Abstract
Synapses are functional units of the nervous system, through which information is transferred between neurons. The development and activity-dependent modification of synapses require temporally and spatially controlled modulation of gene expression. microRNAs (miRNAs) have emerged as essential regulators of gene expression. They are small non-coding RNAs that regulate mRNA stability and translation by interacting with the 3′ untranslated region (3′ UTR) of mRNAs. miRNAs are located to neuronal processes to regulate protein synthesis locally and their expression is regulated by synaptic activity. This article reviews recent findings on the role of miRNAs in synapse development and synaptic plasticity.
Introduction
The development and physiological activities of the nervous system are shaped by the genome and refined by epigenetic mechanisms. microRNAs (miRNAs), a type of small non-coding RNAs, play key roles in the epigenetic, post-transcriptional regulation of gene expression. miRNAs are generated in three-steps: (1) the transcription of miRNA genes into primary miRNAs (pri-miRNAs); (2) the cleavage of pri-miRNAs into precursor miRNAs (pre-miRNAs) by the nuclear RNase III Drosha; (3) the cleavage of pre-miRNAs into miRNA duplexes (miRNA:miRNA*) by the cytoplasmic RNase III Dicer [1]. Mature miRNAs are integrated into the RNA-induced silencing complex (RISC) where they bind to the 3′ untranslated region (UTR) of mRNAs to destabilize mRNAs or inhibit translation [1].
Since the discovery of the first miRNA, Lin-4 in C. elegans [2,3], > 2000 miRNAs have been identified and most of them are expressed in the brain [4]. miRNAs have been implicated in various cellular activities including cell proliferation, differentiation, migration, development, death, etc. Accumulating evidence indicates that in the nervous system, miRNAs play a pivotal role in the regulation of synapses. This review focuses on recent work on the functions of miRNAs in synapse development and synaptic plasticity.
miRNAs are produced and act near synapses
The majority of mammalian synapses are formed between elaborate dendrites and long axons, both of which extend considerate distances from the soma. The long distances pose great challenges for neurons to supply remote synapses with structural and signaling molecules. Local protein synthesis near synapses, therefore, is an effective way to timely and spatially control synaptic compositions. Like mRNAs and polyribosomes, miRNAs are constituents of dendrites, axons and dendritic spines [5]. Using subcellular fractionation, Lugli G et al. found a large number of miRNAs in synaptic fractions, including synaptoneurosomes (enriched for dendritic spines), synaptosomes (predominately comprised of axon terminals with adherent postsynaptic densities) and postsynaptic densities [5]. Moreover, a subset of these miRNAs is expressed at higher levels in synaptic fractions than in the whole cell lysate [5]. Synaptoneurosomes prepared from the rat nucleus accumbens also contain miRNAs [6], but miRNAs enriched in synaptoneurosomes prepared from the nucleus accumbens and the forebrain are different, indicative of a region-specificity of synaptic miRNAs. miRNAs are also present in the axons of rat superior cervical ganglion neurons and in the developing axons of mouse dorsal root ganglion neurons [7,8]. Consistent with the subcellular fractionation experiment, miRNAs are detected in dendrites, dendritic spines, synapses, and axons by in situ hybridization [8-13].
Not only mature miRNAs, pre-miRNAs and Dicer are also located to dendrites and axons [5,14-16], and pre-miRNAs can be cleaved into mature miRNAs near synapses [9,10,15,16]. Current data indicate that NMDA receptors regulate the processing of dendritic pre-miRNAs, but little is known about the underlying mechanism [9,10,15]. A proposed model is that NMDA receptor opening and Ca2+ influx upon synaptic excitation lead to proteolysis and activation of Dicer, which is inactive at the resting state [15]. This model, however, has yet to be experimentally proved. How do miRNAs and pre-miRNAs reach distal synapses? Only pre-miR-134 transportation has been delineated till now. Pre-miR-134 is targeted to dendrites by the DEAH-box helicase DHX36 which binds to its terminal loop [16]. The DHX36-binding site in pre-miR-134, however, is not present in the terminal loops of other dendritic pre-miRNAs, suggesting that the transport mechanism is pre-miRNA specific. Mature miRNAs in neuronal processes can derive from local pre-miRNAs, and can also come from direct delivery from the soma, but dendritic or axonal targeting of mature miRNAs has not been experimentally demonstrated. Given the short length of mature miRNAs, it is less likely that they are targeted to dendrites, axons and synapses through RNA-binding proteins. An advantage of having pre-miRNAs and Dicer in neuronal processes is that pre-miRNAs can be rapidly converted to mature miRNAs locally when needed, therefore serving as a stock of mature miRNAs and a means of localizing specific miRNAs to subcellular structures. What do miRNAs do in neuronal processes? Current data indicate that miRNAs near synapses mainly modulate translation in a neural activity-dependent manner, and the underlying mechanism has been unfolded for several miRNAs. At resting synapses, miR-138 sequesters the Lypla1 mRNA in RISC to prevent it from being accessible to polysomes [17]. Upon NMDA receptor activation, the RISC component MOV10 is degraded, leading to the release of Lypla1 mRNAs from miR-138 sequestration and subsequent translation [17]. miR-129 competes with the RNA-binding protein HuD for binding to mRNAs encoding the dendritic voltage-gated potassium channel Kv1.1 [18]. HuD is stabilized when mTORC1 (an activity-regulated kinase) is inactivated, thereby promoting Kv1.1 mRNA translation by displacing miR-129 [18]. miR-125a inhibits PSD-95 mRNA translation by forming a complex with phospho-FMRP and AGO2 on the PSD-95 mRNA, but this inhibitory complex is disassembled upon FMRP depohosphorylation by mGluR activation [19]. Repression of the AMPA receptor subunit GluA1 translation by miR-501-3p in dendrites is enhanced by NMDA receptor activation during the induction of long-term synaptic depression (LTD) [9]. In these examples, the mRNAs whose translation is locally regulated by miRNAs reside in dendrites and synapses. In addition to these mRNAs, next generation sequencing has identified >2500 mRNAs in dendrites [20]. It is likely that miRNAs regulate the translation of many of these dendritic mRNAs. Activity-dependent regulation of local translation by miRNAs can exert noticeable effect on dendritic spines and synaptic plasticity [9,10,13].
miRNAs in synaptic transmission and synaptic plasticity
The fact that at least 30% human genes are targeted by miRNAs [21] underscores the importance of miRNAs in gene expression regulation. Global attenuation of miRNA biogenesis or miRNA activity has considerable effects on synapses. In heterozygous Dgcr8 (a component of the microprocessor complex cleaving pri-miRNAs into pre-miRNAs) knockout mice, although basal synaptic transmission is intact, 50 Hz stimulation-induced synaptic depression is enhanced and the initial phase of LTP is impaired in the prefrontal cortex [22], but LTP in the hippocampus is increased [23]. Inactivation of Dicer1 in hippocampal excitatory neurons causes increases in dendritic spine length, neural excitability, and post-tetanic potentiation [24-26]. Lower RISC activity following knockdown of Ncoa3, a transcriptional co-activator promoting the transcription of Ago2 (a core component of RISC) results in decreases in the volume of dendritic spines and the amplitude of miniature excitatory postsynaptic currents (mEPSC) [27].
The diversity of genes that each miRNA targets to predicts that different miRNAs have distinct functions. Hence, experiments perturbing only one miRNA at one time have been conducted to tease out individual miRNAs' functions. Such experiments found that several miRNAs including miR-125b, miR-223, miR-137, and miR-146a-5p influence the size of postsynaptic responses by controlling the abundance of postsynaptic glutamate receptors. miR-125b regulates expression of the NMDA receptor subunit GluN2A, and its overexpression in hippocampal neurons causes a decrease in mEPSC amplitude [28]. miR-223 targets the NMDA receptor subunit GluN2B and the AMPA receptor subunit GluA2 [29]. In the hippocampus of miR-223 knockout mice, the mEPSC amplitude is increased and the decay time of NMDA receptor-mediated mEPSCs is prolonged [29]. miR-137 represses expression of the AMPA receptor subunit GluA1 to control AMPA receptor-mediated synaptic currents and the number of functional synapses [30]. miR-146a-5p does not directly target glutamate receptors, but controls the number of AMPA receptors in synapses and synaptic transmission by targeting the microtubule-associated protein 1B which modulates AMPA receptor endocytosis [31].
miRNAs also target signaling and presynaptic proteins. For example, miR-188 regulates dendritic spine number and mEPSC frequency in hippocampal neurons by targeting the semaphorin-3F receptor Nrp-2 [32]; Drosophila miR-1000 suppresses synaptic glutamate release by targeting the vesicular glutamate transporter (Vglut) [33]; and in addition to GluA1, miR-137 also targets proteins in the synaptic vesicle release machinery including complexin-1, N-thylmaleimide-sensitive fusion protein (Nsf) and synaptotagmin-1 to reduce the number of synaptic vesicles proximal to the release site, and frequency facilitation at the mossy fiber-CA3 synapse [34]. miR-132 is also found to modulate synapse number and mEPSCs [28,35,36], but the underlying mechanism is obscure. These findings indicate that miRNAs tune synaptic transmission by targeting the structural and signaling proteins of synapses, and that some miRNAs have both presynaptic and postsynaptic targets.
In addition to basal synaptic transmission, miRNAs have been implicated in various forms of protein synthesis-dependent synaptic plasticity. These miRNAs often change their abundance in response to synaptic stimulation. Among them, miR-26a and miR-384-5p are downregulated in NMDA receptor-dependent LTP to increase the expression of their common target ribosomal S6 kinase 3 (RSK3, a translational regulator), and the resulting translational enhancement is required for long-lasting LTP [13]; miR-124 and miR-22 are reduced in 5-HT induced long-term facilitation (LTF) in Aplysia, leading to increases in the transcription factor CREB and the translational regulator cytoplasmic polyadenylating element binding protein (CPEB), two proteins required to maintain LTF [12,37]; miR-124 is increased by neuronal inhibition to repress GluA2 expression, thereby promoting the expression of calcium-permeable AMPA receptors for homeostatic synaptic plasticity [38]; miR-137 is increased by the mGluR1 agonist DHPG to mediate LTD [30].
Some miRNAs contribute to multiple forms of synaptic plasticity, possibly via different targets. In addition to regulating 5-HT induced LTF and homeostatic synaptic plasticity, miR-124 inhibits NMDA receptor-dependent LTP by controlling Zif268 translation, and knockdown of miR-124 restores LTP in EPAC (an exchange protein directly activated by cAMP) knockout mice [39]. While miR-137 mediates mGluR-dependent LTD [30], it inhibits the induction of mossy fiber LTP in the hippocampus [34]. miR-132 overexpression inhibits both LTP and carbachol-induced LTD in the perirhinal cortex [40], and deleting miR-132 and miR-212 in mice causes aberrant theta burst-induced LTP [41].
miRNAs in the morphogenesis and structural plasticity of neurons
In addition to synaptic function, numerous studies show that miRNAs regulate the development and plasticity of neuronal structures. Neurons need miRNAs to develop and maintain dendrites, axons and dendritic spines. In Ncoa3 knockdown neurons which have a low RISC activity, dendrites become more complex and bear smaller dendritic spines [27]. Deletion of Dicer1 in the adult mouse forebrain shifts dendritic arborization towards distal dendrites and elongates dendritic spines [25]. Specific miRNAs involved in neuronal morphogenesis have begun to be recognized. miR-29a/b (by targeting the actin nucleation factor Arpc3) inhibits the conversion of filopodia into mature dendritic spines [42]; miR-125 promotes the formation of long and thin dendritic protrusions [28]; miR-185 is required for spine formation and dendritic branching and growth [43]; miR-134 (by targeting Limk1), let-7, miR-22, and miR-124 constrain spine width [11,28]; miR-138 (by targeting the depalmitoylation enzyme APT1) inhibits spine volume [44]; and miR-134 increases activity-dependent dendritic complexity by targeting the translational repressor Pumilio2 [45]. miR-132 and miR-181 appear to participate in general neuronal morphogenesis, as miR-132 promotes the extension of DRG neuron axons by targeting the Ras GTPase activator Rasa1, increases dendritic length and induces spine formation by targeting p250GAP, and widens dendritic protrusions [8,35,36], while miR-181 promotes spine formation and attenuates dendritic branching and axonal outgrowth [46].
The number and size of dendritic spines and synapses change not only during development, but also during synaptic plasticity. LTP is accompanied by spine enlargement and formation of new spines, spine shrinkage and elimination are often observed in LTD, and homeostatic synaptic scaling is also associated with elimination or formation of spines and synapses. Translation has been implicated in the long-lasting spine remodeling associated with LTP and LTD requires [10,13], and miRNAs are shown to be key regulators of this process. Using time-lapse imaging of live hippocampal neurons, several miRNAs are found to contribute to the structural plasticity of dendritic spines associated with LTP and LTD. In NMDA receptor-dependent LTP,miR-384-5p is involved in the initiation of spine enlargement, and miR-26a is responsible for maintaining the size of dendritic spines after they become larger [13]. In NMDA receptor-dependent LTD, miR-191 inhibits spine elimination and long-lasting spine shrinkage by repressing the expression of tropomodulin-2 which promotes actin depolymerization, while miR-135 promotes spine remodeling by reducing the expression of complexin1/2 to inhibit AMPA receptor exocytosis [10]. The elimination of spines and synapses occurring in homeostatic synaptic depression is dependent on miRNAs including miR-485 and miR-134, both of which are upregulated by neural activity [47,48]. The function of miR-485 in homeostatic synaptic scaling is conferred by its target the presynaptic protein SV2A, while Pumilio-2 is the responsible target of miR-134 [47,48].
miRNAs in memory
Synaptic plasticity is a cellular mechanism through which the brain encodes and stores information. Synaptic plasticity, therefore, subserves cognitive functions such as learning and memory. In line with their involvement in synaptic plasticity, miRNAs have been recognized as key players in the formation and retrieval of memory. This is first demonstrated in conditional Dicer1 knockout mice which have massive miRNA loss in forebrain excitatory neurons. These mice exhibit enhanced hippocampus-dependent spatial memory in the Morris water maze test and fear memory in the trace and the contextual fear conditioning test [25,26]. Since then, the importance of miRNAs in memory has been confirmed by experiments perturbing individual miRNAs. Using miRNA sponges to inhibit miRNAs in the mouse hippocampus, it is shown that while miR-124 restricts, miR-9 and miR-34 maintain the capacity of spatial learning in the Morris water maze test [49]. miR-132 overexpression in mouse forebrain excitatory neurons impairs novel object recognition memory [50], and overexpressing miR-137 in the dentate gyrus impairs contextual fear memory [34].
Learning can change miRNA abundance, and numerous studies have demonstrated that regulating miRNA expression is necessary for memory formation. Fear-extinction learning in mice leads to increased expression of miR-128b in the infralimbic prefrontal cortex to facilitate the transition from retrieving original fear memory to forming a fear-extinction memory [51]. miR-92 is increased by contextual fear conditioning in the mouse hippocampus to promote contextual fear memory [52]. miR-182 expression is suppressed in the rat amygdala by auditory fear conditioning to support the formation of long-term auditory fear memory [53]. miR-33 in the mouse hippocampus is elevated by contextual fear conditioning to modulate the encoding and retrieval of state-dependent fear [54]. miRNAs in the miR-183/96/182 cluster of mice increase after training with an object recognition task through a protein phosphatase PP1-dependent enhancement of pre-miRNA production [55]. This increase in miRNAs leads to downregulation of the histone deacetylase HDAC9, thereby permitting the formation of long-term object memory [55]. In honeybees, miR-932 is increased by an associative olfactory learning paradigm to regulate long-term memory recall [56].
Concluding remarks
miRNAs in neurons not only modulate translation in the soma as in other types of cells, but also are integral components of the local translational machinery in dendrites and axons. miRNAs target a wide variety of synaptic proteins and translational regulators to fine-tune their expression levels. Mounting evidence indicates that miRNA-mediated mechanisms are essential to establish appropriate synapse number and spine morphology, to adjust synaptic strength and dendritic spines during synaptic plasticity, and to create a permissive cellular environment for cognitive activities.
The number of mammalian miRNAs is in thousands now and still growing. Altering one miRNA at one time is commonly used to study miRNAs as it allows for a clean inspection of individual miRNAs' functions. However, this approach is low throughput so that only a handful of miRNAs have been functionally characterized. Also, since one gene can be targeted by multiple miRNAs, and neural activity usually alters many miRNAs concurrently, technologies capable of manipulating multiple miRNAs at the same time, in the same cell are needed to obtain a comprehensive picture of the miRNA functionality. Future research is also needed to investigate such questions as how miRNAs are targeted to and regulated in neuronal processes, in particular axons, and why the functions of miRNAs are brain region specific.
Figure 1.
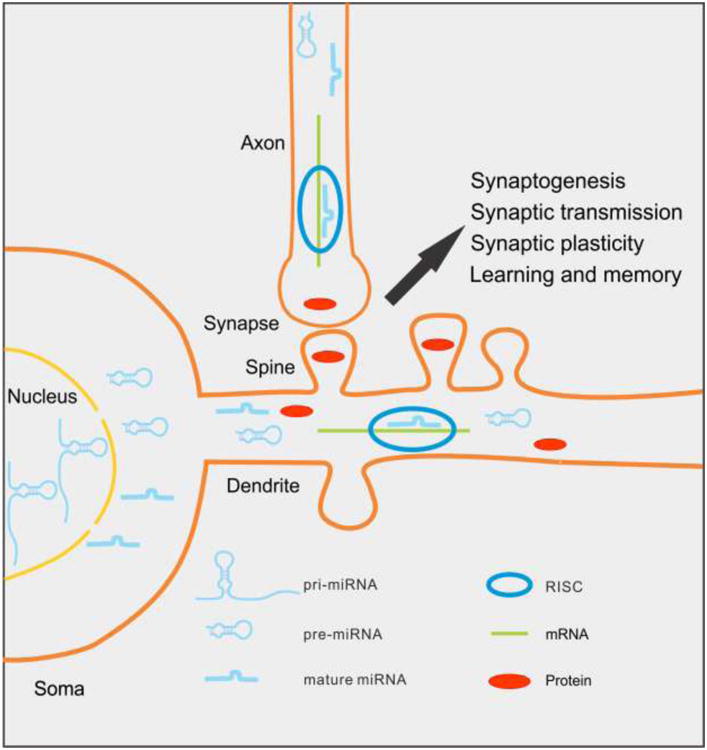
miRNAs are localized in dendrites and axons, and can regulate synapse development and synaptic plasticity locally. Pri-miRNAs are transcribed as long transcripts in the nucleus, and then processed into pre-miRNAs by Drosha and DGCR8. Pre-miRNAs, which form hairpin structures, are cleaved by Dicer to generate mature miRNAs. The pre-miRNAs and mature miRNAs are located in dendrites and axons, and enriched in synaptic fractions. Mature miRNAs are loaded into RISC to regulate gene expression post-transcriptionally, thereby influencing synapse development, synaptic plasticity, and cognition.
Figure 2.
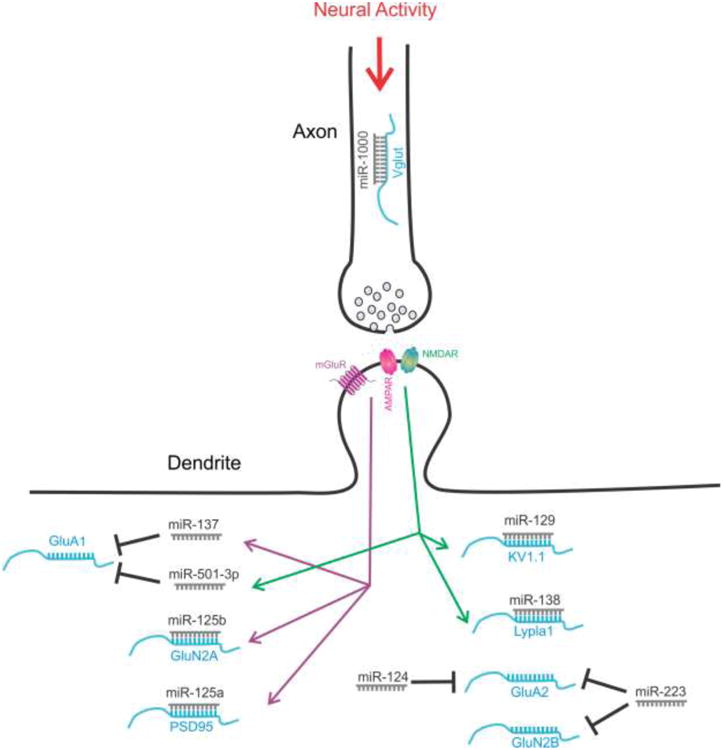
miRNAs are regulated by neural activity. Neural activity-dependent Drosophila miR-1000 regulates glutamate release by targeting Vglut in axons. In dendrites, NMDAR regulates miR-129 (targeting Kv1.1), miR-138 (targeting Lypla1), and miR-501-3p (targeting GluA1); mGluR regulates miR-125a (targeting PSD95), miR-125b (targeting GluN2A), and miR-137 (targeting GluA1); TTX and APV increase miR-124 which suppresses GluA2; miR-223 regulates GluA2 and GluN2B to prevent neuronal excitotoxicity.
Table 1.
Summary of miRNAs including their functions and targets that are discussed in this review.
| miRNA | Target | Function | Species studied | Reference |
|---|---|---|---|---|
| miR-1000 | VGlut | glutamate release, apoptosis | Drosophila | Verma, P., et al., (2015) |
| miR-124 | NA | NMDAR-LTP, spatial learning | Rodent | Yang, Y., et al., (2012) |
| miR-124 | CREB | long-term synaptic facilitation in sensory-motor synapse | Aplysia | Rajasethupathy, P., et al. (2009) |
| miR-124 | GluA2 | calcium-permeable AMPARs and homeostatic plasticity | Rodent | Hou, Q., et al. (2015) |
| miR-124,-9,-34 | NA | spatial learning and working memory | Rodent | Malmevik, J., et al. (2016) |
| miR-125a | PSD-95 | dendritic spine density and dendritic branch | Rodent | Muddashetty, R.S., et al. (2011) |
| miR-125b | GluN2A | dendritic spine morphology | Rodent | Edbauer, D., et al., (2010) |
| miR-128b | RCS | fear extinction | Rodent | Lin, Q., et al., (2011) |
| miR-129 | Kv1.1 | dendrtic regulation of Voltage-gated potassium channel Kv1.1 expression | Rodent | Sosanya, N. M., et al. (2013) |
| miR-132 | p250GAP | synaptic transmission, dendritic spine formation and dendrite morphogenesis | Rodent | Wayman, G. A., et al. (2008), Impey, S., et al. (2010) |
| miR-132 | NA | LTP and LTD in perirhinal cortex, short-term recognition memory | Rodent | Scott, H. L., et al. (2012) |
| miR-132 | NA | short-term synaptic plasticity | Rodent | Lambert, T.J., et al., (2010) |
| miR-132 | Rasa1 | dorsal root ganglion axon growth | Rodent | Hancock, M.L., et al.,(2014) |
| miR-132/212 | NA | basal synaptic transmission and LTP in hippocampus and neocortex | Rodent | Remenyi, J., et al. (2013) |
| miR-134 | Pumilio-2 | dendritic morphology, homeostatic synapse elimination | Rodent | Fiore, R., et al. (2009; 2014) |
| miR-134 | Limk1 | hippocampal dendtic spine morphogenesis | Rodent | Schratt, G. M., et al. (2006) |
| miR-134 | CREB | LTP and contexual fear memory | Rodent | Gao, J., et al. (2010) |
| miR-137 | GluA1 | AMPAR-mediated synaptic transmission, mGluR-LTD | Rodent | Olde Loohuis, N. F., et al. (2015) |
| miR-137 | synaptotagmi n-1 | presynaptic vesicle release, mossy fiber LTP, contextual fear memory | Rodent, human | Siegert, S., et al. (2015) |
| miR-138 | APT1 | morphology of dendritic spines in hippocampal neurons | Rodent | Siegel, G., et al. (2009) |
| miR-146a-5p | Map1b | AMPAR-mediated synaptic transmission, AMPA receptor endocytosis | Rodent | Chen, Y. L. and C. K. Shen (2013) |
| miR-181C | NA | neurite sprouting and synaptogenesis | Rodent | Kos, A., et al. (2016) |
| miR-182 | cortactin, Rac1 | amygdala-dependent long-term memory formation | Rodent | Griggs, E. M., et al. (2013) |
| miR-183/96/182 | HDAC9 | long-term object memory | Rodent | Woldemichael, B.T., et al., (2016) |
| miR-185 | NA | dendrtic spine morphology | Rodent | Xu, B., et al. (2013) |
| miR-188-5p | neuropilin-2 | dendritic spine density, synaptic transmission, | Rodent | Lee, K., et al. (2012) |
| miR-191, miR-135 | Tmod2, Cplx1/2 | NMDAR-LTD and the associated dendritic spine remodeling | Rodent | Hu, Z., et al. (2014) |
| miR-22 | CPEB | long-term synaptic facilitation in sensory-motor synapse | Aplysia | Fiumara, F., et al. (2015) |
| miR-223 | GluN2B and GluA2 | mEPSC, contexual fear memory | Rodent | Harraz, M.M., et al. (2012) |
| miR-26a, miR-384-5p | RSK3 | NMDAR-LTP and the associated dendritic spine remodeling | Rodent | Gu, Q. H., et al. (2015) |
| miR-29a/b | Arpc3 | dendritic spine morphology | Rodent | Lippi, G., et al. (2011) |
| miR-33 | NA | contextual fear memory | Rodent | Jovasevic, V., et al., (2015) |
| miR-485 | SV2A | homoestatic plasticity, dendritic spine density, synaptic transmission | Rodent | Cohen, J. E., et al. (2011) |
| miR-501-3p | GluA1 | dendritic AMPAR expression, dendritic spine remodeling during NMDAR-LTD | Rodent | Hu, Z., et al. (2015) |
| miR-92 | KCC2, CPEB3, MEF2D | contextual fear memory, dendritic spine density | Rodent | Vetere, G., et al. (2014) |
| miR-932 | Act5C | long term memory recall, but not memory acquisition during olfactory learning | Honeybee | Cristino, A. S., et al. (2014) |
Highlights.
miRNAs and RISC are located near synapses
miRNAs regulate local protein synthesis in dendrites and axons
miRNAs play important roles in synapse development and synaptic plasticity
miRNAs are essential for learning and memory
Acknowledgments
This work was supported by NIMH intramural program (#1ZIAMH002881-09).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 4.Shao NY, Hu HY, Yan Z, Xu Y, Hu H, Menzel C, Li N, Chen W, Khaitovich P. Comprehensive survey of human brain microRNA by deep sequencing. BMC Genomics. 2010;11:409. doi: 10.1186/1471-2164-11-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Lugli G, Torvik VI, Larson J, Smalheiser NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem. 2008;106:650–661. doi: 10.1111/j.1471-4159.2008.05413.x. This is the first study that identifies miRNAs and pre-miRNAs in synaptic fractions of the adult mouse forebrain, thereby drawing attention to the local function of miRNAs near synapses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saba R, Storchel PH, Aksoy-Aksel A, Kepura F, Lippi G, Plant TD, Schratt GM. Dopamine-regulated microRNA MiR-181a controls GluA2 surface expression in hippocampal neurons. Mol Cell Biol. 2012;32:619–632. doi: 10.1128/MCB.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010;16:1516–1529. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancock ML, Preitner N, Quan J, Flanagan JG. MicroRNA-132 is enriched in developing axons, locally regulates Rasa1 mRNA, and promotes axon extension. J Neurosci. 2014;34:66–78. doi: 10.1523/JNEUROSCI.3371-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Hu Z, Zhao J, Hu T, Luo Y, Zhu J, Li Z. miR-501-3p mediates the activity-dependent regulation of the expression of AMPA receptor subunit GluA1. J Cell Biol. 2015;208:949–959. doi: 10.1083/jcb.201404092. This study shows that miR-501-3p regulates GluA1 expression locally in dendrites. This miRNA-mediated local regulation of GluA1 is required for long-lasting dendritic spine remodeling during LTD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10••.Hu Z, Yu D, Gu QH, Yang Y, Tu K, Zhu J, Li Z. miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat Commun. 2014;5:3263. doi: 10.1038/ncomms4263. In this study, the authors sequenced the miRNA transcriptome of the mouse hippocampus following the induction of NMDA receptor-dependent LTD. They identified miRNAs that are differentially expressed during LTD, characterized two of these miRNAs, miR-191 and miR-135, in long-lasting dendritic spine restructuring associated with LTD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. This is the first report that miRNAs regulate the morphology of dendritic spines. The authors showed that miR-134 negatively regulates the size of dendritic spines in cultured rat hippocampal neurons by repressing the expression of its target Limk1. [DOI] [PubMed] [Google Scholar]
- 12••.Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Gu QH, Yu D, Hu Z, Liu X, Yang Y, Luo Y, Zhu J, Li Z. miR-26a and miR-384-5p are required for LTP maintenance and spine enlargement. Nat Commun. 2015;6:6789. doi: 10.1038/ncomms7789. This study identified miRNAs that change in expression during NMDA receptor-dependent LTP, and demonstrated that miR-26a and miR-384-5p, two miRNAs downregulated during LTP and both target RSK3, regulate LTP maintenance and long-lasting spine enlargement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HH, Kim P, Phay M, Yoo S. Identification of precursor microRNAs within distal axons of sensory neuron. J Neurochem. 2015;134:193–199. doi: 10.1111/jnc.13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. This study shows, for the first time that Dicer (a cytoplasmic RNase III that cleaves the pre-miRNAs into mature miRNAs), and eIF2c (a component of RNA-induced silencing complex) locate at the somatodendritic compartment of neuron. In addition, Dicer is enriched in dendritic spine and postsynaptic density, and is activated by NMDA recepor activation in a calpain-depdendent manner. [DOI] [PubMed] [Google Scholar]
- 16•.Bicker S, Khudayberdiev S, Weiss K, Zocher K, Baumeister S, Schratt G. The DEAH-box helicase DHX36 mediates dendritic localization of the neuronal precursor-microRNA-134. Genes Dev. 2013;27:991–996. doi: 10.1101/gad.211243.112. This study shows that pre-miR-134 is targeted into the dendrites of hippocampal neurons by the DEAH-box helicase DHX36. DHX36 binds to the terminal loop of pre-miR-134, and this interaction is required for the localization of miR-134 in dendrites and the effect of miR-134 on dendritic spine morphologloy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 18.Sosanya NM, Huang PP, Cacheaux LP, Chen CJ, Nguyen K, Perrone-Bizzozero NI, Raab-Graham KF. Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. J Cell Biol. 2013;202:53–69. doi: 10.1083/jcb.201212089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 22.Fenelon K, Mukai J, Xu B, Hsu PK, Drew LJ, Karayiorgou M, Fischbach GD, Macdermott AB, Gogos JA. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc Natl Acad Sci U S A. 2011;108:4447–4452. doi: 10.1073/pnas.1101219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Earls LR, Fricke RG, Yu J, Berry RB, Baldwin LT, Zakharenko SS. Age-dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J Neurosci. 2012;32:14132–14144. doi: 10.1523/JNEUROSCI.1312-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiorenza A, Lopez-Atalaya JP, Rovira V, Scandaglia M, Geijo-Barrientos E, Barco A. Blocking miRNA Biogenesis in Adult Forebrain Neurons Enhances Seizure Susceptibility, Fear Memory, and Food Intake by Increasing Neuronal Responsiveness. Cereb Cortex. 2016;26:1619–1633. doi: 10.1093/cercor/bhu332. [DOI] [PubMed] [Google Scholar]
- 26.Konopka W, Kiryk A, Novak M, Herwerth M, Parkitna JR, Wawrzyniak M, Kowarsch A, Michaluk P, Dzwonek J, Arnsperger T, et al. MicroRNA loss enhances learning and memory in mice. J Neurosci. 2010;30:14835–14842. doi: 10.1523/JNEUROSCI.3030-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storchel PH, Thummler J, Siegel G, Aksoy-Aksel A, Zampa F, Sumer S, Schratt G. A large-scale functional screen identifies Nova1 and Ncoa3 as regulators of neuronal miRNA function. Embo j. 2015;34:2237–2254. doi: 10.15252/embj.201490643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harraz MM, Eacker SM, Wang X, Dawson TM, Dawson VL. MicroRNA-223 is neuroprotective by targeting glutamate receptors. Proc Natl Acad Sci U S A. 2012;109:18962–18967. doi: 10.1073/pnas.1121288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olde Loohuis NF, Ba W, Stoerchel PH, Kos A, Jager A, Schratt G, Martens GJ, van Bokhoven H, Nadif Kasri N, Aschrafi A. MicroRNA-137 Controls AMPA-Receptor-Mediated Transmission and mGluR-Dependent LTD. Cell Rep. 2015;11:1876–1884. doi: 10.1016/j.celrep.2015.05.040. [DOI] [PubMed] [Google Scholar]
- 31.Chen YL, Shen CK. Modulation of mGluR-dependent MAP1B translation and AMPA receptor endocytosis by microRNA miR-146a-5p. J Neurosci. 2013;33:9013–9020. doi: 10.1523/JNEUROSCI.5210-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee K, Kim JH, Kwon OB, An K, Ryu J, Cho K, Suh YH, Kim HS. An activity-regulated microRNA, miR-188, controls dendritic plasticity and synaptic transmission by downregulating neuropilin-2. J Neurosci. 2012;32:5678–5687. doi: 10.1523/JNEUROSCI.6471-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verma P, Augustine GJ, Ammar MR, Tashiro A, Cohen SM. A neuroprotective role for microRNA miR-1000 mediated by limiting glutamate excitotoxicity. Nat Neurosci. 2015;18:379–385. doi: 10.1038/nn.3935. [DOI] [PubMed] [Google Scholar]
- 34••.Siegert S, Seo J, Kwon EJ, Rudenko A, Cho S, Wang W, Flood Z, Martorell AJ, Ericsson M, Mungenast AE, et al. The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nat Neurosci. 2015;18:1008–1016. doi: 10.1038/nn.4023. The MIR137 gene is genetically associated with the risk for schiziphrenia. The authors observed increased miR-137 expression and impaired pre-synpaptic vesicle trafficking in human neurons derived from patients carrying the disease-associated single nucleotide polymorphism of MIR137. They further showed that overexpressing miR-137 in the mouse dentate gyrus impairs presynaptic plasticity and hippocampus-dependent learning and memory through the miR-137 target synaptotagmin-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Impey S, Davare M, Lesiak A, Fortin D, Ando H, Varlamova O, Obrietan K, Soderling TR, Goodman RH, Wayman GA. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol Cell Neurosci. 2010;43:146–156. doi: 10.1016/j.mcn.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiumara F, Rajasethupathy P, Antonov I, Kosmidis S, Sossin WS, Kandel ER. MicroRNA-22 Gates Long-Term Heterosynaptic Plasticity in Aplysia through Presynaptic Regulation of CPEB and Downstream Targets. Cell Rep. 2015;11:1866–1875. doi: 10.1016/j.celrep.2015.05.034. [DOI] [PubMed] [Google Scholar]
- 38.Hou Q, Ruan H, Gilbert J, Wang G, Ma Q, Yao WD, Man HY. MicroRNA miR124 is required for the expression of homeostatic synaptic plasticity. Nat Commun. 2015;6:10045. doi: 10.1038/ncomms10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Shu X, Liu D, Shang Y, Wu Y, Pei L, Xu X, Tian Q, Zhang J, Qian K, et al. EPAC null mutation impairs learning and social interactions via aberrant regulation of miR-124 and Zif268 translation. Neuron. 2012;73:774–788. doi: 10.1016/j.neuron.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott HL, Tamagnini F, Narduzzo KE, Howarth JL, Lee YB, Wong LF, Brown MW, Warburton EC, Bashir ZI, Uney JB. MicroRNA-132 regulates recognition memory and synaptic plasticity in the perirhinal cortex. Eur J Neurosci. 2012;36:2941–2948. doi: 10.1111/j.1460-9568.2012.08220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remenyi J, van den Bosch MW, Palygin O, Mistry RB, McKenzie C, Macdonald A, Hutvagner G, Arthur JS, Frenguelli BG, Pankratov Y. miR-132/212 knockout mice reveal roles for these miRNAs in regulating cortical synaptic transmission and plasticity. PLoS One. 2013;8:e62509. doi: 10.1371/journal.pone.0062509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lippi G, Steinert JR, Marczylo EL, D'Oro S, Fiore R, Forsythe ID, Schratt G, Zoli M, Nicotera P, Young KW. Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. J Cell Biol. 2011;194:889–904. doi: 10.1083/jcb.201103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell. 2013;152:262–275. doi: 10.1016/j.cell.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kos A, Olde Loohuis N, Meinhardt J, van Bokhoven H, Kaplan BB, Martens GJ, Aschrafi A. MicroRNA-181 promotes synaptogenesis and attenuates axonal outgrowth in cortical neurons. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen JE, Lee PR, Chen S, Li W, Fields RD. MicroRNA regulation of homeostatic synaptic plasticity. Proc Natl Acad Sci U S A. 2011;108:11650–11655. doi: 10.1073/pnas.1017576108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fiore R, Rajman M, Schwale C, Bicker S, Antoniou A, Bruehl C, Draguhn A, Schratt G. MiR-134-dependent regulation of Pumilio-2 is necessary for homeostatic synaptic depression. EMBO J. 2014;33:2231–2246. doi: 10.15252/embj.201487921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malmevik J, Petri R, Knauff P, Brattas PL, Akerblom M, Jakobsson J. Distinct cognitive effects and underlying transcriptome changes upon inhibition of individual miRNAs in hippocampal neurons. Sci Rep. 2016;6:19879. doi: 10.1038/srep19879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen KF, Sakamoto K, Wayman GA, Impey S, Obrietan K. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS One. 2010;5:e15497. doi: 10.1371/journal.pone.0015497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Q, Wei W, Coelho CM, Li X, Baker-Andresen D, Dudley K, Ratnu VS, Boskovic Z, Kobor MS, Sun YE, et al. The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat Neurosci. 2011;14:1115–1117. doi: 10.1038/nn.2891. [DOI] [PubMed] [Google Scholar]
- 52.Vetere G, Barbato C, Pezzola S, Frisone P, Aceti M, Ciotti M, Cogoni C, Ammassari-Teule M, Ruberti F. Selective inhibition of miR-92 in hippocampal neurons alters contextual fear memory. Hippocampus. 2014;24:1458–1465. doi: 10.1002/hipo.22326. [DOI] [PubMed] [Google Scholar]
- 53.Griggs EM, Young EJ, Rumbaugh G, Miller CA. MicroRNA-182 regulates amygdala-dependent memory formation. J Neurosci. 2013;33:1734–1740. doi: 10.1523/JNEUROSCI.2873-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jovasevic V, Corcoran KA, Leaderbrand K, Yamawaki N, Guedea AL, Chen HJ, Shepherd GM, Radulovic J. GABAergic mechanisms regulated by miR-33 encode state-dependent fear. Nat Neurosci. 2015;18:1265–1271. doi: 10.1038/nn.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woldemichael BT, Jawaid A, Kremer EA, Gaur N, Krol J, Marchais A, Mansuy IM. The microRNA cluster miR-183/96/182 contributes to long-term memory in a protein phosphatase 1-dependent manner. Nat Commun. 2016;7:12594. doi: 10.1038/ncomms12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cristino AS, Barchuk AR, Freitas FC, Narayanan RK, Biergans SD, Zhao Z, Simoes ZL, Reinhard J, Claudianos C. Neuroligin-associated microRNA-932 targets actin and regulates memory in the honeybee. Nat Commun. 2014;5:5529. doi: 10.1038/ncomms6529. [DOI] [PubMed] [Google Scholar]


