Abstract
For most neurons to function properly, they need to develop synaptic specificity. This requires finding specific partner neurons, building the correct types of synapses, and fine-tuning these synapses in response to neural activity. Synaptic specificity is common at both a neuron’s input and output synapses, whereby unique synapses are built depending on the partnering neuron. Neuroscientists have long appreciated the remarkable specificity of neural circuits but identifying molecular mechanisms mediating synaptic specificity has only recently accelerated. Here, we focus on recent progress in understanding input and output synaptic specificity in the mammalian brain. We review newly identified circuit examples for both and the latest research identifying molecular mediators including Kirrel3, FGFs, and DGLα. Lastly, we expect the pace of research on input and output specificity to continue to accelerate with the advent of new technologies in genomics, microscopy, and proteomics.
INTRODUCTION
Correctly wiring the mammalian brain is an enormously complex process. In the human brain, billions of individual neurons make thousands of synaptic connections and their connectivity is rarely linear or unidirectional. Instead, neurons often send to and receive information from multiple sources, which is critical for integrating information during cognitive tasks and sensory processing. One way neurons organize, integrate, and partition the flow of neural information is by constructing specific types of synapses with different partner neurons.
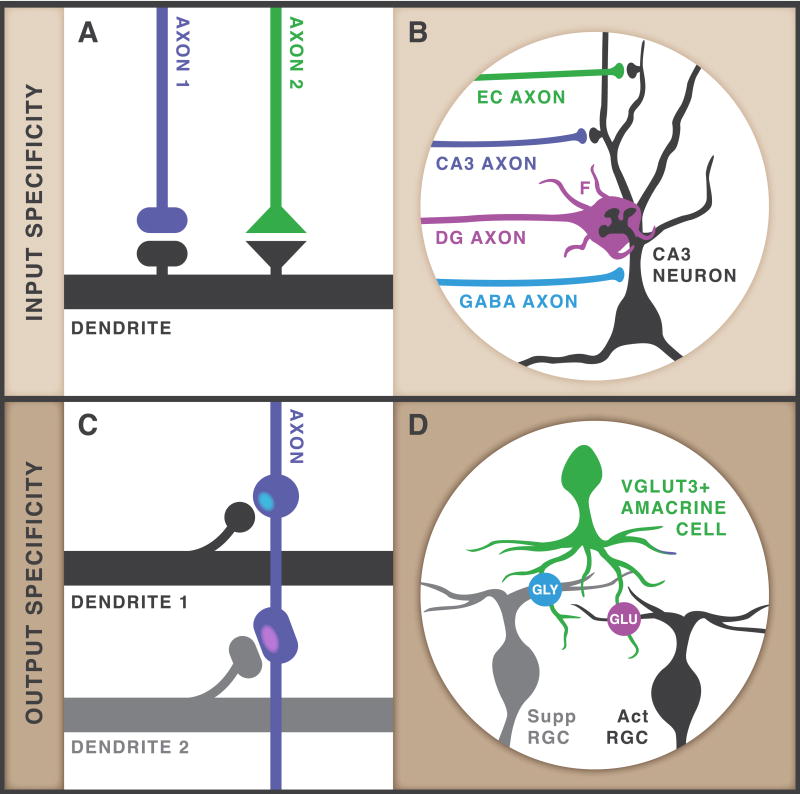
Synaptic specificity can take many forms. Input specificity occurs when one neuron’s dendritic tree makes different types of synapses with different types of axons (Figure 1A). CA3 pyramidal neurons receive many types of axonal inputs and provide an excellent example of input specificity (Figure 1B). Here, inhibitory GABAergic synapses substantially differ from excitatory glutamatergic synapses. In addition to releasing different neurotransmitters, these two types of synapses have a completely distinct cohort of molecular and structural components [1]. In addition, CA3 neurons receive excitatory inputs from dentate granule (DG), other CA3, and entorhinal cortex (EC) neurons and each input forms synapses with distinctive properties. In particular, DG-CA3 mossy fiber synapses are highly specialized synapses with multiple active zones, multi-headed spines, and NMDA receptor independent synaptic plasticity [2].
Figure 1. A Guide to Input and Output Specificity.
(A) In its simplest form, input specificity occurs when one dendrite is innervated by two types of axons and makes different types of synapses with each axon type. These differences can be based on structural, functional, or molecular properties. (B) CA3 pyramidal neurons exemplify input specificity as they have numerous axonal inputs resulting in different types of synapses including inhibitory inputs from GABA interneurons (blue) and excitatory inputs from three different sources, the entorhinal cortex (EC, green), other CA3 neurons (purple), and the dentate gyrus (DG, magenta). Specialized presynanptic mossy fiber filopodia ‘F’ are shown. (C) In its simplest form, output specificity occurs when one axon innervates two types of dendrites and makes different types of synapses with each dendrite type. As for input specificity, these differences can be based on structural, functional, or molecular properties. (D) VGluT3-expressing amacrine cells exemplify output specificity. One amacrine cell makes glutamatergic synapses (purple) when connecting to retinal ganglion cells (RGCs) that are activated (Act) by contrast and motion and glycinergic synapses (blue) when connecting to RGCs that are suppressed (Supp) by contrast and motion.
Output specificity is similar yet distinct from input specificity. Here, one neuron’s axon makes different types of synapses with different types of dendrites (Figure 1C). For example, it was recently discovered VGluT3-expressing amacrine cells (VG3-AC) release glycine or glutamate depending on which type of retinal ganglion cell (RGC) is the postsynaptic partner [3,4]. Using a combination of genetic cell-type labeling, optogenetics, and electrophysiology, it was shown that VG3-ACs form glycinergic synapses with RGCs suppressed by contrast and motion and glutamatergic synapses with RGCs activated by contrast and motion, which include W3 and OFF alpha RGCs (Figure 1D) [3–6]. The molecular mechanisms that establish VG3-AC input specificity remain unknown. However, the immunoglobulin superfamily member sidekick-2 is required for VG3-AC to W3 synapse function [6] and future work will need to determine if this recognition molecule plays a role in VG3-AC synaptic specificity.
New methods in circuit tracing, genetic labeling, imaging, and optogenetics have allowed neuroscientists to observe and describe remarkable instances of input and output specificity throughout the brain, including the examples mentioned above. These studies reveal that synaptic specificity is the norm and not the exception. In addition, many synapse-specific genes and their circuits are linked to neurodevelopmental disorders, but the precise synaptic defects underlying these disorders remain largely unknown [7]. To better understand the cause of neurological disorders and rationally design treatments, it is critical to identify the molecular pathways and mechanisms mediating synaptic specificity. The goal of this focused review is to discuss the most recent advances in our understanding of input and output specificity in the mammalian brain by examining how neurons find specific synaptic partners, build different types of synapses, and fine-tune their communication over time.
FINDING NEURONAL PARTNERS
One key step in the development of input and output specificity is target recognition. At some point, a neuron needs to identify what type of axon or dendrite is in contact. It is logical to propose that target recognition occurs prior to synapse formation but it could also occur during or after synapse formation. Most synapses form so rapidly, within minutes to hours [8–10], that it is difficult to experimentally determine when target recognition occurs. Regardless, target recognition requires communication between two neurons. Transmembrane cell adhesion molecules are uniquely suited to this role because they can interact with the surface of nearby neurons via extracellular domains and relay this information inside the cell via intracellular domains. Moreover, the completion of large-scale in situ hybridization, mRNA sequencing, and synaptic proteomic screens firmly established that cell adhesion molecules are numerous and differentially-expressed throughout the brain [11–19]. Thus, different neuron cell types and even individual neurons express unique combinations of cell adhesion molecules, which may provide a molecular code for target recognition.
Though it is long hypothesized that cell adhesion molecules regulate synaptic specificity, it is only recently that strong experimental evidence substantiated the importance of cell adhesion molecules for input and output specificity. One example of input specificity is mediated by the leucine-rich repeat protein netrin-G ligand-2 (NGL-2) [20]. NGL-2 is expressed in hippocampal CA1 pyramidal neurons, which have an apical dendrite spanning two synaptic layers. In the stratum radiatum (SR) layer, CA1 neurons receive synaptic input from CA3 neurons. In the stratum lacunosum moleculare (SLM) layer, they receive synaptic input from entorhinal cortex neurons. Loss of NGL-2 selectively reduces spine and synapse formation in the SR layer but not the SLM layer [20]. Further evidence suggests NGL-2 functions only at CA3-CA1 SR synapses via a mechanism of selective localization that likely requires interaction with its presynaptic ligand Netrin-G2, which is expressed by CA3 axons but not entorhinal cortex axons [20,21].
Leucine-rich repeat proteins also play a role in CA1 neuron output specificity. CA1 axons form specific types of synapses with different types of GABAergic interneurons. CA1 neurons form facilitating synapses with a low release probability onto oriens-lacunosum moleculare (O-LM) interneurons and depressing synapses with a high release probability onto parvalbumin-positive (PV) interneurons [22]. Elfn1 is a leucine-rich repeat containing transmembrane protein selectively expressed by O-LM neurons [22]. Loss of Elfn1 specifically disrupts short-term facilitation at CA1 to O-LM synapses and, remarkably, its expression is sufficient to turn depressing CA1-PV synapses into mildly facilitating synapses [22]. Currently, the mechanism by which postsynaptic Elfn1 communicates with presynaptic CA1 axons remains unknown.
Leucine-rich repeat proteins are not the only class of adhesion molecules shown to regulate input and output specificity. Kirrel3 is a member of the Immunoglobulin superfamily and was recently shown to regulate output specificity of hippocampal DG axons [11]. DG axons synapse onto and directly excite two main target types; (i) CA3 pyramidal neurons via a giant bouton and (ii) GABAergic interneurons via filopodial synapses (Figure 1B). Kirrel3 is a homophilic cell adhesion molecule that, in the hippocampus, is only expressed by DG neurons and some GABA neurons, suggesting Kirrel3 functions specifically at DG-GABA and not DG-CA3 synapses. In support, Kirrel3 knockout mice have a reduced density of DG axon filopodia and increased activity in CA3 neurons after DG stimulation [11]. This functional phenotype is consistent with a loss of DG-GABA synapses that normally produce feed-forward inhibition to constrain CA3 activity. These results suggest Kirrel3 may locate selectively to mossy fiber filopodia but selective localization of Kirrel3 remains to be tested.
In addition to cell adhesion molecules, new research demonstrates that secreted axon guidance molecules such as the Semaphorin (Sema) family can also mediate synaptic specificity but with a twist. Instead of promoting synapse formation, Sema signaling may regulate synaptic specificity by inhibiting synapse formation. Signaling via the secreted Sema3F and its membrane bound co-receptors (Neuropilin-2 and Plexin-A3) is selectively required to restrict spine formation on apical, but not basal, dendrites of layer V cortical neurons [23]. Similar to the NGL-2 mechanism, this may be mediated by selective localization to the affected layer; in this case the apical dendrites [23]. Sema signaling also regulates output specificity of thalamostriatal axons [24]. Here, Sema3E is expressed by thalamic axons that synapse onto striatal medium spiny neurons of the so-called “direct” and “indirect” pathways. Direct and indirect striatal neurons are physically intermixed but they perform distinct functions [25]. Only direct pathway neurons express the Sema3E receptor Plexin-D1, and both Sema3E and Plexin-D1 knockout mice have increased thalamic synapses onto direct (but not indirect) pathway neurons compared to wildtype mice [24]. This suggests Sema3E may remain associated with thalamocortical axons and interacts with postsynaptic Plexin-D1 to restrict the number of thalamic inputs made onto direct pathway striatal neurons.
Taken together, an emerging body of work suggests differentially-expressed cell recognition systems play a major role in guiding input and output synaptic specificity across the brain. Moreover, the high fidelity of neural connections likely requires concerted mechanisms of positive and negative synaptic specificity cues.
BUILDING DIFFERENT TYPES OF SYNAPSES
How does one neuron build specific types of synapses with different partner neurons? There is still little known about how an individual neuron constructs distinct types of synapses and understanding these mechanisms is crucial as more and more specificity molecules are implicated in neurological disorders [7]. However, more progress has been made in comparing excitatory and inhibitory synapse formation. Many genes have been identified that function exclusively at either excitatory or inhibitory synapses [1]. Here, we will not focus on listing these genes because, in most cases, they were primarily studied in cell culture and little is known about their mechanism of specificity. Instead, we briefly discuss new evidence that two groups of molecules, the neuroligins and fibroblast growth factors (FGFs), balance formation of excitatory and inhibitory synapses.
Neuroligins are postsynaptic cell adhesion proteins with an established, but sometimes controversial, role in synapse formation. Most evidence suggests neuroligin-1 (NL1) mediates excitatory synapse formation and neuroligin-2 (NL2) mediates inhibitory synapse formation [26]. NL1 and NL2 are thought to orchestrate excitatory or inhibitory postsynapse construction via intracellular interactions. NL1 interacts with the scaffolding protein PSD-95 and recruits glutamate receptors to excitatory synapses. Conversely, NL2 interacts with gephryin and GABA receptors at inhibitory synapses [27]. However, both NL1 and NL2 share the same PSD-95 and gephryin interacting domains and therefore both have potential to induce excitatory and inhibitory synapses. Recent studies suggest their ability to establish an excitatory versus inhibitory synapse depends upon post-translational modifications and the expression of their presynaptic binding partner neurexin. For example, Neurexin-1β (Nrx1β) is predominantly expressed in excitatory axons and binds NL1 with higher affinity than NL2. However, overexpression of Nrx1β and NL1 in connected GABA neuron-CA1 pairs increases inhibitory input into the CA1 neuron. This suggests NL1 induces inhibitory connections if its preferred presynaptic partner is expressed in inhibitory neurons [28]. Thus, NL1 may not exclusively mediate excitatory synapse formation but also inhibitory synapses based on context. In another study, binding of Nrx1β to NL1 was shown to induce NL1 phosphorylation, which leads to preferential binding with PSD-95 over gephyrin [29]. Moreover, neurexins, neuroligins, or both may be expressed at limiting amounts, creating competition for synapse formation [28,30]. Thus, synaptic specificity could be partially determined by the expression levels of trans-synaptic proteins in competing partner neurons. Taken together, new work suggests that intracellular targeting of kinases and phosphatases, and the availability of different binding partners, may govern whether an excitatory or inhibitory synapse will be built.
Another emerging group of molecules that moderate excitatory versus inhibitory synapse formation are FGFs. FGF22 and FGF7 are released from CA3 dendrites and promote excitatory and inhibitory synapse formation, respectively [31–33]. FGFs localize to unique locations along the CA3 dendrite due to their co-transport with either excitatory or inhibitory postsynaptic proteins on separate kinesin motor proteins. More specifically, FGF22 co-transports with the excitatory postsynaptic scaffolding protein SAP102, whereas FGF7 co-transports with gephryin [32]. Thus, input specificity in CA3 dendrites is partially determined by the subcellular localization of FGFs during postsynapse construction. On the presynaptic side, input specificity is reinforced by different FGF receptors. In DG neurons, Fgfr2b and Fgfr1b are both required for FGF22 signaling, whereas in GABA neurons Fgfr2b but not Fgfr1b is required for FGF7 signaling [33]. Moreover, some of the downstream signaling mechanisms for building the presynaptic inputs are beginning to be elucidated for this pathway. For example, Fgfr2b signals via FRS2 and P13K to promote excitatory synapse formation [33]. Future work will need to determine whether similar or unique signaling mechanisms mediate the construction of FGF-dependent inhibitory presynapses.
FINE-TUNING SYNAPTIC SPECIFICITY
For effective neuron communication, synapses must adapt to new information. Adding to the layers of synaptic specificity present in the brain, different types of input and output synapses can undergo specific forms of functional and structural plasticity. Moreover, mechanisms vary greatly depending on the cellular and molecular players involved [34]. An example of functional output specificity occurs in the basal nucleus of the amygdala. Here, cholecystokinin (CCK)-positive basket cell interneurons make synapses onto two different types of principal neurons; one that mediates high fear responses and one that mediates low fear responses. The CCK neurons make very similar synapses onto these two principal neuron types; however, they develop unique plasticity properties with each type of target neuron. Synapses made onto low-fear neurons, but not high-fear, undergo activity-dependent suppression, which is correlated with increased levels of the endocannabinoid-synthesizing enzyme DGLα on the postsynaptic side [35]. Future experiments will need to determine whether DGLα is required for functional plasticity and define upstream mechanisms that instruct low fear neurons to increase DGLα expression. This example of functional synaptic specificity is unique because differing functional plasticity is the primary specificity determinant in this circuit. In other systems, differing plasticity mechanisms more commonly result from a myriad of initial structural and molecular differences.
The basal amygdala also provides an example of input specific plasticity. In addition to CCK input synapses, principal amygdala neurons also receive synapses from parvalbumin (PV)-positive interneurons. Each of these inputs are differentially regulated during fear extinction. During fear extinction, some principal amygdala neurons that were active during fear conditioning are silenced while others remain active. Silenced principal neurons receive more PV perisomatic synapses. In contrast, primary neurons that remain active have altered CCK perisomatic synapses with increased levels of cannabinoid receptor type 1 (CB1R) [36]. Because CB1R blocks GABA release [37] this change decreases CCK synapse inhibition to the primary neuron. Thus, a unique type of plasticity occurs at each type of GABA input to principle amygdala neurons. These examples from the basal amygdala emphasize how input and output specific plasticity are critical for circuit function and behavior.
CONCLUSIONS AND OUTLOOK
In this review, we examined recent progress made in identifying molecular mechanisms that establish and maintain input and output synaptic specificity. Though progress is ongoing, we still know very little about the intracellular signaling mechanisms that organize and build specific synapse types along a neuron. However, a list of emerging technologies is rapidly advancing to combat this knowledge gap. Imaging of single neurons using cutting-edge in vivo microscopy techniques recently revealed inhibitory synapses are more structurally dynamic than excitatory synapses [38]. In addition, super-resolution microscopy enables 3D imaging of synapse structure at higher volumes with greater speed and molecular information than is afforded with electron microscopy. For example, super-resolution microscopy recently revealed markedly different PSD95 structures along a single dendrite, suggesting presynaptic input is critical in determining the size and complexity of PSD-95 complexes [39]. Lastly, advances in proteomics are enabling the identification of novel molecular markers and mediators of synaptic specificity. Targeting horseradish peroxidase to the synaptic cleft allows for near-neighbor biotinylation, streptavidin purification, and mass spectrometry identification of synaptic cleft proteins [40]. This has provided a list of proteins at excitatory and inhibitory synaptic clefts and future studies can take advantage of this technique to elucidate the subtle differences between the molecular profiles of more closely related synapses within specific circuits. Together, these new technologies will allow for the identification and precise location of novel regulators of synaptic specificity.
HIGHLIGHTS.
Cell adhesion systems are critical determinants of synaptic specificity.
Neuroligins and FGFs contribute to the balance of inhibitory to excitatory synapses.
Synaptic plasticity mechanisms can differ for different synaptic inputs and outputs.
New tools are advancing our molecular understanding of synaptic specificity.
Acknowledgments
We would like to thank the entire Williams lab for critical reading of the manuscript. Research in the Williams lab is funded by the NIMH 1R01MH105426, Autism Speaks, and the Alfred P. Sloan, Edward Mallinckrodt Jr., and Whitehall Foundations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gatto CL, Broadie K. Genetic controls balancing excitatory and inhibitory synaptogenesis in neurodevelopmental disorder models. Front Synaptic Neurosci. 2010;2:4. doi: 10.3389/fnsyn.2010.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicoll RA, Schmitz D. Synaptic plasticity at hippocampal mossy fibre synapses. Nat Rev Neurosci. 2005;6:863–876. doi: 10.1038/nrn1786. [DOI] [PubMed] [Google Scholar]
- **3.Lee S, Zhang Y, Chen M, Zhou ZJ. Segregated Glycine-Glutamate Co-transmission from vGluT3 Amacrine Cells to Contrast-Suppressed and Contrast-Enhanced Retinal Circuits. Neuron. 2016;90:27–34. doi: 10.1016/j.neuron.2016.02.023. Using optogenetics and dual patch-clamping, Lee et al. demonstrate that VGluT3 expressing amacrine cells (VG3-ACs) release glycine onto RGCs that are suppressed by contrast and release glutamate onto RGCs that are enhanced by contrast. This work, along with Tien et al. 2016, demonstrates for the first time that VG3-ACs release two different neurotransmitters in a target-specific manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **4.Tien N-W, Kim T, Kerschensteiner D. Target-Specific Glycinergic Transmission from VGluT3-Expressing Amacrine Cells Shapes Suppressive Contrast Responses in the Retina. Cell Rep. 2016;15:1369–1375. doi: 10.1016/j.celrep.2016.04.025. In an exquisite set of experiments, Tien et al. demonstrate both optogenetically and by circuit reconstruction that VG3-ACs release glycine and form inhibitory synapses with suppressed by contrast (SbC) RGCs. In addition, they genetically ablate VG3-ACs and show a reduction in the inhibition of SbC-RGCs. This work along with Lee et al. 2016 demonstrates for the first time that VG3-ACs are dual neurotransmitter neurons that release glycine to regulate contrast processing in the retina. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S, Chen L, Chen M, Ye M, Seal RP, Zhou ZJ. An unconventional glutamatergic circuit in the retina formed by vGluT3 amacrine cells. Neuron. 2014;84:708–715. doi: 10.1016/j.neuron.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krishnaswamy A, Yamagata M, Duan X, Hong YK, Sanes JR. Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature. 2015;524:466–470. doi: 10.1038/nature14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Washbourne P. Synapse Assembly and Neurodevelopmental Disorders. Neuropsychopharmacology. 2014;40:4–15. doi: 10.1038/npp.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman HV, Bresler T, Garner CC, Ziv NE. Assembly of new individual excitatory synapses: time course and temporal order of synaptic molecule recruitment. Neuron. 2000;27:57–69. doi: 10.1016/s0896-6273(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 9.Ahmari SE, Buchanan J, Smith SJ. Assembly of presynaptic active zones from cytoplasmic transport packets. Nat Neurosci. 2000;3:445–451. doi: 10.1038/74814. [DOI] [PubMed] [Google Scholar]
- 10.Bresler T, Shapira M, Boeckers T, Dresbach T, Futter M, Garner CC, Rosenblum K, Gundelfinger ED, Ziv NE. Postsynaptic density assembly is fundamentally different from presynaptic active zone assembly. J Neurosci. 2004;24:1507–1520. doi: 10.1523/JNEUROSCI.3819-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **11.Martin EA, Muralidhar S, Wang Z, Cervantes DC, Basu R, Taylor MR, Hunter J, Cutforth T, Wilke SA, Ghosh A, et al. The intellectual disability gene Kirrel3 regulates target-specific mossy fiber synapse development in the hippocampus. eLife. 2015;4:e09395. doi: 10.7554/eLife.09395. Martin, Muralidhar, et al. uncover a potential mechanism for output specificity at DG mossy fiber synapses. DG presynapses are giant boutons that primarily connect with CA3 neurons but also send out filopodial extension that connect with GABA interneurons. The homophilic cell adhesion molecule Kirrel3 was specifically required for filopodial development but not giant bouton formation, suggesting that Kirrel3 may be a molecular mechanism by which a single neuron signals the construction of markedly different synapses made by the same axon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams ME, Wilke SA, Daggett A, Davis E, Otto S, Ravi D, Ripley B, Bushong EA, Ellisman MH, Klein G, et al. Cadherin-9 regulates synapse-specific differentiation in the developing hippocampus. Neuron. 2011;71:640–655. doi: 10.1016/j.neuron.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craig AM, Kang Y. Neurexin-neuroligin signaling in synapse development. Curr Opin Neurobiol. 2007;17:43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Wit J, Sylwestrak E, O’Sullivan ML, Otto S, Tiglio K, Savas JN, Yates JR, Comoletti D, Taylor P, Ghosh A. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko J, Fuccillo MV, Malenka RC, Südhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64:791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo J, Kwon S-K, Choi S, Kim S, Lee J-R, Dunah AW, Sheng M, Kim E. Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat Neurosci. 2009;12:428–437. doi: 10.1038/nn.2279. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Burette A, Chung HS, Kwon S-K, Woo J, Lee HW, Kim K, Kim H, Weinberg RJ, Kim E. NGL family PSD-95-interacting adhesion molecules regulate excitatory synapse formation. Nat Neurosci. 2006;9:1294–1301. doi: 10.1038/nn1763. [DOI] [PubMed] [Google Scholar]
- 18.Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Südhof TC. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- 19.Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nat Rev Neurosci. 2007;8:206–220. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeNardo LA, de Wit J, Otto-Hitt S, Ghosh A. NGL-2 regulates input-specific synapse development in CA1 pyramidal neurons. Neuron. 2012;76:762–775. doi: 10.1016/j.neuron.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimura-Akiyoshi S, Niimi K, Nakashiba T, Itohara S. Axonal netrin-Gs transneuronally determine lamina-specific subdendritic segments. Proc Natl Acad Sci U S A. 2007;104:14801–14806. doi: 10.1073/pnas.0706919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sylwestrak EL, Ghosh A. Elfn1 regulates target-specific release probability at CA1-interneuron synapses. Science. 2012;338:536–540. doi: 10.1126/science.1222482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tran TS, Rubio ME, Clem RL, Johnson D, Case L, Tessier-Lavigne M, Huganir RL, Ginty DD, Kolodkin AL. Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature. 2009;462:1065–1069. doi: 10.1038/nature08628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding JB, Oh W-J, Sabatini BL, Gu C. Semaphorin 3E-Plexin-D1 signaling controls pathway-specific synapse formation in the striatum. Nat Neurosci. 2011;15:215–223. doi: 10.1038/nn.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wichmann T, DeLong MR. Functional and pathophysiological models of the basal ganglia. Curr Opin Neurobiol. 1996;6:751–758. doi: 10.1016/s0959-4388(96)80024-9. [DOI] [PubMed] [Google Scholar]
- 26.Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bemben MA, Shipman SL, Nicoll RA, Roche KW. The cellular and molecular landscape of neuroligins. Trends Neurosci. 2015;38:496–505. doi: 10.1016/j.tins.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Futai K, Doty CD, Baek B, Ryu J, Sheng M. Specific trans-synaptic interaction with inhibitory interneuronal neurexin underlies differential ability of neuroligins to induce functional inhibitory synapses. J Neurosci. 2013;33:3612–3623. doi: 10.1523/JNEUROSCI.1811-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giannone G, Mondin M, Grillo-Bosch D, Tessier B, Saint-Michel E, Czöndör K, Sainlos M, Choquet D, Thoumine O. Neurexin-1β binding to neuroligin-1 triggers the preferential recruitment of PSD-95 versus gephyrin through tyrosine phosphorylation of neuroligin-1. Cell Rep. 2013;3:1996–2007. doi: 10.1016/j.celrep.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 30.Kwon H-B, Kozorovitskiy Y, Oh W-J, Peixoto RT, Akhtar N, Saulnier JL, Gu C, Sabatini BL. Neuroligin-1-dependent competition regulates cortical synaptogenesis and synapse number. Nat Neurosci. 2012;15:1667–1674. doi: 10.1038/nn.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terauchi A, Johnson-Venkatesh EM, Toth AB, Javed D, Sutton MA, Umemori H. Distinct FGFs promote differentiation of excitatory and inhibitory synapses. Nature. 2010;465:783–787. doi: 10.1038/nature09041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *32.Terauchi A, Timmons KM, Kikuma K, Pechmann Y, Kneussel M, Umemori H. Selective synaptic targeting of the excitatory and inhibitory presynaptic organizers FGF22 and FGF7. J Cell Sci. 2015;128:281–292. doi: 10.1242/jcs.158337. Intracellular mechanisms for how synapse specificity is established are largely unknown. This study provides evidence that distinct transport mechanisms lead to differential subcellular targeting of synaptogenic FGFs. Thus, the targeted transport of FGFs may determine the segregation of excitatory and inhibitory synapse formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *33.Dabrowski A, Terauchi A, Strong C, Umemori H. Distinct sets of FGF receptors sculpt excitatory and inhibitory synaptogenesis. Development. 2015;142:1818–1830. doi: 10.1242/dev.115568. Dabrowski et al. identify the presynaptic receptors that interact with FGFs during excitatory and inhibitory synapse formation. They also identify downstream signaling mechanisms involved in building FGF-dependent excitatory synapses. This is an exciting advance as much synaptogenesis research is focused on trans-synaptic interactions and we still know very little about intracellular signaling mechanisms that orchestrate the construction of synapses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsen RS, Sjöström PJ. Synapse-type-specific plasticity in local circuits. Curr Opin Neurobiol. 2015;35:127–135. doi: 10.1016/j.conb.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *35.Vogel E, Krabbe S, Gründemann J, Cusulin JIW, Lüthi A. Projection-Specific Dynamic Regulation of Inhibition in Amygdala Micro-Circuits. Neuron. 2016;91:644–651. doi: 10.1016/j.neuron.2016.06.036. This study identifies a circuit where functional plasticity establishes output specificity. CCK interneurons in the basal amygdala form similar synapses onto low-fear and high-fear principal neurons but they develop different plasticity properties. Vogel et al. found that increased levels of the endocannabinoid-synthesizing enzyme DGLα are associated with the low-fear synapses that acquire activity-dependent suppression. [DOI] [PubMed] [Google Scholar]
- 36.Trouche S, Sasaki JM, Tu T, Reijmers LG. Fear extinction causes target-specific remodeling of perisomatic inhibitory synapses. Neuron. 2013;80:1054–1065. doi: 10.1016/j.neuron.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katona I, Rancz EA, Acsady L, Ledent C, Mackie K, Hajos N, Freund TF. Distribution of CB1 cannabinoid receptors in the amygdala and their role in the control of GABAergic transmission. J Neurosci. 2001;21:9506–9518. doi: 10.1523/JNEUROSCI.21-23-09506.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **38.Villa KL, Berry KP, Subramanian J, Cha JW, Oh WC, Kwon H-B, Kubota Y, So PTC, Nedivi E. Inhibitory Synapses Are Repeatedly Assembled and Removed at Persistent Sites In Vivo. Neuron. 2016;89:756–769. doi: 10.1016/j.neuron.2016.01.010. Villa et al. examined the dynamics of inhibitory synapses in vivo. This is significant because inhibitory synapses typically form on the shaft and, unlike excitatory synapses that form on spines, do not have an obvious structural component for imaging. Thus, the authors heroically combined single cell fluorescent labeling of inhibitory markers with daily in vivo imaging. Intriguingly, they discovered that inhibitory synapses are more dynamic than excitatory spine synapses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broadhead MJ, Horrocks MH, Zhu F, Muresan L, Benavides-Piccione R, DeFelipe J, Fricker D, Kopanitsa MV, Duncan RR, Klenerman D, et al. PSD95 nanoclusters are postsynaptic building blocks in hippocampus circuits. Sci Rep. 2016;6:24626. doi: 10.1038/srep24626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **40.Loh KH, Stawski PS, Draycott AS, Udeshi ND, Lehrman EK, Wilton DK, Svinkina T, Deerinck TJ, Ellisman MH, Stevens B, et al. Proteomic Analysis of Unbounded Cellular Compartments: Synaptic Clefts. Cell. 2016;166:1295–1307. e21. doi: 10.1016/j.cell.2016.07.041. This study represents a major technical advance for comparing the molecular content of different types of synapses. The authors conduct a proteomic screen of excitatory and inhibitory synapses by targeting new enzymatic tools selectively to the synaptic clefts of excitatory or inhibitory synapses. Future work can target proteomics to other synaptic compartments to compare synapses that are more closely related and have thus far remained difficult to parse out molecularly. [DOI] [PMC free article] [PubMed] [Google Scholar]