrapper-/- (C) and wrapper-/-;wrapper-GAL4/UAS-cont (D, E). Note the restoration of Nrx IV asymmetric localization (white arrowheads, D′, E′) upon Cont expression in MG (white arrowheads, D″ and E″). Higher magnification confocal image of MG in lateral view revealed localization of Cont in MG (white arrowhead, E″, E‴) and endogenous localization of Cont in channel glia (red arrowhead, E″, E‴). Anterior is to the top in B-E.
Conclusion
In this study we provide insights into the structural and functional aspects of Drosophila MG cells with the help of a newly engineered transgenic wrapper-GAL4 fly line. Using the wrapper-GAL4 driver line, we highlight a variety of genetic manipulations that could be done to understand MG biology in Drosophila. We also provide data that reiterate the importance of two essential genes, Nrx IV and Wrapper in neuron-glial interactions and ensheathment of commissural axons at the CNS midline. Future studies using the wrapper-GAL4 driver will be highly instrumental to explore MG cell structure and function in vivo, and in carrying out genetic screens to identify new genes that might improve our understanding of the process of axonal ensheathment and vertebrate myelination during development, health, and disease.
Supplementary Material
Highlights.
Generation of midline glia-specific wrapper-GAL4
Useful in cell biological analysis of midline glia through out development
Useful in gene expression profiling of midline glia
Useful in genetic and functional manipulation of midline glia
Acknowledgments
We thank the BAC Engineering Core at the University of North Carolina at Chapel Hill for generating the wrapper-GAL4 construct, GenetiVision (Houston) for wrapper-GAL4 transgenic flies, UT Health EM Core Facilities for electron microscopy and members of the Bhat laboratory for helpful discussions. This work was supported by grants from the National Institutes of Health (NS050356), the Zachry Foundation and the University of Texas Health at San Antonio.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Awasaki T, Lee T. New tools for the analysis of glial cell biology in Drosophila. Glia. 2011;59(9):1377–1386. doi: 10.1002/glia.21133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Blauth K, Rogers SL, Bhat MA. Drosophila Neurexin IV is Required for Repulsive Midline Axon Guidance and Functions in Concert with Roundabout and Slit. Journal of Neuroscience. 2010;(30):5653–5667. doi: 10.1523/JNEUROSCI.6187-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Pillai AM, Paik R, Li J, Bhat MA. Axonal Ensheathment and Septate Junction Formation in the Peripheral Nervous System of Drosophila. Journal of Neuroscience. 2006a;26(12):3319–3329. doi: 10.1523/JNEUROSCI.5383-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Sousa AD, Bhat MA. Organization and function of septate junctions: an evolutionary perspective. Cell Biochem Biophys. 2006b;46(1):65–77. doi: 10.1385/CBB:46:1:65. [DOI] [PubMed] [Google Scholar]
- Baumgartner S, Littleton JT, Broadie K, Bhat MA, Harbecke R, Lengyel JA, Chiquet-Ehrismann R, Prokop A, Bellen HJ. A Drosophila neurexin is required for septate junction and blood-nerve barrier formation and function. Cell. 1996;87(6):1059–1068. doi: 10.1016/s0092-8674(00)81800-0. [DOI] [PubMed] [Google Scholar]
- Bhat MA. Molecular organization of axo-glial junctions. Current Opinion in Neurobiology. 2003;13(5):552–559. doi: 10.1016/j.conb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Buttermore ED, Thaxton C, Bhat MA. Organization and Maintenance of Molecular Domains in Myelinated Axons. J Neurosci Res. 2013;91(5):603–622. doi: 10.1002/jnr.23197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira-Rosario A, Scoggin S, Shalaby NA, Williams ND, Hiesinger PR, Buszczak M. Recombineering homologous recombination constructs in Drosophila. J Vis Exp. 2013;(77):e50346. doi: 10.3791/50346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles P, Tait S, Faivre-Sarrailh C, Barbin G, Gunn-Moore F, Denisenko-Nehrbass N, Guennoc AM, Girault JA, Brophy PJ, Lubetzki C. Neurofascin Is a Glial Receptor for the Paranodin/Caspr-Contactin Axonal Complex at the Axoglial Junction. Current Biology. 2002;12(3):217–220. doi: 10.1016/s0960-9822(01)00680-7. [DOI] [PubMed] [Google Scholar]
- Crews ST. Axon-glial interactions at the Drosophila CNS midline. Cell Adh Migr. 2010;4(1):67–71. doi: 10.4161/cam.4.1.10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson BJ, Zou Y. Navigating intermediate targets: the nervous system midline. Cold Spring Harb Perspect Biol. 2010;2(8):a002055. doi: 10.1101/cshperspect.a002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenfeld G, Stork T, Klambt C. Neuron-glia interaction in the insect nervous system. Curr Opin Neurobiol. 2005;15(1):34–39. doi: 10.1016/j.conb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Ejsmont RK, Ahlfeld P, Pozniakovsky A, Stewart AF, Tomancak P, Sarov M. Recombination-mediated genetic engineering of large genomic DNA transgenes. Methods Mol Biol. 2011;772:445–458. doi: 10.1007/978-1-61779-228-1_26. [DOI] [PubMed] [Google Scholar]
- Faivre-Sarrailh C, Banerjee S, Li J, Hortsch M, Laval M, Bhat MA. Drosophila contactin, a homolog of vertebrate contactin, is required for septate junction organization and paracellular barrier function. Development. 2004;131(20):4931–4942. doi: 10.1242/dev.01372. [DOI] [PubMed] [Google Scholar]
- Freeman MR. Drosophila central nervous system glia. Cold Spring Harb Perspect Biol. 2015;7(11) doi: 10.1101/cshperspect.a020552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulkerson E, Estes PA. Common motifs shared by conserved enhancers of Drosophila midline glial genes. J Exp Zool B Mol Dev Evol. 2011;316(1):61–75. doi: 10.1002/jez.b.21382. [DOI] [PubMed] [Google Scholar]
- Giangrande A. Development and organization of glial cells in Drosophila melanogaster. Int J Dev Biol. 1996;40(5):917–927. [PubMed] [Google Scholar]
- Hartenstein V. Structure and development of glia in Drosophila. Glia. 2011;59(9):1237–1252. doi: 10.1002/glia.21162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert AL, Monk KR. Advances in myelinating glial cell development. Curr Opin Neurobiol. 2016;42:53–60. doi: 10.1016/j.conb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo A. Neuron-glia interactions during axon guidance in Drosophila. Biochem Soc Trans. 2003;31:50–55. doi: 10.1042/bst0310050. [DOI] [PubMed] [Google Scholar]
- Jacobs JR. The midline glia of Drosophila: a molecular genetic model for the developmental functions of glia. Prog Neurobiol. 2000;62(5):475–508. doi: 10.1016/s0301-0082(00)00016-2. [DOI] [PubMed] [Google Scholar]
- Ito K, Urban J, Technau GM. Distribution, classification and development of Drosophila glial cells in the late embryonic and early larval ventral nerve cord. Roux's Arch Dev Biol. 1995;204:284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila. Cell. 1999;96(6):785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Lane S, McDermott K, Dockery P, Fraher J. The developing cervical spinal ventral commissure of the rat: a highly controlled axon-glial system. J Neurocytol. 2004;33(5):489–501. doi: 10.1007/s11068-004-0512-x. [DOI] [PubMed] [Google Scholar]
- Nave KA. Myelination and the trophic support of long axons. Nat Rev Neurosci. 2010;11(4):275–283. doi: 10.1038/nrn2797. [DOI] [PubMed] [Google Scholar]
- Neuhaus-Follini A, Bashaw GJ. Crossing the embryonic midline: molecular mechanisms regulating axon responsiveness at an intermediate target. Wiley Interdiscip Rev Dev Biol. 2015;4(4):377–389. doi: 10.1002/wdev.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer JN, Kopczynski CC, Fetter RD, Bland KS, Chen WY, Goodman CS. Wrapper, a Novel Member of the Ig Superfamily, Is Expressed by Midline Glia and Is Required for Them to Ensheath Commissural Axons in Drosophila. Neuron. 1998;21(5):991–1001. doi: 10.1016/s0896-6273(00)80618-2. [DOI] [PubMed] [Google Scholar]
- O'Donnell M, Chance RK, Bashaw GJ. Axon growth and guidance: receptor regulation and signal transduction. Annu Rev Neurosci. 2009;32:383–412. doi: 10.1146/annurev.neuro.051508.135614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlou O, Theodorakis K, Falk J, Kutsche M, Schachner M, Faivre-Sarrailh C, Karagogeos D. Analysis of interactions of the adhesion molecule TAG-1 and its domains with other immunoglobulin superfamily members. Mol Cell Neurosci. 2002;20(3):367–381. doi: 10.1006/mcne.2002.1105. [DOI] [PubMed] [Google Scholar]
- Scholz H, Sadlowski E, Klaes A, Klambt C. Control of midline glia development in the embryonic Drosophila CNS. Mech Dev. 1997;64:137–151. doi: 10.1016/s0925-4773(97)00078-6. [DOI] [PubMed] [Google Scholar]
- Simpson JH, Bland KS, Fetter RD, Goodman CS. Short-range and long-range guidance by Slit and its Robo receptors: a combinatorial code of Robo receptors controls lateral position. Cell. 2000;103(7):1019–1032. doi: 10.1016/s0092-8674(00)00206-3. [DOI] [PubMed] [Google Scholar]
- Slováková J, Carmena A. Canoe functions at the CNS midline glia in a complex with Shotgun and Wrapper-Nrx-IV during neuron-glia interactions. Development. 2011;138(8):1563–1571. doi: 10.1242/dev.056192. [DOI] [PubMed] [Google Scholar]
- Sonnenfeld MJ, Jacobs JR. Apoptosis of the midline glia during Drosophila embryogenesis: a correlation with axon contact. Development. 1995;121:569–578. doi: 10.1242/dev.121.2.569. [DOI] [PubMed] [Google Scholar]
- Stollewerk A, Klambt C, Cantera R. Electron microscopic analysis of Drosophila midline glia during embryogenesis and larval development using beta-galactosidase expression as endogenous cell marker. Microsc Res Tech. 1996;35(3):294–306. doi: 10.1002/(SICI)1097-0029(19961015)35:3<294::AID-JEMT8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Stork T, Thomas S, Rodrigues F, Silies M, Naffin E, Wenderdel S, Klambt C. Drosophila Neurexin IV stabilizes neuron-glia interactions at the CNS midline by binding to Wrapper. Development. 2009;136(8):1251–61. doi: 10.1242/dev.032847. [DOI] [PubMed] [Google Scholar]
- Thaxton C, Bhat M. Myelination and Regional Domain Differentiation of the Axon. In: Koenig E, editor. Cell Biology of the Axon. Springer; Berlin Heidelberg: 2009. pp. 65–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaxton C, Pillai AM, Pribisko AL, Dupree JL, Bhat MA. Nodes of Ranvier Act as Barriers to Restrict Invasion of Flanking Paranodal Domains in Myelinated Axons. Neuron. 2011;69(2):244–257. doi: 10.1016/j.neuron.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314(5806):1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, Bellen HJ, Hoskins RA. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6(6):431–4. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler SR, Banerjee S, Blauth K, Rogers SL, Bhat MA, Crews ST. Neurexin IV and Wrapper interactions mediate Drosophila midline glial migration and axonal ensheathment. Development. 2009;136(7):1147–1157. doi: 10.1242/dev.030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DW, Tyrer M, Shepherd D. Tau and tau reporters disrupt central projections of sensory neurons in Drosophila. J Comp Neurol. 2000;428:630–640. doi: 10.1002/1096-9861(20001225)428:4<630::aid-cne4>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Xiao H, Hrdlicka LA, Nambu JR. Alternate functions of the single-minded and rhomboid genes in development of the Drosophila ventral neuroectoderm. Mech Dev. 1996;58:65–74. doi: 10.1016/s0925-4773(96)00559-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


