Abstract
Many key regulatory proteins, including members of the Ras family of GTPases, are modified at their C terminus by a process termed prenylation. This processing is initiated by the addition of an isoprenoid lipid, and the proteins are further modified by a proteolytic event and methylation of the C-terminal prenylcysteine. Although the biological consequences of prenylation have been characterized extensively, the contributions of prenylcysteine methylation to the functions of the modified proteins are not well understood. This reaction is catalyzed by the enzyme isoprenylcysteine carboxyl methyltransferase (Icmt). Recent genetic disruption studies have provided strong evidence that blocking Icmt activity has profound consequences on oncogenic transformation. Here, we report the identification of a selective small-molecule inhibitor of Icmt, 2-[5-(3-methylphenyl)-1-octyl-1H-indol-3-yl]acetamide (cysmethynil). Cysmethynil treatment results in inhibition of cell growth in an Icmt-dependent fashion, demonstrating mechanism-based activity of the compound. Treatment of cancer cells with cysmethynil results in mislocalization of Ras and impaired epidermal growth factor signaling. In a human colon cancer cell line, cysmethynil treatment blocks anchorage-independent growth, and this effect is reversed by overexpression of Icmt. These findings provide a compelling rationale for development of Icmt inhibitors as another approach to anticancer drug development.
Keywords: cell transformation, protein isoprenylation, protein methylation, Ras signaling
A C-terminal CaaX motif, where C is cysteine, the a's are aliphatic amino acids, and X can be any of a number of amino acids, targets a variety of eukaryotic proteins to a series of posttranslational modifications important for their localization and function (1, 2). This processing is initiated by the covalent attachment of a 15-carbon farnesyl or a 20-carbon geranylgeranyl lipid to the cysteine of the CaaX motif, a reaction catalyzed by protein farnesyltransferase (FTase) or protein geranylgeranyltransferase type I (3). After prenylation, the C-terminal three amino acids (i.e., the -aaX) are removed by a specific CaaX protease termed Rce1 (4, 5) and the now C-terminal prenylcysteine is methylated by isoprenylcysteine carboxyl methyltransferase (Icmt; refs. 6–8). As polytopic membrane proteins localized to the endoplasmic reticulum, both Rce1 and Icmt are unusual in their respective classes (9).
Proteins that terminate in a –CaaX motif regulate a number of pathways important in oncogenesis. The best studied example is the central role of the Ras family of proteins in growth factor activation of the MAP kinase signaling cascade (10, 11). Constitutive activation of this pathway is transforming in a wide variety of cell types, and activating mutations in Ras have been found in almost 30% of all cancers, including 50% of colon cancers and up to 90% of pancreatic cancers (12). In addition, many cancers contain alterations upstream of Ras, and the resultant hyperactivation of Ras is thought to contribute to tumorigenesis in these cancers as well (13, 14). Many other CaaX proteins have also been implicated in oncogenesis and tumor progression, and these proteins most likely require processing via the prenylation pathway for function (2, 15).
Both the membrane targeting and the transforming abilities of Ras require processing through the prenylation pathway (16, 17). For this reason, the protein prenyltransferases, most notably FTase, have been targets of major drug discovery programs for the last decade (18, 19). Presently, several FTase inhibitors are being evaluated in clinical trials (15, 19). These experimental agents have shown significant activity in a number of clinical trials, but the overall response rates in patients have been less than initially hoped. One possible explanation for this lack of efficacy is the process of alternate prenylation that allows some FTase substrates to be modified by geranylgeranyltransferase type I when FTase activity is limiting (20–22). Recent studies using genetic disruption of Icmt have demonstrated that Ras proteins are significantly mislocalized and tumorigenesis is markedly impaired in cells that lack Icmt (23, 24). After this discovery, CaaX protein methylation has gained attention as a target in oncogenesis (25).
With emerging evidence for the importance of Icmt-catalyzed CaaX protein methylation in oncogenesis, there is a clear need for specific pharmacological agents to target this process. However, the only such agents available to date have been analogs of the substrate prenylcysteine or the product S-adenosylhomocysteine; all of these analogs have been reported to have pleiotropic effects (26–28). Here, we report the discovery of an indole-based small-molecule inhibitor of Icmt. Treatment of cancer cells with this compound that we have named 2-[5-(3-methylphenyl)-1-octyl-1H-indol-3-yl]acetamide (cysmethynil), results in a decrease in Ras carboxylmethylation, mislocalization of Ras, and impaired signaling through Ras pathways. Cysmethynil treatment blocks anchorage-independent growth in a human colon cancer cell line, and this effect is reversed by overexpression of Icmt. These findings, together with the findings from genetic disruption of this enzyme, suggest that Icmt inhibitors may have significant therapeutic potential.
Materials and Methods
Materials. Farnesyl pyrophosphate was from Biomol, the chemical library was from PPD Discovery (Research Triangle Park, NC), streptavidin-Sepharose beads were from Amersham Pharmacia, puromycin and S-adenosylmethionine (AdoMet) were from Sigma, and S-(5′-adenosyl)-l-homocysteine was from Fluka. Epidermal growth factor was from EMD Bioscience. [methyl-3H]methionine and [methyl-3H]AdoMet were from PerkinElmer. CellTiter 96 Aqueous One solution cell proliferation assay was from Promega. pEGFP and pLPCX were from Clontech. Sf9 membranes containing recombinant Rce1 and Icmt, termed Rce1 membranes and Icmt membranes, respectively, were made in our laboratory as described (5). Farnesylated K-Ras was made by in vitro modification of bacterially expressed K-Ras with purified FTase as described (5). Biotin-S-farnesyl-l-cysteine (BFC) was synthesized as detailed elsewhere (R.A.B. and P.J.C., unpublished work). Mouse embryonic fibroblast cell lines were established and maintained as described (29). Madin–Darby canine kidney (MDCK) cells stably expressing GFP-H-Ras, GFP-K-Ras and GFP-N-Ras were a generous gift of M. Philips (New York University Medical School, New York).
Icmt Assay: Screening. The small-molecule screen was performed in 96-well multiscreen filtration plates (Millipore) by using a Beckman Biomek FX robot. Briefly, Sf9 membranes containing Rce1 and Icmt were suspended in 100 mM Hepes, pH 7.4/5 mM MgCl2 (Buffer A) such that 40 μl of suspension had 1 μg of Rce1 membrane protein and 0.2 μg of Icmt membrane protein; protein values are of total membrane protein. This membrane suspension was dispensed into wells, to which was added the library compounds in DMSO (1.3 μl) to yield a final concentration of 15–30 μM. After 15 min at room temperature, 10 μl of buffer A containing 13 μM [3H]AdoMet (5 Ci/mmol; 1 Ci = 37 GBq) and 5 μM farnesylated K-Ras were added. After another 30 min, the reaction was quenched with 100 μl of 6% SDS/45% trichloroacetic acid (TCA). Precipitated proteins were recovered on the membranes of the plate wells by vacuum filtration of the entire plate, and the wells were washed three times with 200 μl of 6% TCA. Scintillation fluid [Microscint 20 (Packard), 50 μl per plate per well] was added, the plates were sealed as per manufacturer's instructions, and radioactivity in each well was determined by using a Packard TopCount NXT microplate scintillation counter.
Icmt Assay: Secondary Analysis. Secondary assays and kinetic analyses were by adding recombinant Icmt (0.5 μg of Sf9 membrane protein) to an assay mixture containing 4 μM BFC, 5 μM [3H]AdoMet (1.2 Ci/mmol), and either inhibitor or DMSO in a total volume of 45 μl of Buffer A. Reactions were incubated for 20 min at 37°C, terminated with 5 μl of 10% Tween 20, and then 10 μl of streptavidin beads in 500 μl of 20 mM NaH2PO4, pH 7.4/150 mM NaCl (Buffer B) were added. The interaction between biotin and streptavidin was allowed to proceed overnight at 4°C under gentle agitation, whereupon the beads were harvested by centrifugation, washed three times with 0.5 ml of buffer B, and resuspended in 100 μl of the same buffer for radioactivity determination. In the experiments to measure potential time-dependent inhibition of Icmt, the same assay was used except that the Icmt membrane suspension was first mixed with inhibitor in buffer A; this solution was incubated for 30 min at 37°C, whereupon the remaining components of the reaction mixture were added and the reactions were incubated an additional 20 min at 37°C before product isolation and radioactivity determination.
Cell Growth Determination. Cell growth assays were performed in a 96-well plate format. Briefly, ≈1,000 cells were plated into each well of the plate. After 24 h, the media were replaced with media containing either compound or vehicle. Media and drug were replaced every 24 h. Cell determinations were made at the times indicated by adding 19 μl of CellTiter 96 Aqueous One solution to each well followed by incubating the plates in the dark at 37°C for 2 h, after which the absorbance at 490 nm was read. Background absorbance from blank wells containing only media with compound or vehicle were subtracted from each test well.
Localization of GFP Proteins in MDCK Cells. MDCK cells stably expressing GFP-H-Ras, GFP-K-Ras, or GFP-N-Ras were grown on 35-mm coverslips treated with poly-d-lysine. MDCK cells expressing Yes-GFP were prepared by transient transfection of the Yes-GFP construct (30), using Superfect reagent (Qiagen, Valencia, CA) following the manufacturer's instructions. Cells were grown in media containing 10% FBS to ≈25% confluence and then treated with 1% DMSO or cysmethynil at the indicated concentrations. Cells were imaged 72 h after drug treatment (24 h for transiently transfected cells) on an Olympus inverted microscope with an UltraView spinning-disk confocal (PerkinElmer LAS) and a krypton/argon laser with a 488-nm line, attached to a cooled charge-coupled device camera (Hamamatsu). Initial image acquisition and manipulation was performed with metamorph software (Universal Imaging, Downingtown, PA).
Phosphoprotein Analysis. Cells were grown for three days in media containing 1% FBS with cysmethynil or vehicle as indicated. Half of the wells were then treated with EGF (10 ng/ml) and half with vehicle for 10 min, whereupon cells were rinsed with PBS and harvested. Total cell lysates (30 μg protein) were resolved on 4–20% Tris-glycine gels (Invitrogen). Proteins were transferred to nitrocellulose and probed with a mix of anti-phospho-Akt and anti-phospho-p42/44 mitogen-activated protein kinase (MAPK) antibodies, or with anti-tubulin or anti-phospho NFκB antibody (Cell Signaling Technology, Beverly, MA) as indicated. Visualization was performed with alkaline phosphatase (Promega) as per the manufacturer's instructions.
Generation of Stable Cell Lines Expressing GFP or GFP-Icmt. Full-length human ICMT was cloned into pEGFP after restriction endonuclease digestion with BamHI and XhoI. Retroviral constructs were generated by cloning EGFP or GFP-ICMT into the pLPCX retroviral vector. These constructs, along with the helper plasmid 467, were transfected into human embryonic kidney 293 cells. Virus was harvested 48 h after transfection and used to infect DKOB8 cells. Cells were treated with virus for 24 h, allowed to recover for another 24 h, selected in 0.5 μg/ml puromycin for ≈3 weeks, and then sorted for expression of GFP on a Becton Dickinson FACSVantageSE cell sorter. GFP-positive cells selected in this manner were cultured as per normal DKOB8 cells (31).
Soft Agar Growth Assay. Soft agar culture media was prepared with 10% FBS in 1× minimum essential medium α. Bottom agar (0.5 ml 2.4% noble agar in 1.5 ml of soft agar culture media) was plated in each cell culture dish. Cells were harvested by trypsinization at ≈80% confluence, mixed into top agar (10,000 cells per plate, 0.3% noble agar in soft agar culture media) and poured onto prepared plates. Cysmethynil or DMSO was included at the indicated concentrations in both the top and bottom agar layers. For each condition, triplicate samples were prepared. Plates were fed twice a week with 300 μl of 1× MEM containing 10% FBS and the appropriate concentrations of cysmethynil or DMSO. After 3 weeks, plates were stained by addition of 300 μl of 10 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (Sigma-Aldrich) in PBS followed by incubation at 37°C in an incubator with 5% CO2 for 3 h. Plates were then treated with 0.4 M HCl in 300 μl of isopropyl alcohol and incubated overnight before imaging.
Results
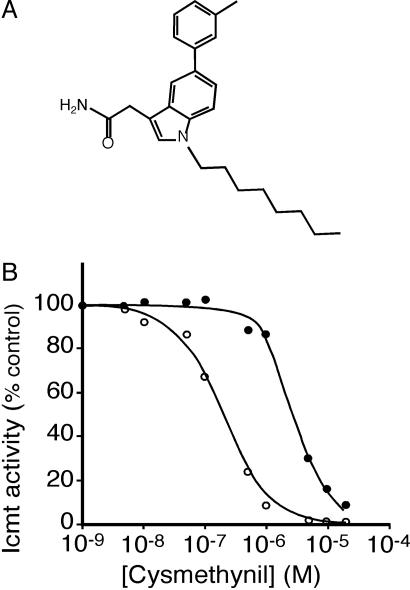
Identification of an Indole-Based Selective Inhibitor of Icmt. To identify small-molecule inhibitors of Icmt, we screened a diverse chemical library of ≈10,000 compounds. The library contained 70+ subfamilies derived from unique scaffolds. We used an in vitro screen in which Icmt activity was measured as the incorporation of a [3H]methyl group into a farnesylated, Rce1-proteolyzed, K-Ras substrate (see Materials and Methods). Compounds that showed >50% inhibition at 50 μM were subjected to a secondary screen by using a small-molecule substrate of Icmt, BFC. From this screen, we identified a group of compounds with an indole core structure that had significant activity against Icmt. The most potent of these compounds was what we term cysmethynil (Fig. 1A). This compound was independently synthesized and characterized to confirm identity and purity (see Supporting Text and Scheme 1, which are published as supporting information on the PNAS web site), and all studies described below were performed by using the independently synthesized compound.
Fig. 1.
Cysmethynil, a small-molecule inhibitor of Icmt. (A) Structure of the indole-based compound, cysmethynil. (B) Time-dependent inhibition of Icmt by cysmethynil. Icmt activity was measured as the incorporation of [3H] from [3H]AdoMet into the Icmt substrate BFC. The assay was performed either with (○) or without (•) preincubating Icmt with cysmethynil as described in Materials and Methods.
In the initial in vitro assay using BFC as the prenylcysteine substrate, the IC50 for Icmt inhibition by cysmethynil was determined to be 2.4 μM (Fig. 1B). In this assay, the substrates and the inhibitor were premixed, and the reaction was initiated by the addition of enzyme. However, when the enzyme was premixed with inhibitor and AdoMet for 15 min before initiation of the reaction with BFC, a dramatic increase in inhibitor potency was observed with a measured IC50 of <200 nM (Fig. 1B). These data suggest that cysmethynil is a time-dependent inhibitor of Icmt. Importantly, even at concentrations up to 50 μM, cysmethynil did not inhibit the other enzymes in the prenylation pathway (FTase, geranylgeranyltransferase type I, and Rce1), nor did it inhibit an AdoMet-dependent DNA methyltransferase or an unrelated protein methyltransferase (the SssI DNA methyltransferase and PCMT1 protein methyltransferase, respectively) (data not shown).
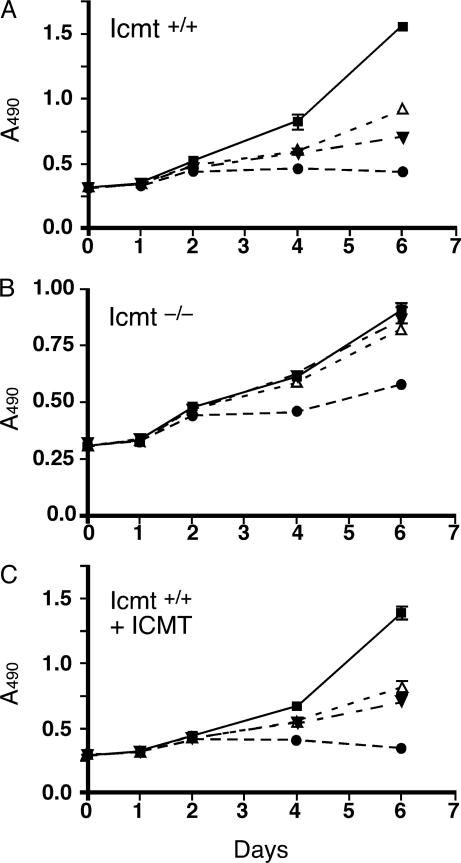
Cysmethynil Treatment Impacts Cell Growth in an Icmt-Dependent Fashion. To evaluate the potential cellular activity of cysmethynil, we took advantage of a cell model of Icmt deficiency developed from the gene-disruption studies (32). Reasoning that cells that had adapted to grow in the absence of Icmt activity should be resistant to the effects of the inhibitor, we treated Icmt–/– mouse embryonic fibroblasts and matched wild-type cells with increasing concentrations of the compound and monitored cell growth for 6 days (Fig. 2). Treatment with cysmethynil resulted in a dose-dependent inhibition of growth wild-type cells (Fig. 2 A), but Icmt–/– cells were largely unaffected (Fig. 2B). Furthermore, when the human ICMT gene was stably expressed in Icmt–/– cells, the reconstituted cell line regained sensitivity to cysmethynil (Fig. 2C). These results provide strong evidence for an antiproliferative activity of cysmethynil that is mechanism-based, i.e., directly due to an impact on Icmt activity.
Fig. 2.
Icmt-dependent growth inhibition of mouse embryonic fibroblasts by using cysmethynil. Mouse embryonic fibroblasts grown in media containing 8% serum were treated with DMSO (▪) or cysmethynil at concentrations of 15 (▵), 20 (▾), or 30 (•) μM. Media and drug were replaced daily, and cell growth was monitored for 6 days as described in Materials and Methods. Data represent the mean and SD of four replicate wells. (A) Icmt+/+ cells. (B) Icmt–/– cells. (C) Icmt–/– cells stably transfected with a gene expressing human ICMT (Icmt–/–/ICMT cells).
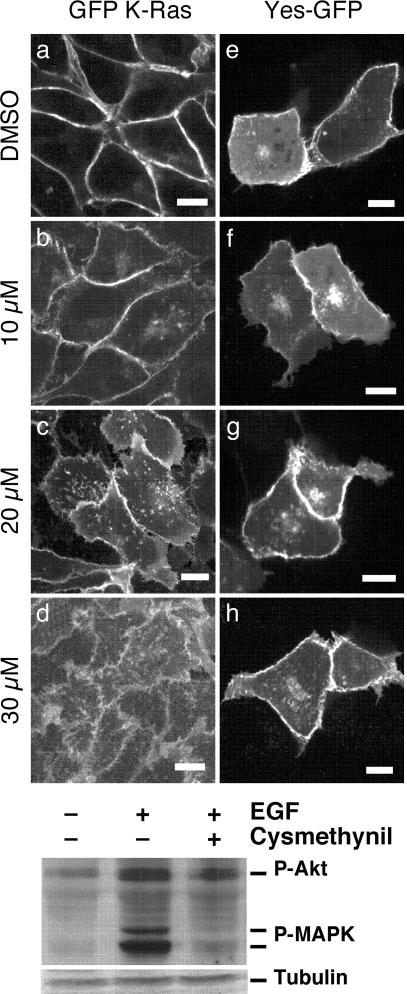
Cysmethynil Treatment of Cells Results in Mislocalization of Ras and Impairment of Growth Factor Signaling. Carboxylmethylation is important for proper plasma membrane localization of Ras (24). Based on this observation, we predicted that treatment of cells with an Icmt inhibitor would lead to a loss of Ras from the plasma membrane. To test this hypothesis, MDCK cells stably expressing GFP-tagged K-Ras were treated with increasing concentrations of cysmethynil for 72 h before imaging by confocal fluorescence microscopy. As shown in Fig. 3 a–d, cysmethynil treatment led to a dose-dependent mislocalization of GFP-K-Ras. Similar effects were noted for GFP-H-Ras and GFP-N-Ras in these cells and for all three Ras isoforms expressed in mouse embryonic fibroblasts (data not shown). As a control, MDCK cells expressing a fusion of GFP to the N terminus of Yes, a protein kinase localized to the plasma membrane by N-terminal myristoylation and palmitoylation (30), were treated under the same conditions. Cysmethynil treatment did not affect the plasma membrane localization of Yes-GFP (Fig. 3 e–h), indicating that the compound does not globally disrupt trafficking to the plasma membrane.
Fig. 3.
Impact of cysmethynil treatment on Ras localization and signaling. (Upper) Mislocalization of GFP Ras in cysmethynil-treated cells. MDCK cells expressing GFP-tagged K-Ras (a–d) or Yes-GFP (e–h) were treated with 1% DMSO (a and e) or 10 (b and f), 20 (c and g), or 30 (d and h) μM cysmethynil. Live cells were imaged on a confocal microscope as described in Materials and Methods.(Lower) Impact of cysmethynil treatment on EGF-stimulated protein phosphorylation. Wild-type mouse embryonic fibroblasts were grown for 3 days in media containing 1% serum in the presence of DMSO or 1 μm cysmethynil as described in Materials and Methods. Where indicated, cells were treated with EGF for the final 10 min before harvesting. Cell lysates containing equal amounts of protein were resolved on a 13% SDS-polyacrylamide gel and probed with antiphospho-Akt, antiphospho-p42/44 MAPK, or anti-β-tubulin antibody as indicated.
Growth factor signaling to MAPK involves CaaX proteins, again most notably Ras, and inhibition of CaaX protein methylation has been reported to impair EGF-mediated phosphorylation and activation of MAPK (29, 33). To evaluate the effect of cysmethynil treatment on EGF-mediated activation of MAPK and other signaling proteins, DKOB8 colon cancer cells were grown under low serum (1%) conditions in the presence of either vehicle or 1 μM cysmethynil. After 3 days of treatment, the cells were treated with either EGF or additional vehicle. Cell lysates were separated on SDS/PAGE and immunoblotted with a mixture of phospho-specific antibodies for Akt and p42/44 MAPK, or with an antibody for β-tubulin. As shown in Fig. 3 Lower, the level of activated Akt increased ≈3-fold, and that of activated p42/44 MAPK nearly 10-fold in EGF-treated cells. The EGF-induced increase in MAPK was almost completely blocked by cysmethynil treatment, whereas the increase in Akt phosphorylation was partially but not completely attenuated. This observation supports the hypothesis that cysmethynil treatment impacts on signaling through Ras-dependent pathways, because the activation of MAPK by EGF occurs primarily via the Ras pathway; corresponding activation of Akt involves both Ras-independent and Ras-dependent processes (13). Also, it is interesting to note that under low (1%) serum conditions, cysmethynil affects cellular processes at 5- to 10-fold lower concentrations than when the cells are grown in higher (8–10%) serum, suggesting that this compound, like many pharmacological agents, is buffered by serum.
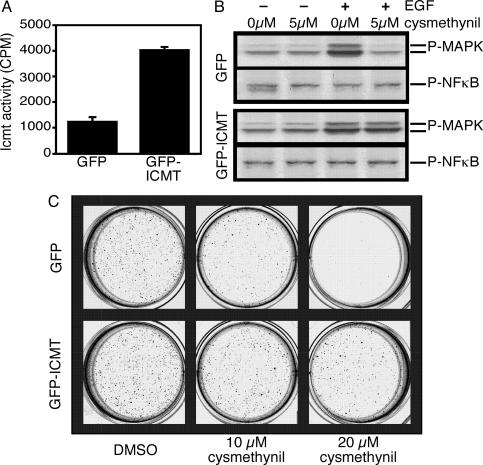
Impact of Cysmethynil Treatment on the Transformed Phenotype of Colon Cancer Cells. The data detailed above all point to a potential impact of Icmt inhibition on blocking the transformed phenotype of cancer cells. One of the classic methods to assess transformation of cells is by measuring their ability to grow in soft agar (34), and genetic disruption of Icmt in cells has been shown to block mouse anchorage-independent growth triggered by activated Ras (23). To directly assess the role of Icmt in any ability of cysmethynil to block the transformed phenotype of cancer cells, we first set out to engineer the DKOB8 colon cancer cells to stably overexpress Icmt; conferring resistance to a pharmacological agent by overexpression of the target is a classic means to confirm the mechanism of action of the agent. Through this approach, we were able to create a line of DKOB8 cells expressing GFP-Icmt in which the level of Icmt activity was elevated 4-fold compared with a parallel line in which GFP alone was expressed (Fig. 4A). Although modest, this level of Icmt overexpression was sufficient to protect the cells from cysmethynil blockade of EGF-stimulated MAPK activation (Fig. 4B), providing further confirmation that this effect of the inhibitor is because of its ability to impact on Icmt activity in the cells.
Fig. 4.
Overexpression of Icmt rescues EGF-stimulated MAPK activation and anchorage-independent growth in cysmethynil-treated cells. (A) Creation of cell lines stably overexpressing Icmt. DKOB8 cells were engineered to stably express either GFP alone or a GFP–Icmt fusion protein as described in Materials and Methods. Membrane fractions from GFP- and GFP-ICMT-expressing lines were assayed for Icmt activity by using the BFC assay. (B) Overexpression of Icmt restores EGF-stimulated MAPK activation in cysmethynil-treated cells. DKOB8 cells stably expressing GFP or GFP-ICMT were either left untreated or treated with 5 μM cysmethynil for 3 days in reduced-serum media as described in Materials and Methods. Where indicated, cells were treated with EGF for the final 10 min before harvesting. Cell lysates containing equal amounts of protein were resolved on a 13% SDS-polyacrylamide gel and probed with antiphospho-p42/44 MAPK, or as a control, antiphospho NFκB antibody as indicated. (C) Impact of cysmethynil on anchorage-independent growth of DKOB8 colon cancer cells. DKOB8 cells stably expressing GFP (first row) or GFP-ICMT (second row) were suspended in 0.3% noble agar and plated on a base of 0.6% noble agar; both the top and bottom layers contained either 1% DMSO or 10 or 20 μM cysmethynil as indicated. After 3 weeks of growth, colonies were stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide and imaged.
Having established that overexpression of Icmt conferred resistance to cysmethynil, we then evaluated both cell lines for the effect of cysmethynil on anchorage-independent growth. DKOB8/GFP and DKOB8/GFP-Icmt cells growing in soft agar were treated with either vehicle or increasing concentrations of cysmethynil. After 3 weeks, the plates were stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide to identify viable cells and imaged. As shown in Fig. 4C, treatment with cysmethynil significantly impaired the ability of the DKOB8/GFP cells to grow in soft agar, with a concentration of 20 μM dramatically reducing the ability of cells to form colonies. However, elevated Icmt activity was sufficient to rescue the ability of the cells to form colonies in soft agar in the presence of 20 μM cysmethynil (Fig. 4C, second row of wells). These results provide compelling evidence that cysmethynil blockade of Icmt is responsible for the ability of the compound to impact on both growth factor signaling pathways and on anchorage-independent growth of these cancer cells.
Discussion
The functional consequence of carboxylmethylation of CaaX proteins has been a matter of speculation since the prenylation pathway was first identified (6, 7). Icmt-catalyzed methylation is clearly essential for some biologies, as evidenced by the embryonic lethal phenotype when the gene encoding this enzyme is disrupted in mice (32). However, the contribution of carboxylmethylation to specific biological processes has been difficult to address. Although much work has been performed by using prenylcysteine analogs or agents that elevate S-(5′-adenosyl)-l-homocysteine to inhibit Icmt activity in cells (29, 35–38), the nonspecific nature of these compounds has made it difficult to attribute specific outcomes to an inhibition of Icmt (27, 28). The establishment of cell lines lacking Icmt has greatly helped the field (23, 39), but researchers are restricted to these few cell lines.
Even with these limitations, there are exciting hints about the involvement of Icmt in a number of biological systems. Increasing evidence suggests that Icmt-catalyzed methylation impacts signaling through Ras, and more importantly, that a lack of Icmt can slow or even stop cellular transformation (23, 29, 33). In addition, several studies have linked Icmt inhibition to significant effects on endothelial cells, including increased permeability and apoptosis (35, 36, 40). Inhibitors of Icmt might therefore have significant utility as anti-cancer agents. In fact, there is evidence that one existing anti-cancer drug, methotrexate, targets Icmt through an elevation of its product inhibitor S-(5′-adenosyl)-l-homocysteine (29).
Although much of the work on Icmt has centered on the consequences of carboxylmethylation of Ras proteins, some intriguing findings have been reported for other CaaX proteins processed by Icmt. Methylation of RhoA plays a major role in stability of the protein (23, 41), and the effects of Icmt inhibition on endothelial cells noted above have been suggested to be due to impact on carboxylmethylation of RhoA in these cells (35, 36). Outside the family of GTPases, methylation of lamin B clearly influences its interaction with the nuclear envelope (39). The identification of cysmethynil as an inhibitor of Icmt provides a selective pharmacological tool to probe the potential functional consequences of CaaX protein methylation in cellular systems and also the involvement of Icmt in biologies that are important in both normal and pathological cellular processes.
Supplementary Material
Acknowledgments
We thank James Otto (Duke University) for recombinant human Rce1 and Icmt. This work was supported by National Institutes of Health Grants GM46372 (to P.J.C.) and AR050200 and HL076839 (to S.G.Y.), a Howard Hughes Medical Institute Predoctoral Fellowship (to A.M.W.-V.), and a fellowship from l'Association Pour la Recherche Contre le Cancer (to R.A.B.). M.O.B. was supported by grants from the Swedish Cancer Society and the Swedish Research Council.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Icmt, isoprenylcysteine carboxyl methyltransferase; cysmethynil, 2-[5-(3-methylphenyl)-1-octyl-1H-indol-3-yl]acetamide; Rce1, CaaX protease; AdoMet, S-adenosylmethionine; BFC, biotin-S-farnesyl-l-cysteine; MAPK, mitogen-activated protein kinase; MDCK, Madin–Darby canine kidney; FTase, farnesyltransferase.
References
- 1.Zhang, F. L. & Casey, P. J. (1996) Annu. Rev. Biochem. 65, 241–269. [DOI] [PubMed] [Google Scholar]
- 2.Kloog, Y. & Cox, A. D. (2004) Semin. Cancer Biol. 14, 253–261. [DOI] [PubMed] [Google Scholar]
- 3.Casey, P. J. & Seabra, M. C. (1996) J. Biol. Chem. 271, 5289–5292. [DOI] [PubMed] [Google Scholar]
- 4.Boyartchuk, V. L., Ashby, M. N. & Rine, J. (1997) Science 275, 1796–1800. [DOI] [PubMed] [Google Scholar]
- 5.Otto, J. C., Kim, E., Young, S. G. & Casey, P. J. (1999) J. Biol. Chem. 274, 8379–8382. [DOI] [PubMed] [Google Scholar]
- 6.Clarke, S., Vogel, J. P., Deschenes, R. J. & Stock, J. (1988) Proc. Natl. Acad. Sci. USA 85, 4643–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hrycyna, C. A., Sapperstein, S. K., Clarke, S. & Michaelis, S. (1991) EMBO J. 10, 1699–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai, Q., Choy, E., Chiu, V., Romano, J., Slivka, S. R., Steitz, S. A., Michaelis, S. & Philips, M. R. (1998) J. Biol. Chem. 273, 15030–15034. [DOI] [PubMed] [Google Scholar]
- 9.Young, S. G., Ambroziak, P., Kim, E. & Clarke, S. (2000) in The Enzymes, eds. Tamanoi, F. & Sigman, D.G. (Academic, San Diego), Vol. 21, pp. 156–213. [Google Scholar]
- 10.Malumbres, M. & Barbacid, M. (2003) Nat. Rev. Cancer 3, 459–465. [DOI] [PubMed] [Google Scholar]
- 11.Shields, J. M., Pruitt, K., McFall, A., Shaub, A. & Der, C. J. (2000) Trends Cell Biol. 10, 147–154. [DOI] [PubMed] [Google Scholar]
- 12.Bos, J. L. (1989) Cancer Res. 49, 4682–4689. [PubMed] [Google Scholar]
- 13.Gschwind, A., Fischer, O. M. & Ullrich, A. (2004) Nat. Rev. Cancer 4, 361–370. [DOI] [PubMed] [Google Scholar]
- 14.Schlessinger, J. (2000) Cell 103, 211–225. [DOI] [PubMed] [Google Scholar]
- 15.Doll, R. J., Kirschmeier, P. & Bishop, W. R. (2004) Curr. Opin. Drug Discov. Devel. 7, 478–486. [PubMed] [Google Scholar]
- 16.Hancock, J. F., Magee, A. I., Childs, J. E. & Marshall, C. J. (1989) Cell 57, 1167–1177. [DOI] [PubMed] [Google Scholar]
- 17.Kato, K., Cox, A. D., Hisaka, M. M., Graham, S. M., Buss, J. E. & Der, C. J. (1992) Proc. Natl. Acad. Sci. USA 89, 6403–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibbs, J. B., Oliff, A. & Kohl, N. E. (1994) Cell 77, 175–178. [DOI] [PubMed] [Google Scholar]
- 19.Karp, J. E., Kaufmann, S. H., Adjei, A. A., Lancet, J. E., Wright, J. J. & End, D. W. (2001) Curr. Opin. Oncol. 13, 470–476. [DOI] [PubMed] [Google Scholar]
- 20.James, G. L., Goldstein, J. L. & Brown, M. S. (1995) J. Biol. Chem. 270, 6221–6226. [DOI] [PubMed] [Google Scholar]
- 21.Whyte, D. B., Kirschmeier, P., Hockenberry, T. N., Nunez-Oliva, I., James, L., Catino, J. J., Bishop, W. R. & Pai, J. K. (1997) J. Biol. Chem. 272, 14459–14464. [DOI] [PubMed] [Google Scholar]
- 22.Sebti, S. M. & Der, C. J. (2003) Nat. Rev. Cancer 3, 945–951. [DOI] [PubMed] [Google Scholar]
- 23.Bergo, M. O., Gavino, B. J., Hong, C., Beigneux, A. P., McMahon, M., Casey, P. J. & Young, S. G. (2004) J. Clin. Invest. 113, 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergo, M. O., Leung, G. K., Ambroziak, P., Otto, J. C., Casey, P. J. & Young, S. G. (2000) J. Biol. Chem. 275, 17605–17610. [DOI] [PubMed] [Google Scholar]
- 25.Clarke, S. & Tamanoi, F. (2004) J. Clin. Invest. 113, 513–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang, P. K., Gordon, R. K., Tal, J., Zeng, G. C., Doctor, B. P., Pardhasaradhi, K. & McCann, P. P. (1996) FASEB J. 10, 471–480. [PubMed] [Google Scholar]
- 27.Ma, Y. T., Shi, Y. Q., Lim, Y. H., McGrail, S. H., Ware, J. A. & Rando, R. R. (1994) Biochemistry 33, 5414–5420. [DOI] [PubMed] [Google Scholar]
- 28.Scheer, A. & Gierschik, P. (1993) FEBS Lett. 319, 110–114. [DOI] [PubMed] [Google Scholar]
- 29.Winter-Vann, A. M., Kamen, B. A., Bergo, M. O., Young, S. G., Melnyk, S., James, S. J. & Casey, P. J. (2003) Proc. Natl. Acad. Sci. USA 100, 6529–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCabe, J. B. & Berthiaume, L. G. (1999) Mol. Biol. Cell 10, 3771–3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habets, G. G., Knepper, M., Sumortin, J., Choi, Y. J., Sasazuki, T., Shirasawa, S. & Bollag, G. (2001) Methods Enzymol. 332, 245–260. [DOI] [PubMed] [Google Scholar]
- 32.Bergo, M. O., Leung, G. K., Ambroziak, P., Otto, J. C., Casey, P. J., Gomes, A. Q., Seabra, M. C. & Young, S. G. (2001) J. Biol. Chem. 276, 5841–5845. [DOI] [PubMed] [Google Scholar]
- 33.Chiu, V. K., Silletti, J., Dinsell, V., Wiener, H., Loukeris, K., Ou, G., Philips, M. R. & Pillinger, M. H. (2003) J. Biol. Chem. 279, 7346–7352. [DOI] [PubMed] [Google Scholar]
- 34.Clark, G. J., Cox, A. D., Graham, S. M. & Der, C. J. (1995) Methods Enzymol. 255, 395–412. [DOI] [PubMed] [Google Scholar]
- 35.Kramer, K., Harrington, E. O., Lu, Q., Bellas, R., Newton, J., Sheahan, K. L. & Rounds, S. (2003) Mol. Biol. Cell 14, 848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu, Q., Harrington, E. O., Hai, C. M., Newton, J., Garber, M., Hirase, T. & Rounds, S. (2004) Circ. Res. 94, 306–315. [DOI] [PubMed] [Google Scholar]
- 37.Roullet, J. B., Xue, H., Chapman, J., McDougal, P., Roullet, C. M. & McCarron, D. A. (1996) J. Clin. Invest. 97, 2384–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kowluru, A., Seavey, S. E., Li, G., Sorenson, R. L., Weinhaus, A. J., Nesher, R., Rabaglia, M. E., Vadakekalam, J. & Metz, S. A. (1996) J. Clin. Invest. 98, 540–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maske, C. P., Hollinshead, M. S., Higbee, N. C., Bergo, M. O., Young, S. G. & Vaux, D. J. (2003) J. Cell Biol. 162, 1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, H., Yoshizumi, M., Lai, K., Tsai, J. C., Perrella, M. A., Haber, E. & Lee, M. E. (1997) J. Biol. Chem. 272, 25380–25385. [DOI] [PubMed] [Google Scholar]
- 41.Backlund, P. S., Jr. (1997) J. Biol. Chem. 272, 33175–33180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.