Abstract
Neutral sphingomyelinases sphingomyelin phosphodiesterase (SMPD)2 and -3 hydrolyze sphingomyelin to phosphocholine and ceramide. smpd2 is expressed ubiquitously, and smpd3 is expressed predominantly in neurons of the CNS. Their activation and the functions of the released ceramides have been associated with signaling pathways in cell growth, differentiation, and apoptosis. However, these cellular responses remain poorly understood. Here we describe the generation and characterization of the smpd3–/– and smpd2–/–smpd3–/– double mutant mouse, which proved to be devoid of neutral sphingomyelinase activity. SMPD3 plays a pivotal role in the control of late embryonic and postnatal development: the smpd3-null mouse develops a novel form of dwarfism and delayed puberty as part of a hypothalamus-induced combined pituitary hormone deficiency. Our studies suggest that SMPD3 is segregated into detergent-resistant subdomains of Golgi membranes of hypothalamic neurosecretory neurons, where its transient activation modifies the lipid bilayer, an essential step in the Golgi secretory pathway. The smpd3–/– mouse might mimic a form of human combined pituitary hormone deficiency.
Sphingomelinases (SMases, EC3.1.4.12) [sphingomyelin (SM) phosphodiesterases (SMPD)] initiate the catabolism of SM, a major component of the lipid bilayer of subcellular membranes. SMases differ in their enzymatic properties, regulation, tissue distribution, and subcellular localization. SM is degraded constitutively by acid SMase (SMPD1) in lysosomes (1–6). Genetic defects of smpd1 lead to SM storage. SMPD1-deficient mouse mutants generated by gene targeting, mimic the fatal human Niemann–Pick disease, type A (7, 8). SMPD1 activity exceeds that of nonlysosomal neutral SMases (nSMases) in all tissues except brain (9). Their function is poorly understood. Ligand cell-surface receptor-mediated activation of nSMases is believed to trigger a regulated breakdown of SM to ceramide regarded as a lipid-signaling molecule in cellular pathways ranging from antiapoptosis and cell survival and proliferation to cell senescence (10, 11). Recently, several observations have challenged the notion that ceramide plays a role as mediator in these reactions (12–19). This development underlines the need for a molecular dissection of the functions of nSMases.
We have initiated genetic and molecular studies on the role of nSMases in cellular processes in their genuine environment. Two Mg2+-dependent nSMases, SMPD2 and -3, were cloned (13) and characterized (14–16). Smpd2 is ubiquitously expressed, SMPD3, the most prominent nSMase, particularly in brain.
Here we describe the generation and phenotype of two null mutants, the SMPD3-deficient (smpd3–/–) and the smpd2–/– smpd3–/– double mutant mouse. The double mutant completely lacked nSMase activity but developed neither a SM storage disease nor impaired apoptosis during a lifespan of >2 years.
Mutant smpd3–/– mice developed an unexpected pleiotropic phenotype with severe retardation of late embryonic and postnatal growth, a new hypothalamic form of combined pituitary hormone deficiency (CPHD), different from the Ames and Snell dwarfs, respectively (17–19), and the proconvertase null mouse (20, 21). Our studies revealed a pivotal function of SMPD3 in the control of the hypothalamus–pituitary growth axis.
Materials and Methods
Generation of the smpd3–/– and smpd2–/– × smpd3–/– Double Mutant Mouse Line. The smpd3 targeting construct in the pPNT vector used for the electroporation of CJ7 ES cells contained a 2-kb 5′ fragment with 1.4-kb exon1, interrupted by the neocassette, and a 4-kb 3′ fragment, isolated from a mouse genomic λ-phage library (Fig. 1). Selection of ES cell clones, blastocyst injection, and the generation of the smpd3–/– mouse line, Southern blot hybridization analysis, and PCR primers used in the genotypic characterization of the smpd3 and the smpd2/smpd3 double knockout mouse mutants are described in Supporting Text, which is published as supporting information on the PNAS web site.
Fig. 1.
smpd3-knockout strategy, genotype analysis, and absence of smpd3 expression. (A) Targeted disruption of the smpd3 locus. (a) wt murine smpd3 gene locus. (b) Targeting construct. (c) Targeted smpd3 locus. (B) Southern blot hybridization analysis. SstI-restriction fragment length polymorphisms of DNA of ES clones and tail biopsies of control (+/+), smpd3+/–, and smpd3–/– siblings of F1 and F2 generation. Diagnostic fragments: wt DNA, 4.6 kb; and smpd3–/–, 6.4 kb. (D) Northern blot hybridization of total brain RNA of +/+, +/–, and –/– of 3-wk-old mice. Probe: smpd3 cDNA, β-actin cDNA loading control. (E) Western blot analysis of brain protein extract (p14). (C) Genotyping of smpd2–/–smpd3–/– double mutant by PCR. Tail DNA from wt, smpd2±smpd3–/–, and smpd2–/–smpd3–/– mice was used as template for PCR with primer pairs listed under Methods and Materials. Lanes: a, wt smpd2 sibling; b, smpd2+/–; c, smpd2–/–. The 3-kb fragment indicates the wt–, the 3.9-kb fragment, the targeted smpd2 allele. Lanes: a′, PCR fragment of the smpd3+/+ and c′, the smpd3–/– allele. The 1.3-kb fragment monitors the wt smpd3+/+ allele, the 2.9-kb fragment, and the smpd3–/– allele. (D) Northern blot analysis. (E) Western blot analysis.
Northern Blot Analysis. Total RNA was extracted from kidney, liver, and brain by using TRIzol (Life Technologies, Grand Island, NY) and poly(A)+ mRNA by using the Oligotex mRNA Midi Kit (Qiagen, Chatsworth, CA), for Northern blot analysis. Radioactive signals were detected by phosphoimaging.
Semiquantitative RT-PCR. Quality-controlled RNA was transcribed by using mouse leukemia virus reverse transcriptase (Stratagene). Quantitative PCR amplification was optimized for each primer pair at 15, 20, 25, and 30 cycles to ensure the linear range and carried out in the presence of 1 μCi of (1 Ci = 37 GBq) [α-32P]dCTP (22).
EGFP-N2–SMPD3 Fusion Construct. Full-length smpd3 cDNA, inserted into the pcDNA 3.1 myc/his vector (16), was amplified with primers NSM2 5′XhoI-s (5′-CTC GAG ATG GTT TTG TAC ACG ACC CCC TTT CTT-3′) and NSM2 3′KpnI as (5′-GGT ACC ACG CCT CCT CTT CCC CTG CAG ACA CCA-3′) and ligated into the XhoI-KpnI restricted pEGFP-N2 vector in frame with EGFP.
SMase Assay. Mg2+-dependent neutral SMase activity in protein fractions of Triton X-100 solubilized membrane fractions of total brain and human embryonic kidney (HEK)293 cells was determined by using [N-14CH3] SM, as described (14).
Subcellular Fractionation and Protein Analysis. Golgi fraction from mouse brain and HEK293 cells were isolated by discontinuous density gradient centrifugation and of detergent insoluble membrane subdomains (DIMs) through an Optiprep gradient (23) and as outlined in Supporting Text.
Lipid Analysis. Total lipids of tissues, cells, and subcellular fractions were extracted (24) and analyzed as described (25).
Transfection. HEK293 cells were grown in DMEM, supplemented with 5% FCS and 10% horse serum. The mouse smpd3 and smpd3-egfp mammalian expression vectors were introduced into HEK293 cells by electroporation and grown to 80% confluency.
Western Blot Analysis and Immunohistochemistry. Western blot analysis of proteins of membrane extracts from wt and mock- and stably msmpd3-overexpressing HEK293 cells and tissues of 3-mo-old CD1 and immunohistochemical procedures are described in Supporting Text.
Results
smpd3–/– and smpd2–/–smpd3–/– Double Nullizygous Mouse Mutants. We designed a targeting vector for the disruption of smpd3 by inserting a neocassette into exon I, which encodes the two putative transmembrane domains and part of the catalytic site of SMPD3 (Fig. 1A). The 4.6- and 6.4-kb SstI restriction fragment length polymorphism of ES cell and tail DNA in Southern blot hybridization monitored the wt and correctly targeted smpd3 locus, respectively (Fig. 1B). The absence of the 6-kb smpd3 specific mRNA in brain, liver, and thymus of smpd3–/– mice, probed with full-length smpd3 cDNA in Northern blot analysis, indicated the absence of smpd3 expression (Fig. 1D). The failure of mouse SMPD3-specific antibodies to recognize the 70-kDa full-length protein or truncated fragments in total protein and membrane fractions of brain in Western blot analysis proved we had generated a SMPD3-deficient mouse (Fig. 1D).
Crossing smpd2–/– and smpd3–/– mice yielded smpd2–/– smpd3–/– double mutant animals. The mutated smpd2–/– (13) and smpd3–/– gene loci of double mutant mice were confirmed by their respective restriction fragment length polymorphisms in Southern blot analysis and PCR, which yielded fragments of 3.9 kb (smpd2–/–) (Fig. 1E, lane c) and 2.9 kb (smpd3–/–) (Fig. 1E, lane c′).
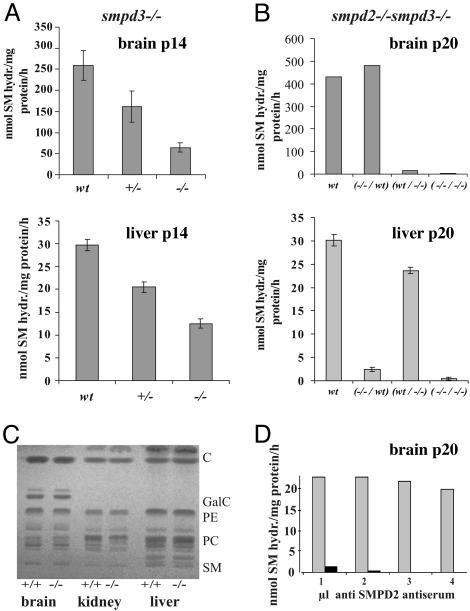
smpd2–/–smpd3 –/– Double Mutant Mice Lack Neutral SMase Activity and Show No Signs of SM Storage Disease. SMase assays in lysates and membrane fractions of brain and liver of postnatal day 14 (p14) and p42 wt, smpd3+/–, smpd3–/–, and smpd2–/–smpd3–/– mice demonstrated that the residual nSMase activity in organs of the smpd3–/– mouse is due to SMPD2 activity. The double mutant is completely devoid of neutral SMase activity. The sensitive radioactive SMase assay detected no enzymatic activity in brain and liver of the smpd2–/–smpd3–/– mice (Fig. 2 A and B).
Fig. 2.
The enzymatic activity of only two nSMases, SMPD2 and -3, is detectable in the mouse. (A and B) Assay in tissue homogenates of brain and liver of p14 smpd3–/– mice, n = 6 (A); and brain and liver of p20 smpd2–/–smpd3–/– double mutant, n = 6(B). Notice the different scale of the enzyme activity (y axis) in brain and liver. (D) Inhibition of nSMase activity in brain extracts of smpd3–/– mice with anti-SMPD2 antiserum, n = 3. Gray bars, residual total nSMase activity in brain extracts; black bars, nSMase activity after addition of antiserum. (C) smpd3–/– mice develop no SM storage disease. Total lipid extracts of brain, kidney, and liver of wt and smpd3–/– mice (12 months of age), separated by high-performance thinlayer chromatography, revealed no differences in their lipid pattern.
SMPD2 activity is inhibited by SMPD2-specific antibodies (15). Anti-SMPD2 antibodies inhibited the residual activity in brain and liver extracts of the smpd3–/– mouse in a concentration-dependent manner to completion (shown for brain in Fig. 2D).
We concluded from these observations that SMPD2 and -3 account for the total neutral SMase activity in mouse tissue.
Unlike the smpd1–/– mutant (Niemann–Pick mouse) with massive lysosomal SM storage predominantly in the reticuloendothelial system and brain (7, 8), the smpd3–/– and smpd2–/–smpd3–/– mice developed no accumulation of SM in neurons, thymocytes, hepatocytes, splenocytes, and kidney cells. Quantitative thin-layer chromatographic analysis of lipid extracts of brain, liver, and kidney showed no difference in the pattern of the main lipid classes cholesterol, phosphatidylethanolamine, phosphatidylcholine, SM, and cerebrosides and sulfatides in brain extracts of wt and smpd3–/– mice, respectively (Fig. 2C). Histology of tissue sections revealed no lipid deposits (not shown). These findings excluded a SM–cholesterol storage defect in smpd3 null as well as the smpd2–/–smpd3–/– animals.
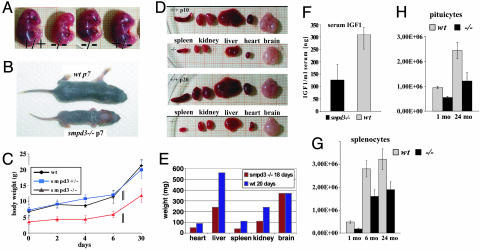
SMPD3 Deficiency Causes a Dwarf Phenotype. smpd3–/– mice unexpectedly developed severe growth retardation at approximately embryonic day (e)14 (Fig. 3A), which persisted to the end of development (around p70) and through a lifespan of 2 years. The growth rate of newborn nullizygous mice was less than half of their wt littermates (Fig. 3 B and C) and could be attributed to a corresponding hypoplasia of all organs except brain (Fig. 3 D and E).
Fig. 3.
The SMPD3-deficient mouse develops dwarfism. (A) Comparison of e14 embryos smpd3–/–, smpd+/–, and smpd+/+ siblings. (B) Severe postnatal growth retardation of p7 wt male smpd3–/– and wt p7 male littermates. (C) Growth curves of wt, smpd3+/–, and smpd3–/– male (n = 10) and female (n = 10) littermates during postnatal development. (D) Hypoplasia of spleen, kidney, liver, heart, and brain of –/– and +/+ p10 and p20 mice. (E) Weight of organs of control +/+ and smpd3–/– of p10 and p20 mice. Generalized hypoplasia in the smpd3–/– mouse. (F–H) Serum IGF1 concentration of wt and smpd3–/– male siblings, n = 10 (F); cell count of splenocytes (n = 4 control and 4 smpd3–/– male mice) at 1, 6, and 24 mo of age (G); and cell count of pituicytes of control (n = 4) and smpd3–/– (n = 4) male mice at 1 and 24 mo of age (H).
Next we tested whether the dwarfism could be caused by an impairment of the hypothalamus-pituitary–insulin-like growth factor (IGF)1 growth axis. Growth hormone (GH) and IGF1 regulate postnatal growth by common and independent pathways (24–26). Serum IGF1 concentration is a stable indicator of GH secretion during the postnatal growth phase, which regulates the cell cycle (24). The G2 phase in dividing cells of IGF1 null mice is prolonged four times (25).
In developing smpd3–/– mice (p10-p70,) serum IGF1 concentration was only approximately one-third the concentration of control sera (Fig. 3F). The hypoplasia of organs is due to a reduced cell number. The total number of splenocytes and of pituicytes of 1-, 6-, and 24-mo-old smpd3–/– and C57BL/6 control mice was reduced proportionate to the weight of spleen and pituitary (Fig. 3 G and H), but the cell morphology was indistinguishable from the corresponding wt tissues (not shown).
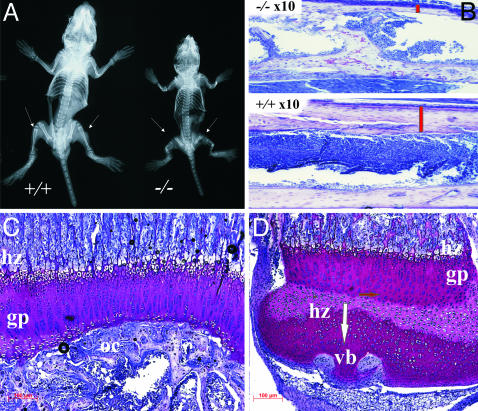
Growth retardation of the smpd3–/– mouse was most notably manifested in delayed ossification of long bones. X-ray imaging of the skeleton of wt and smpd3–/– mice (p42) revealed the short stature and deformations of long bones (Fig. 4A). Bone histology of smpd3-null mice underscored the immature architecture and the retarded ossification of bone cortices: only few vascular buds, the initial step of ossification, instead of the extended secondary ossification centers of age matched wt littermates, and small hypertrophic chondrocyte zones, the center of longitudinal growth (Fig. 4 B–D).
Fig. 4.
Growth retardation and deformations of the smpd3–/– skeleton. (A) X-ray image of 5-wk-old wt and homozygous smpd3–/– littermates. The long bones of homozygous mice are proportionally shorter, and bone structures are more translucent. Arrows, multiple deformations of femur, tibia, and fibula. (B) Cortices of tibia shafts of male wt and smpd3–/– mice, 6 wk of age. The red bar points to the different thickness of cortices. (C and D) Proximal tibia of smpd+/+ (C) and smpd3–/– (D) mice, ×40, 7-μm sections. gp, growth plate; hz, hypertrophic zone; oc, secondary ossification center; vb, vascular buds.
We therefore concluded that low IGF1 serum concentration in smpd3–/– mice might be an essential factor that contributes to the hypoplasia by a prolongation of the cell cycle and thereby growth retardation.
Growth Retardation Is Part of a CPHD. The pituitary gland of the smpd3–/– mutant showed a remarkable hypoplasia (Fig. 5A). The number of pituicytes is reduced (see Fig. 3H), and their nuclei appear small, dense, and shrunken. In addition to the dwarf phenotype, the onset of reproductive age of male and female smpd3–/– mice was delayed by 6–8 weeks. Homozygous crossings yielded on average about four to six litters. Genotyping of 25 litters from heterozygous crossings with 147 live births yielded non-Mendelian ratios (41 +/+, 78 +–, and 27 –/– pups). The distorted ratio indicated low embryonic lethality of smpd3–/– embryos.
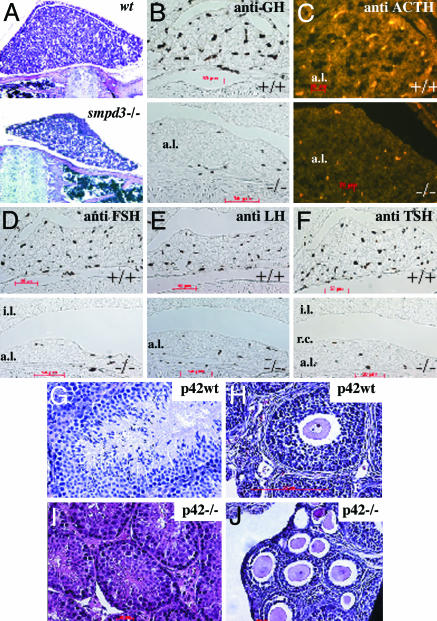
Fig. 5.
Histology and immunocytochemistry of pituitary. (A) Hematoxylin/eosin staining of sagittal sections of the pituitary of 8-wk-old male wt and smpd3–/– mice, ×120. Immunocytochemistry of paraffin-embedded sagittal sections of the pituitary of wt and smpd3–/– mice (p20) is shown. Antibodies stained only their specific peptide hormone antigen in the anterior pituitary lobe. (B–F) Primary antibody: anti-GH (B); anti-ACTH (C); anti-follicle-stimulating hormone (FSH) (D); anti-luteinizing hormone (E); anti-TSH (F). (B–D and F) Secondary antibodies: horseradish peroxidase-conjugated anti-rabbit IgG (B, D and F); Alexa-conjugated anti-rabbit IgG (C). a.l., anterior lobe; i.l., intermediate lobe. (Upper) Control +/+.(Lower) smpd3–/–. Light microscopy of cross sections: hematoxylin/eosin-stained testis in p42 wt (G) and p42 smpd3–/– (I) male mouse; p42 ovary of control wt (H) and smpd3–/– (J) female mouse.
We probed the functional state of the smpd3–/– pituitary by immunohistochemical comparison of the expression of GH, ACTH (corticotropin), thyroid-stimulating hormone (TSH), follicle-stimulating hormone (FSH), and luteinizing hormone (LH) in the anterior lobe of pituitary of control and smpd3–/– mouse brains. The number of anti-GH-labeled somatotrophic, anti-ACTH-labeled corticotrophic, anti-FSH- and LH-labeled gonadotrophic, and anti-TSH labeled thyreotrophic pituicytes in the anterior lobe of smpd3–/– pituitary were strongly reduced (Fig. 5 B–F). Histology of testes and ovaries of control and smpd3–/– siblings (p42) supported the immunohistochemical results (Fig. 5 G–J). Size and maturation of male and female gonads in smpd3-null mice lagged behind considerably. Seminiferous tubules of testes of p42 male controls contained mature sperm, but the round secondary spermatocytes in smpd3–/– testes indicated their early maturation stage (Fig. 5 G and I). Smpd3-null ovaries were small, with only few primordial and primarily preantral follicles with only two or a few layers of granulosa cells surrounding the zona pellucida, whereas wt ovary were filled with antral and preovulatory follicles embedded in a multilayer of granulosa cells (Fig. 5 H and J), which morphologically underscores the delayed puberty of smpd3–/– mice.
Preliminary determinations of T3/T4 and cortisol concentrations in pooled sera of cohorts of five animals each revealed that serum T3/T4 in adult (3 mo) smpd3–/– male mice (2.8 and 1.9 pg/ml) were not significantly different from age-matched control mice (3.3 and 1.8 pg/ml), but p20 mutant males had significantly reduced T3/T4 serum concentrations (1.8 and 1.2 pg/ml). Serum cortisol concentrations of p20 female smpd3–/– mice were 39 ng/ml and of male 32 ng/ml but 62 ng/ml in control male mice when measured at midphotophase.
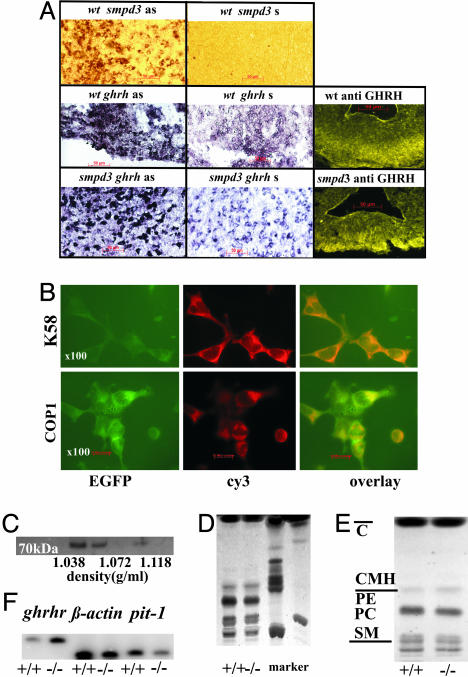
Smpd3 in the Regulation of the Hypothalamus Pituitary Axis. Smpd3 is predominantly expressed in CNS neurons. The CPHD phenotype and the immunohistochemical results of the pituitary of smpd3–/– mice suggested a perturbed hypothalamic secretion of releasing hormones, causing an impaired synthesis and secretion of anterior pituitary hormones. We focused our studies paradigmatically on the expression of smpd3 and ghrh in the hypothalamus by in situ hybridization and immunohistochemistry of coronal sections of brain of p10 to p20 wt and smpd3–/– mice by using smpd3 and ghrh digoxigenin-labeled antisense cRNA probes. Smpd3 specific mRNAs were strongly expressed in wt but not in smpd3-null neurons of the hypothalamic region (Fig. 6A). Neurons of the arcuate nucleus and paraventricular region of wt and smpd3–/– mice were not only heavily stained with antisense ghrh-specific cRNA, absent when hybridized with sense cRNA, but also in immunostaining using fluorescence (cy3) labeled anti-GH-releasing hormone (GHRH) antibodies (Fig. 6A).
Fig. 6.
smpd3 and ghrh expression in hypothalamic neurosecretory neurons. (A) In situ hybridization of coronal sections of wt (p20) mouse hypothalamus using digoxigenin-labeled antisense smpd3- and ghrh-specific cRNA probes, respectively, and immunofluorescence staining (cy3) using anti-GHRH antibodies. Extensive labeling of neurons expressing smpd3 and ghrh in neurosecretory hypothalamic neurons located in the paraventricular region and the arcuate nucleus is shown. (B) Colocalization of the SMPD3-EGFP fusion protein and Golgi markers K58 and coat protein I (COPI). (C) SMPD3 is segregated into DIMs of Golgi membranes. Western blot of proteins of Triton X-100 solubilized Golgi membrane fractions of three pooled WT mouse brains, Optiprep gradient fractions (density d = 1.038–1.072 g/ml). The affinity-purified anti-SMPD3 antibody was used. (D) High-performance thin-layer chromatography analysis of total lipid extract of Golgi membranes. (E) DIMs isolated from Golgi membranes. Markers: PC, phosphatidylcholine; PE, phosphatidylethanolamine; CMH, ceramide monohexoside; and C, cholesterol. (F) Expression of Pit1 and GHRH receptor in pituicytes of p20 control and smpd3–/– mice. Semiquantitative RT-PCR of brain and pituitary RNA was carried out as described in Supporting Text.
The specific labeling of hypothalamic neurosecretory neurons and the unaltered GHRH expression in semiquantitative RT-PCR (not shown) is consistent with the inhibition of GHRH secretion, which leads to down-regulation of GH synthesis and reduced secretion in pituitary somatotrophs smpd3-null mice.
We next studied the expression of ghrh receptor in somatotrophs by semiquantitative RT-PCR of pituitary RNA of p20 control and smpd3–/– mice. We observed a 4-fold up-regulation of the expression of ghrh receptor (Fig. 6F). This strongly suggested that the reduced circulating GHRH and GH synthesis caused a feedback response in somatotrophs of the pituitary of the mutant mouse.
SMPD3 Is Integrated into Triton X-100 Insoluble Segregated Subdomains of Golgi Membranes. To investigate the mechanism underlying the growth promoting role of the lipid modifying enzyme SMPD3, we established first the subcellular localization of SMPD3 in brain of p4 to p10 wt and smpd3–/– mice and in a HEK293 cell line stably expressing smpd3 and smpd3-egfp.
Immunohistochemical analysis demonstrated that the SMPD3–EGFP fusion protein colocalized with Golgi markers K58 and COP1 in HEK293 cells stably overexpressing smpd3-egfp (Fig. 6B). The EGFP tag did not interfere in the SMPD3 enzyme assay.
Purified Golgi membrane fractions from whole brain of p4–p10 wt and smpd3–/– mice and of HEK cells were isolated by gradient centrifugation for the biochemical analysis. Most of the SMPD3 activity separated with the Golgi fraction as shown by Western blot analysis (Fig. 6C). We dissociated purified Golgi membranes from brain of control and smpd3–/– mice as well as of spmd3-overexpressing HEK cells with Triton X-100 and separated the DIM fraction from the Triton X-100 soluble fraction by gradient centrifugation. The total SMPD3 activity of the Golgi fraction was concentrated in the DIM fraction. Western blot analysis detected SMPD3 as 70-kDa protein only in DIMs (Fig. 6D). Lipid extracts of the Golgi membranes and of Golgi derived DIMs indicated the enrichment of cholesterol and sphingolipids in these fractions (Fig. 6 D and F).
The colocalization of the neutral SMase SMPD3 and its substrate SM in these defined membrane subdomains of the Golgi suggests that SMPD3 might play a critical role in the regulated Golgi secretory vesicle trafficking of neuroendocrine cells of hypothalamic nuclei.
Discussion
We have studied the role of neutral SMases in two null allelic mouse mutants, the smpd3–/– mutant, deficient in the main representative among the SMPD, and in the double mutant smpd2–/–smpd3–/–, in which the two nSMases smpd2 and smpd3 were deleted. The double mutant turned out to be completely devoid of neutral SMase activity.
The first unexpected result was that, unlike the smpd1–/–, or Niemann–Pick mouse, which exhibits massive SM storage in lysosomes of cells of the reticuloendothelial system, smpd3–/– and smpd2–/–smpd3–/– mice developed no SM storage abnormalities. SMPD3 is integrated in enzymatically inactive form in Golgi membrane subdomains and requires a regulated cell-specific activation, the mechanism(s) of which remains to be examined.
SMPD3 Deficiency Causes Dwarfism and Delayed Puberty. Two dominant phenotypic signs govern the smpd3–/– and the smpd2–/– smpd3–/– mouse lines: the growth retardation of the mutants recognizable around e14 and postnatally through all stages of development and delayed gonadal maturation.
Hypothalamic GHRH regulates pituitary GH secretion, the fundamental determinant of body size and weight. GH stimulates the hepatic synthesis and secretion of IGF1, the mediator of GH action on all target tissues (for review, see refs. 26 and 27). This dogma underwent considerable modifications when liver-specific IGF1 deletion suggested that nonhepatic tissue derived circulating IGF1 is sufficient for growth and development (28, 29). IGF1 regulates the cell cycle, as demonstrated by the igf1-null mutant mouse (28, 30).
Low-serum IGF1 leads to the profound prolongation of the cell cycle in developing smpd3–/– mice, which causes the reduced number of cells in most tissues as shown here paradigmatically for splenocytes and pituicytes.
The function of IGF1 in the regulation of skeletal growth and development is well known (28, 31). Reduced pituitary GH synthesis and IGF1 serum concentration in smpd3–/– mice explain the retardation of chondrocyte proliferation, differentiation in the epiphyseal growth plate and secondary ossification centers, and of the calcification in metaphyseal long bone.
Isolated GH deficiency results from the disruption of the growth axis by mutations of critical components at different sites, e.g., of the GH receptor gene (ghr) causing Laron syndrome (32) of the GHRH receptor gene in the “little mouse” (33, 34) or the deletion of IGF1 expression and of the IGF1 receptor (28, 30).
Two spontaneous mouse mutants, the Snell mouse (17) and the Ames dwarf mouse (18), originate from mutations of pituitary-specific transcription factors, Pit1 and Prop1, which regulate the expression of the ghrh receptor and of the GH, βTSH, and PRL genes in embryonic lineage development of pituicytes (19).
smpd3–/– mutant mice show a normal developmentally dependent expression of Prop1 and Pit1, as documented by quantitative RT-PCR of e10 to e18 and adult pituitary mRNA (data not shown). In p20 mutant pituitary prop1 expression was no longer detectable, as expected (18), pit1 expression in control and mutant mice was indistinguishable, but ghrh receptor expression was up-regulated 4-fold in the smpd3–/– pituitary (Fig. 6F).
A striking phenotypic similarity exists also between the smpd3–/– mouse and the proprotein convertases phosphatidylcholine-(PC)1/3 (SPC3) deficient mouse (20). Convertases are endoproteolytic enzymes of the regulated secretory pathway in hypothalamic neuroendocrine and the extraneuronal endocrine system. PC1/3 disruption in the mouse causes a lack of circulating mature GHRH and low pituitary GH and IGF1 serum concentrations (21). The regulated release of hypothalamic releasing hormones maintains the structure and function of the pituitary.
Our in situ hybridization studies combined with immunohistochemistry expression studies and semiquantitative RT-PCR demonstrated that ghrh expression and synthesis in neurosecretory neurons of the hypothalamic arcuate nucleus and paraventricular area of wt as well as smpd3-null mice was not inhibited (Fig. 6A). These results are consistent with an inhibition of GHRH secretion. Low circulating GHRH triggers a feedback response in somatotrophs to an elevated expression of ghrhr (Fig. 6F).
The smpd3–/– mouse develops a CPHD. Not only was the number of GH expressing somatotrophic cells strongly reduced but also ACTH, follicle-stimulating hormone, luteinizing hormone, and TSH synthesizing pituicytes were markedly reduced in number and function, leading to the hypoplasia of the pituitary gland of the smpd3-null mutant (Fig. 5 A–F), as the molecular basis of the combined pituitary hormone deficiency due to a defective release mechanism. Reduced serum T3/T4 and cortisol concentration in developing smpd3–/– mice compared with control mice support these immunohistochemical, morphological, and gene expression results.
A Mechanistic View of the Role of SMPD3 in the Golgi of Neurosecretory Neurons. How can we envisage the function of brain-specific SMPD3, a membrane lipid-modifying enzyme, in the control of postnatal development?
A clue to the function of SMPD3 came from topological studies of SMPD3 in the subdomains of Golgi membranes. The total SMPD3 of the Golgi segregated with Triton X-100 DIMs, a fraction of which is enriched in cholesterol, SM, and glycosphingolipid, respectively. We concluded that SMPD3 and its substrate SM are segregated together in the sphingolipid-enriched Golgi DIMs. The putative topology of SMPD3, integrated in the Golgi membrane with two transmembrane domains (amino acids 15–37 and 57–79), each 23 amino acid residues long, suggests an orientation of the catalytic C-terminal part toward the lumen of the Golgi (16). The bilayer dimensions of the DIMs favor the selective sorting of SMPD3 into their liquid-ordered bilayer (47- to 56-Å spacing), segregated from phospholipid-rich cholesterol-poor liquid-disordered domains (35–40 Å) (36, 37). SM hydrolysis induces the segregation of cholesterol from the SM/cholesterol clusters, thereby rearranging the bilayer between the different domains. These dynamics might initiate or facilitate the budding of the phospholipid-rich SM-depleted coat protein I (COPI) vesicles (38) and leave SM/cholesterol remnant domains of the trans-Golgi network for vesicular transport to the plasma membrane. SM synthase (phosphatidylcholine ceramide phosphocholine transferase) is also localized in the Golgi compartment (39, 40). The association and cooperation of both SMPD3 and the SM synthase might be required for the maintenance of the domain structures of the Golgi secretory pathway.
The two null mouse models allowed the assessment of the role of SMPD3 in the Golgi secretory pathway of hypothalamic neurosecretory neurons and provide an aspect of a previously undescribed function of a lipid-modifying enzyme in Golgi vesicle trafficking.
Our studies strongly support the role of smpd3 as a regulator of postnatal development. The observed CPHD reflects the perturbed secretion of releasing hormones, which lead to the hypoplasia of the anterior pituitary. Further studies of the smpd3–/– and smpd2–/–smpd3–/– mutants may reveal still other functions of its regulated activation. Promoter studies might elucidate the tissue specific and development dependent regulation of smpd3 expression.
Interruption of the insulin/IGF1-like daf2 pathway in Caenorhabditis elegans extends its lifespan by 30–100% (41). Pit1 (Ames) and Prop1 (Snell) hypopituitary dwarf mice are mammalian mutants with prolonged longevity, 50% in male and 64% in female, which has been attributed to so-far-unknown intrinsic mechanisms in GH/IGF1 deficiency (42, 43). In view of these observations, it will be interesting to follow the lifespan of smpd3–/– mice.
Finally, the phenotype of smpd3–/– mice might have important clinical implications for understanding the genetics of human CPHDs. Beyond the mutations of transcription factor Pit1 and Prop1, a molecular understanding of the major group of CPHD with short stature-delayed puberty is missing. The studies reported here might give access to the molecular and genetic basis of one of the several forms of dwarfism with unknown etiology.
Supplementary Material
Acknowledgments
This project was supported by the Center for Molecular Medicine, Cologne (CMMC), the Deutsche Forschungsgemeinschaft (Sto32/38-2), and the Günther and Arina Lauffs Foundation.
Author contributions: W.S. designed research; W.S. and M.Z. performed research; J.K. contributed new reagents/analytic tools; W.S., B.J., B.B., and M.Z. analyzed data; and W.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SM, sphingomyelin; SMPD, SM phosphodiesterase; SMase, sphingomyelinase; nSMase, neutral SMase; CPHD, combined pituitary hormone deficiency; DIM, detergent insoluble membrane subdomain; HEK, human embryonic kidney; pn, postnatal day n; en, embryonic day n; GH, growth hormone; TSH, thyroid-stimulating hormone; IGF, insulin-like growth factor; GHRH, GH-releasing hormone.
References
- 1.Kanfer, J. N., Young, O. M., Shapiro, D. & Brady, R. O. (1966) J. Biol. Chem. 241, 1081–1084. [PubMed] [Google Scholar]
- 2.Barnholz, Y., Roitman, A. & Gatt, S. (1966) J. Biol. Chem. 241, 3731–3737. [PubMed] [Google Scholar]
- 3.Quintern, L. E., Weitz, G., Nehrkorn, H., Tager, J. M., Schram, A. W. & Sandhoff, K. (1987) Biochim. Biophys. Acta 922, 323–336. [DOI] [PubMed] [Google Scholar]
- 4.Quintern, L. E., Schuchman, E. H., Levran, O., Suchi, M., Ferlinz, K., Reinke, H., Sandhoff, K. & Desnick, R. J. (1989) EMBO J. 8, 2469–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newrzella, D. & Stoffel, W. (1992) Biol. Chem. Hoppe-Seyler 373, 1233–1238. [DOI] [PubMed] [Google Scholar]
- 6.Newrzella, D. & Stoffel, W. (1996) J. Biol. Chem. 271, 32089–32095. [DOI] [PubMed] [Google Scholar]
- 7.Otterbach, B. & Stoffel, W. (1995) Cell 81, 1053–1061. [DOI] [PubMed] [Google Scholar]
- 8.Horinouchi, K., Erlich, S., Perl, D. P., Ferlinz, K., Bisgaier, C. L., Sandhoff, K., Desnick, R. J., Stewart, C. L. & Schuchman, E. H. (1995) Nat. Genet. 10, 288–293. [DOI] [PubMed] [Google Scholar]
- 9.Spence, M. W. & Burgess, J. K. (1978) J. Neurochem. 30, 917–919. [DOI] [PubMed] [Google Scholar]
- 10.Kolesnick, R. N. & Kronke, M. (1998) Annu. Rev. Physiol. 60, 643–665. [DOI] [PubMed] [Google Scholar]
- 11.Hannun, Y. A. & Obeid, L. M. (2002) J. Biol. Chem. 277, 25847–25850. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann, K. & Dixit, V. M. (1998) Trends Biochem. Sci. 23, 374–377. [DOI] [PubMed] [Google Scholar]
- 13.Zumbansen, M. & Stoffel, W. (2002) Mol. Cell. Biol. 22, 3633–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomiuk, S., Hofmann, K., Nix, M., Zumbansen, M. & Stoffel, W. (1998) Proc. Natl. Acad. Sci. USA 95, 3638–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomiuk, S., Zumbansen, M. & Stoffel, W. (2000) J. Biol. Chem. 275, 5710–5717. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann, K., Tomiuk, S., Wolff, G. & Stoffel, W. (2000) Proc. Natl. Acad. Sci. USA 97, 5895–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, S., Crenshaw, E. B., 3rd, Rawson, E. J., Simmons, D. M., Swanson, L. W. & Rosenfeld, M. G. (1990) Nature 347, 528–533. [DOI] [PubMed] [Google Scholar]
- 18.Sornson, M. W., Wu, W., Dasen, J. S., Flynn, S. E., Norman, D. J., O'Connell, S. M., Gukovsky, I., Carriere, C., Ryan, A. K., Miller, A. P., et al. (1996) Nature 384, 327–333. [DOI] [PubMed] [Google Scholar]
- 19.Radovick, S., Nations, M., Du, Y., Berg, L. A., Weintraub, B. D. & Wondisford, F. E. (1992) Science 257, 1115–1118. [DOI] [PubMed] [Google Scholar]
- 20.Furuta, M., Yano, H., Zhou, A., Rouille, Y., Holst, J. J., Carroll, R., Ravazzola, M., Orci, L., Furuta, H. & Steiner, D. F. (1997) Proc. Natl. Acad. Sci. USA 94, 6646–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu, X., Zhou, A., Dey, A., Norrbom, C., Carroll, R., Zhang, C., Laurent, V., Lindberg, I., Ugleholdt, R., Holst, J. J. & Steiner, D. F. (2002) Proc. Natl. Acad. Sci. USA 99, 10293–10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson, P. A. & Melton, D. A. (1994) Curr. Biol. 4, 676–686. [DOI] [PubMed] [Google Scholar]
- 23.Simons, M., Kramer, E. M., Thiele, C., Stoffel, W. & Trotter, J. (2000) J. Cell Biol. 151, 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bligh, E. & Dyer, W. (1959) Can. J. Biochem. Physiol. 37, 911–917. [DOI] [PubMed] [Google Scholar]
- 25.Zumbansen, M. & Stoffel, W. (1997) J. Biol. Chem. 272, 10904–10909. [DOI] [PubMed] [Google Scholar]
- 26.Daughaday, W. H. & Rotwein, P. (1989) Endocr. Rev. 10, 68–91. [DOI] [PubMed] [Google Scholar]
- 27.Rubin, R. & Baserga, R. (1995) Lab. Invest. 73, 311–331. [PubMed] [Google Scholar]
- 28.Lupu, F., Terwilliger, J. D., Lee, K., Segre, G. V. & Efstratiadis, A. (2001) Dev. Biol. 229, 141–162. [DOI] [PubMed] [Google Scholar]
- 29.Butler, A. A. & Le Roith, D. (2001) Annu. Rev. Physiol. 63, 141–164. [DOI] [PubMed] [Google Scholar]
- 30.Efstratiadis, A. (1998) Int. J. Dev. Biol. 42, 955–976. [PubMed] [Google Scholar]
- 31.Ohlsson, C., Bengtsson, B. A., Isaksson, O. G., Andreassen, T. T. & Slootweg, M. C. (1998) Endocr. Rev. 19, 55–79. [DOI] [PubMed] [Google Scholar]
- 32.Amselem, S., Duquesnoy, P., Attree, O., Novelli, G., Bousnina, S., Postel-Vinay, M. C. & Goossens, M. (1989) N. Engl. J. Med. 321, 989–995. [DOI] [PubMed] [Google Scholar]
- 33.Kopchick, J. J. & Laron, Z. (1999) Mol. Genet. Metab. 68, 232–236. [DOI] [PubMed] [Google Scholar]
- 34.Lin, S. C., Lin, C. R., Gukovsky, I., Lusis, A. J., Sawchenko, P. E. & Rosenfeld, M. G. (1993) Nature 364, 208–213. [DOI] [PubMed] [Google Scholar]
- 35.Gaylinn, B. D., Dealmeida, V. I., Lyons, C. E., Jr., Wu, K. C., Mayo, K. E. & Thorner, M. O. (1999) Endocrinology 140, 5066–5074. [DOI] [PubMed] [Google Scholar]
- 36.Bretscher, M. S. & Munro, S. (1993) Science 261, 1280–1281. [DOI] [PubMed] [Google Scholar]
- 37.Hanada, K., Nishijima, M., Akamatsu, Y. & Pagano, R. E. (1995) J. Biol. Chem. 270, 6254–6260. [DOI] [PubMed] [Google Scholar]
- 38.Brugger, B., Sandhoff, R., Wegehingel, S., Gorgas, K., Malsam, J., Helms, J. B., Lehmann, W. D., Nickel, W. & Wieland, F. T. (2000) J. Cell Biol. 151, 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Futerman, A. H., Stieger, B., Hubbard, A. L. & Pagano, R. E. (1990) J. Biol. Chem. 265, 8650–8657. [PubMed] [Google Scholar]
- 40.Jeckel, D., Karrenbauer, A., Birk, R., Schmidt, R. R. & Wieland, F. (1990) FEBS Lett. 261, 155–157. [DOI] [PubMed] [Google Scholar]
- 41.Kenyon, C., Chang, J., Gensch, E., Rudner, A. & Tabtiang, R. (1993) Nature 366, 461–464. [DOI] [PubMed] [Google Scholar]
- 42.Baker, J., Liu, J. P., Robertson, E. J. & Efstratiadis, A. (1993) Cell 75, 73–82. [PubMed] [Google Scholar]
- 43.Carter, C. S., Ramsey, M. M., Ingram, R. L., Cashion, A. B., Cefalu, W. T., Wang, Z. Q. & Sonntag, W. E. (2002) J. Gerontol. A 57, B177–B188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.