Abstract
The mechanisms by which retinal neurons are patterned along the dorsal/ventral axis remain largely unknown, yet this patterning is integral for the topographic mapping of visual space. With an interest in elucidating the mechanisms that regulate the development of this retinal axis, we have characterized a T-box family transcription factor, Tbx2b, during zebrafish retinogenesis. Tbx2b is expressed throughout all phases of retinal development with a striking asymmetry of distribution highest dorsally to lowest ventrally. To examine Tbx2b function during retinal development, two morpholino antisense oligonucleotides were created; one blocking the translational start site of Tbx2b and the other interfering with Tbx2b mRNA splicing. Injection of either of these morpholinos resulted in profound defects in the development of the dorsal retina. By using molecular markers for neuronal subtypes, the ventral retina contained all cell types, whereas in the dorsal retina, only retinal ganglion cells expressed markers of differentiation. The cells of the dorsal retina were postmitotic, however, as demonstrated by a lack of BrdUrd incorporation during the normal periods of retinal differentiation. Markers for dorsal and ventral retinal compartments were also expressed normally in Tbx2b morphants. Combined, these observations suggest that the cellular mechanisms regulating neuronal differentiation within the retina are asymmetric about the dorsal/ventral axis and that Tbx2b mediates this process within the dorsal retina.
Keywords: T-box, retinal development
The division of the retina into dorsal and ventral compartments is crucial for the appropriate mapping of visual space. Retinal ganglion cells (RGCs), the output neurons of the retina, are endowed with a topographic spatial code relating their position within these retinal compartments onto the higher visual centers of the brain (1). Whereas much has been learned about the molecular mechanisms underlying axonal pathfinding from the retina to the brain, the mechanisms by which retinal neurons are patterned along the dorsal/ventral (D/V) axis still remain largely unknown. Functional perturbations in a variety of model systems have demonstrated that retinoic acid (RA), bone morphogenetic proteins (BMPs), and Sonic hedgehog each play important roles in regulating D/V retinal development (2). For example, manipulation of RA levels in zebrafish have demonstrated that RA is essential for the formation of the ventral retina: abrogation of RA signaling results in ventral retina deletion, whereas increased RA levels result in ventral retina duplication (3, 4). Importantly, no such factor or determinant has yet been identified to specifically regulate the formation of the dorsal retina.
The mechanisms that regulate the morphological differentiation of retinal neurons have also recently begun to be elucidated. In zebrafish, the young (yng) gene encodes a Brahma-related SWI-SNF chromatin remodeling factor, brg-1, that is required nonautonomously by all retinal neurons for their ultimate differentiation (5). yng mutants express early markers for retinal neuron subtypes but do not undergo terminal differentiation and morphogenesis, resulting in a nonlaminated retina with no morphologically distinguishable retinal neurons. This defect is manifest in part by a concomitant loss of mitogen-activated protein kinase activity or levels in yng mutants (6).
With an interest in the mechanisms that regulate D/V retinal development, we have begun to characterize several candidate molecules that might participate in patterning this retinal axis. Of particular interest are members of the T-box family of transcriptional regulators, several of which are expressed asymmetrically about the D/V axis (7–9). T-box transcription factors are characterized by their homology to the DNA-binding domain of Brachyury, the founding member of the T-box gene family (10). The importance of T-box genes in development is underscored by their involvement in a variety of human congenital pathologies, including Holt–Oram syndrome, DiGeorge syndrome, and ulnar-mammary syndrome (11).
This paper reports the functional characterization of Tbx2b (formerly known as tbx-c; ref. 7) during zebrafish retinogenesis and a surprising link between D/V retinal development and neuronal morphogenesis. Interestingly, loss of Tbx2b function results in a block to neuronal differentiation in the dorsal retina with no disruption to compartmentally restricted gene products in the dorsal or ventral retinas. This finding strongly suggests that not only are there differences in the upstream signaling components that regulate the formation and patterning of the dorsal and ventral compartments of the retina but also that there are distinct cellular mechanisms that asymmetrically facilitate neuronal differentiation about this axis.
Materials and Methods
Animals. Embryos were raised at 28.5°C by using the methods of Westerfield (12) and staged according to Kimmel et al. (13).
Morpholino Antisense Knockdown and Controls. Two morpholino antisense oligonucleotides (MASOs; GeneTools) were designed to perturb Tbx2b function. Tbx2b-ATG was designed to –2 to +23 to block translation from the predicted start site. Tbx2b-SP was designed to the exon 3 splice-donor site, based on the predicted genomic structure of Tbx2b from Ensembl. One-cell-stage Oregon AB embryos were injected with 6 ng of either of the Tbx2b-MASOs, or a standard control MASO, along with 40-kDa FITC-dextran. For Tbx2b-ATG target-site controls, synthetic oligonucleotides were annealed together, recapitulating the Tbx2b-ATG target site along with BamHI and NcoI complementary overhangs. This double-stranded oligonucleotide was then ligated in-frame into BamHI/NcoI-digested pCS2-EGFP. A second EGFP fusion construct was made where the Tbx2b-ATG target site was engineered with five base mismatches. These constructs were linearized with NotI, purified, and used as a template to generate in vitro-transcribed 5′-capped mRNAs with the mMESSAGE mMACHINE kit (Ambion). mRNA (0.1 ng) was coinjected with 4 ng of MASO, and EGFP fluorescence was assayed at 8 h postfertilization (hpf). To determine Tbx2b-SP efficacy, 20 embryos at each time point were solubilized in TRIzol and 2 μg of total RNA used in oligo(dT)-primed RT-PCRs. cDNA (4 μl) was used in subsequent PCRs with primers designed to the 5′ and 3′ ends of the full-length Tbx2b coding sequence and in a second reaction with primers designed to the 3′ end of exon 2 and the 5′ end of exon 4. The Tbx2b-SP splice product amplified by the exon2-exon4 primers was also subcloned into pGEM-T and sequenced bidirectionally to verify alternative splicing of the transcript.
Histology. Histology was performed as described in Gross et al. (14).
Immunohistochemistry. Immunohistochemistry was performed essentially as described in Link et al. (5). Slides were mounted in Vectashield mounting medium containing DAPI (Vector Laboratories) and imaged on a Zeiss 510 laser-scanning confocal microscope. Three to five optical sections (1 μm in thickness) were collected and projected by using Zeiss confocal software. The following antibodies and dilutions were used: Islet 1/39.4d5 (1:10, Developmental Studies Hybridoma Bank), Hu/16A11 (1:20, Chemicon), 1d1 (1:30), 5e11 (1:100), zn5 (1:100), zpr1 (1:20), and goat anti-mouse Cy3 secondary (1:500, Jackson ImmunoResearch).
BrdUrd Assays. Tbx2b-SP-injected embryos were dechorionated and incubated in fish water with 10 mM BrdUrd (Sigma) for defined time periods and immediately killed thereafter. Embryos were processed for immunohistochemistry as above with the addition of a 10-min incubation in 4 M HCl at 37° before blocking to relax chromatin and facilitate BrdUrd detection. Rat anti-BrdUrd (Immunologicals Direct) and Cy3 rabbit anti-rat were used at 1:100 dilutions.
Riboprobes and in Situ Hybridization. Hybridizations were performed essentially as described by Jowett and Lettice (15) by using digoxigenin-labeled antisense RNA probes. Tbx2b was amplified from cDNA generated from 28-hpf embryos and subcloned into PGEM-T (details are available upon request). Tbx2b was linearized with ApaI, and antisense RNA probes were transcribed with SP6. Probe synthesis constructs for the listed genes were generously provided by the following researchers: ephrinB2A (C.-B. Chien, University of Utah School of Medicine, Salt Lake City), msx-c (M. Ekker, Ottawa Health Research Institute, Ottawa), tbx5.1 (D. Garrity, Cardiovascular Research Center/Massachusetts General Hospital, Boston), pax2a (B. Riley, Texas A & M University, College Station), and vax2 (S. Wilson, University College London, London).
Results
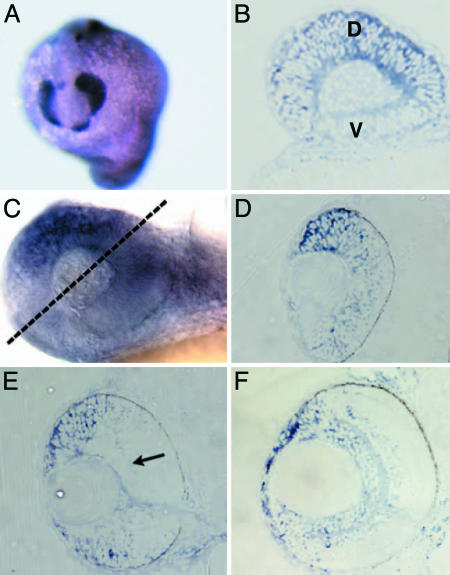
The temporal and spatial aspects of Tbx2b expression and distribution were determined by in situ hybridization. Early aspects of Tbx2b distribution in zebrafish up to 22 hpf have been described (7, 8), so this study largely focused on later time points during which eye development was progressing (Fig. 1). Tbx2b is expressed throughout the optic primordium at 15 hpf (Fig. 1A). By 24 hpf, Tbx2b distribution is markedly asymmetric with higher levels dorsally than ventrally (Fig. 1B). This asymmetry proceeds through 30 hpf (Fig. 1 C and D) and 48 hpf (Fig. 1E). At 48 hpf, expression is largely confined to the proliferative marginal zones at the retinal periphery, although faint expression can be observed in the ganglion cell layer (Fig. 1E, arrow). By 72 hpf, Tbx2b expression remains strongest in the dorsal retinal periphery and is also distributed throughout the inner retina (Fig. 1F). Other regions of Tbx2b expression include the heart, epiphysis, otic vesicles, and excretory system (7, 8).
Fig. 1.
Tbx2b expression during retinal development. (A) At 15 hpf, Tbx2b is expressed throughout the retinal neuroepithelium. (B) Sagittal section through a 24-hpf eye. Tbx2b is expressed throughout the retinal neuroepithelium, with highest levels in the dorsal retina. (C) Whole-mount and 3-μm (D–F) sections showing Tbx2b distribution throughout eye development. (C and D) Tbx2b expression at 30 hpf is prominent in the dorsal retina and decreases into the ventral retina. Section at the plane is indicated by dashed line. (E) At 48 hpf, strong Tbx2b expression is observed in the dorsal retinal periphery, and fainter expression is observed in the ventral periphery and throughout the ganglion cell layer (arrow). (F) Expression at 72 hpf is also strong in the dorsal periphery, and expression extends through the inner retina. D, dorsal; V, ventral.
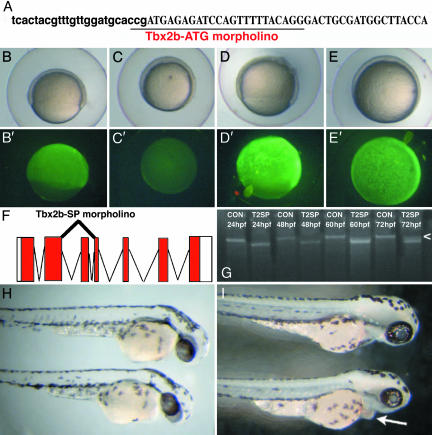
To directly assess Tbx2b function, two MASOs were created and microinjected into one-cell-stage embryos (16). The first MASO, Tbx2b-ATG, targeted the translational start site of Tbx2b (Fig. 2A), and the second MASO, Tbx2b-SP, targeted the exon 3 splice-donor site (Fig. 2F and ref. 17). Controls for Tbx2b-ATG specificity entailed fusing the 25-bp Tbx2b-ATG target site in-frame to EGFP and microinjecting this mRNA into one-cell-stage embryos. Coinjection with a standard control MASO did not affect EGFP expression (Fig. 2 B and B′), whereas coinjection of Tbx2b-ATG prevented EGFP expression (Fig. 2 C and C′). A second fusion was created where 5 bp of the 25-bp Tbx2b-ATG target site were mutated such that Tbx2b-ATG, if specifically targeting the translational start site of Tbx2b, should not be able to bind, and, therefore, EGFP expressed normally. This result was indeed the case; expression of mismatch-EGFP mRNA, along with a control MASO (Fig. 2 D and D′) or with Tbx2b-ATG (Fig. 2 E and E′), resulted in similar levels of EGFP expression, demonstrating that Tbx2b-ATG is selective for its 25-bp target site. Injection of Tbx2b-SP, targeting the exon 3 splice-donor site of Tbx2b, results in the excision of exon 3 from the Tbx2b transcript. Exon 3 of Tbx2b encodes amino acids 211–259, a significant portion of the 3′ end of the T-box DNA-binding domain. Thus, excision of this region should prevent Tbx2b from functioning normally in regulating transcription of target genes. RT-PCR for full-length Tbx2b message indicated that Tbx2b-SP was effective in blocking normal Tbx2b splicing, resulting in exon 3 excision (Fig. 2G). RT-PCR was also performed with primers flanking exon 3, and the resulting splice variant was sequenced bidirectionally to confirm the nature of the Tbx2b-SP splice product (data not shown). RT-PCR on injected samples from 24 to 72 hpf showed that Tbx2b-SP was effective up to 48 hpf, but, by 60 hpf, a small amount of normally spliced Tbx2b was present, the levels of which increased by 72 hpf (Fig. 2G, arrowhead).
Fig. 2.
MASO perturbations to Tbx2b function. (A) Location of the translation-blocking MASO (Tbx2b-ATG). Letters for the 5′ UTR are in lowercase, and letters for the coding sequence are in uppercase. (B–E) Efficacy of Tbx2b-ATG MASO. (B and B′) EGFP was fused to the 25-bp target site of Tbx2b-ATG, and mRNA was microinjected into one-cell embryos along with a control MASO. EGFP was expressed normally (B′). (C and C′) Coinjection of the Tbx2b-ATG target-EGFP mRNA with Tbx2b-ATG prevents EGFP expression, indicating that the MASO is functional. (D and E) Expression of a 5-bp-mismatched Tbx2b target site fused to EGFP. EGFP is expressed (D′), even when Tbx2b-ATG is coinjected (E′), indicating that the morpholino is relatively specific for this 25-bp Tbx2b target site. (F) Design of a splice-blocking MASO (Tbx2b-SP) that overlaps the exon 3 splice-donor site, resulting in an excision of exon 3, which encodes 49 aa (residues 211–259) of the distal portion of the T-box DNA-binding domain. (G) RT-PCR of Tbx2b expression to determine Tbx2b-SP efficacy. Splicing out of exon 3 removes 147 bp, which can be resolved by gel electrophoresis. Tbx2b-SP functions until 60 hpf, at which point some normally spliced Tbx2b mRNA can be observed. By 72 hpf, an increasing amount of normally spliced Tbx2b is present (arrowhead). (H and I) Morphology of 6 ng of Tbx2b-ATG-injected embryo at 48 hpf (H) and 6 ng of Tbx2b-SP-injected embryo at 72 hpf (I). A control (8 ng) MASO-injected embryo is above the Tbx2b-MASO-injected sibling in each image. Injection of either Tbx2b-MASO results in smaller eyes and pericardial edema obvious by 48 hpf (H) and lasting through 72 hpf (arrow in I).
Gross morphological phenotypes obtained from the injection of either Tbx2b-MASO were quite similar. By 36 hpf, Tbx2b morphants began to show cardiac edema and blood pooling ventrally (data not shown). Cardiac edema was more obvious at 48 hpf in Tbx2b morphants, and they also displayed somewhat smaller eyes than did their control-injected siblings (Fig. 2H). At 72 hpf, ventral blood pooling and cardiac edema was still apparent (Fig. 2I, arrow) but varied in severity between individual morphants, suggesting that some recovery was occurring. Obvious defects in otic development were also observed (data not shown), but other aspects of development and morphogenesis in Tbx2b morphants appeared indistinguishable from controls (Fig. 2I). In particular, no obvious defects in midline or notochord structures were observed as reported to result from dominant-negative tbx-c mRNA overexpression (7). A closer inspection of Tbx2b morphants and the use of molecular markers, however, might reveal more subtle defects in these structures.
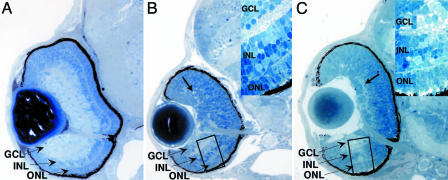
We first sought to determine the gross histological effects on retinal development of Tbx2b MASO injection (Fig. 3). At 72 hpf, control MASO-injected embryos were indistinguishable from wild-type siblings. All of the principal cellular and synaptic laminae of the retina were well formed with an intact lens and retinal pigment epithelium (Fig. 3A). Tbx2b morphants, however, displayed profound defects in the formation of their dorsal retinas at 72 hpf (Fig. 3 B and C). Tbx2b-ATG-injected embryos displayed a well formed ventral retina with all of the appropriate differentiated neuronal cells types, as evaluated by established morphological criteria for retinal cell type identification (Fig. 3B Inset and refs. 18 and 19). The neurons of the dorsal retina in these morphants showed little overt morphological differentiation. Small patches of inner plexiform layer are evident, which is indicative of RGC or amacrine cell formation, but no other morphologically obvious cell types or layers could be identified (Fig. 3B, arrow, and ref. 20). Tbx2b-ATG morphants form an optic nerve, the dorsal and ventral RGC axonal composition of which is currently unknown. In Tbx2b-SP morphants, obvious morphological differentiation of dorsal retinal neurons was similarly impaired, but ventral retina development was also slightly disrupted when compared with Tbx2b-ATG morphants, perhaps reflecting a dominant-negative effect of the mis-spliced Tbx2b protein (Fig. 3C). Many Tbx2b-SP morphants also showed an increased region of undifferentiated cells at their ventral retinal periphery. Morphological differentiation of all retinal neurons was apparent within the ventral retina, however, although it was confined to a smaller region of the compartment (Fig. 3C). The severity of effects varied between Tbx2b-SP morphants with some morphants forming a larger region of differentiated cells in their ventral retinas than others (data not shown). Furthermore, Tbx2b-SP morphants showed somewhat less severe defects in dorsal retinal development, because larger regions of inner plexiform layer were often observed in these embryos (Fig. 3C, arrow). By 4 days postfertilization, embryos injected with either Tbx2b-MASO recovered, and their dorsal retinas contained all differentiated neural cell types. Photoreceptor development was usually delayed but recovered fully by 5–6 days postfertilization such that, beyond their slightly smaller size, these were indistinguishable from control injected embryos (data not shown). Because of Tbx2b mRNA turnover, this observation most likely reflects a decrease in morpholino levels, allowing for recovery, an observation supported by the Tbx2b-SP-splicing RT-PCR assays (Fig. 2G). Nonetheless, these results suggest that Tbx2b is involved in regulating neuronal morphogenesis about the D/V axis of the zebrafish retina.
Fig. 3.
Tbx2b morphants display abnormal retinal development along their D/V axis. (A) Control MASO-injected embryos at 72 hpf. At this point of retinal development, morphological differentiation of RGCs (GCL), amacrine cells, bipolar cells (INL), and photoreceptors (ONL) is obvious, as are their synaptic plexiform projection layers. An optic nerve composed of RGC axons grossly separates dorsal from ventral retinal territories. Injection of Tbx2b-ATG (B) or Tbx2b-SP (C) results in a block to morphological differentiation of neurons in the dorsal retina, leaving the ventral retina largely unaffected at 72 hpf. (B and C Insets) High-magnification views of the ventral retina of each morphant. (B) Tbx2b-ATG injection results in a ventral retina with all cell types present (Inset) but no obvious morphological differentiation of neurons in the dorsal retina. Small plexiform regions can be observed in the dorsal retina, indicating that some RGC differentiation is likely to have occurred (arrow). (C) Tbx2b-SP injection results in a similar retinal phenotype. Neuronal differentiation is severely impaired in the dorsal retina, but larger regions of inner plexiform are often observed within the dorsal compartment (arrow). Additionally, a larger region of undifferentiated cells is observed in the ventral retinal periphery of these morphants, but neuronal differentiation within the ventral retina is unaffected (Inset). Dorsal is up in all images.
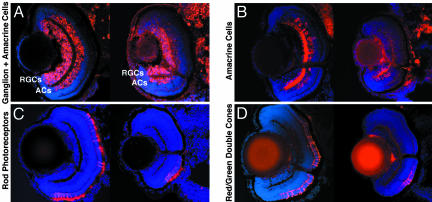
To further characterize the Tbx2b morphant phenotype, molecular markers for neuronal cell types in the retina were used such that the cellular composition of the dorsal and ventral retinas could be unambiguously determined (Fig. 4). Because we could assay the efficacy of Tbx2b interference in each batch of Tbx2b-SP morphants, immunohistochemistry from this group of morphants is shown; however, identical results were obtained from Tbx2b-ATG morphants (data not shown). Several markers for RGC differentiation were first examined: HuC/D, a neuronal RNA-binding protein (21); Islet 1, a LIM homeodomain transcription factor (22), and zn5, an antigen expressed during later periods of RGC differentiation. Each of these markers was expressed in Tbx2b-SP morphants, indicating that RGCs had differentiated at least partially, both ventrally and dorsally (Fig. 4A and data not shown). In addition to RGC expression, HuC/D is also expressed in amacrine cells at 72 hpf. In Tbx2b-SP morphants, HuC/D was expressed only in amacrine cells of the ventral retina (Fig. 4A). The 5e11 antigen is expressed during later periods of amacrine cell differentiation. Similar to HuC/D, 5e11 was only expressed in the amacrine cells of the ventral retina. No dorsal cellular expression was observed, although faint expression within the dorsal IPL patches could be seen (Fig. 4B). In addition to RGCs, Islet 1 is also expressed by subsets of differentiating amacrine, bipolar, and horizontal cells at 72 hpf (5, 23). Control morphants expressed Islet 1 in each of these cell types, whereas in Tbx2b-SP morphants, Islet 1 was only detected within these cells of the ventral retina (data not shown). Photoreceptor differentiation was assayed by using the 1d1 antigen, which is expressed by rods (Fig. 4C), and the zpr-1 antigen, which is expressed by red/green double cones (Fig. 4D). Both antigens were present throughout the dorsal and ventral retina of control embryos, but neither was expressed in the dorsal retina of Tbx2b-SP morphants, indicating that these photoreceptor subtypes were also absent.
Fig. 4.
Tbx2b-SP morphants do not express most markers for most neuronal cell types in their dorsal retina. Central, transverse retinal cryosections were stained for cell-specific neuronal markers at 72 hpf (see text for details). Control (at Left in each image) and Tbx2b-SP injected (at Right in each image) retinas are presented, dorsal is up in all images. (A) The Hu antigen (16A11) is expressed early in differentiating RGCs and amacrine cells (ACs). In Tbx2b-SP morphants, Hu is detected only in the RGCs of the dorsal retina. (B) The 5e11 antigen is expressed in amacrine cells and their processes. In Tbx2b-SP morphants, expression is observed only in a patch of amacrine cells in the ventral retina. The 1d1 antigen, expressed by rod photoreceptors (C), and the zpr-1 antigen, expressed by red/green double cones (D), are distributed throughout the retina in control embryos, but expression is not observed in the dorsal retina of Tbx2b-SP morphants.
Lineage studies have shown that retinal cell subtype specification occurs at, or shortly after, the terminal mitosis (24). Thus, that cells of the dorsal retina in Tbx2b-SP morphants, other than RGCs, do not express neuronal markers raises the possibility that these cells have not exited the cell cycle and remain in an unspecified progenitor-like state. To identify proliferative cells, BrdUrd pulses were performed in control and Tbx2b-SP morphants between 48 and 54 hpf and 48 and 60 hpf. If cells of the dorsal retina remain proliferative in Tbx2b-SP morphants, they will incorporate BrdUrd as they divide to a greater extent than control-injected embryos. After both durations of exposure, this result was not the case. The amount and location of proliferative cells in both control and Tbx2b-SP morphants were indistinguishable, indicating that dorsal retinal cells in Tbx2b-SP morphants were indeed postmitotic, and thus, likely impaired in morphological differentiation (Fig. 7, which is published as supporting information on the PNAS web site).
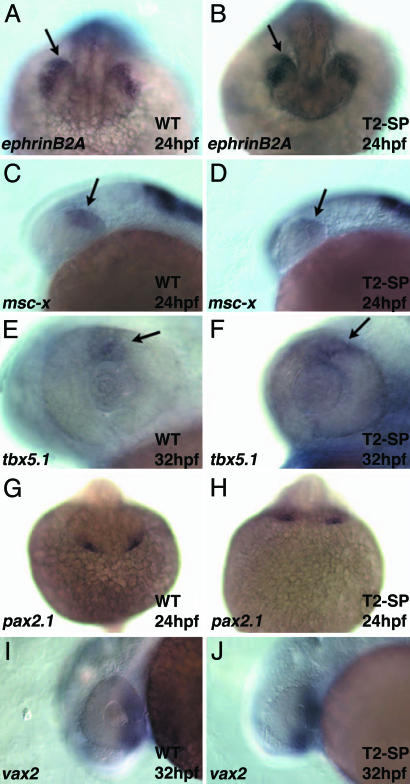
Several distinct territorial markers have been identified as general markers for the dorsal and ventral retinal compartments. These markers include dorsally: ephrinB2A (25), msx-c (26), tbx5.1 (27), and BMP4 (28), and ventrally: pax2a (29) and vax2 (30). Whereas the direct functions of most of these molecules have yet to be well characterized within the retina, they are thought to play a role in compartment patterning, and serve as useful markers for the gross subdivision of the retina into dorsal and ventral compartments. To determine the extent to which the D/V axis was disrupted upon Tbx2b functional interference, the expression of several of these markers was assayed in Tbx2b-SP morphants (Fig. 5). No differences in expression of dorsal markers (Fig. 5 A–F) or ventral markers (Fig. 5 G–J) were observed between wild-type and Tbx2b-SP morphants. Thus, functional perturbation to Tbx2b appears not to significantly affect marker gene expression in the dorsal or ventral retinal compartments, but rather, to directly affect the ability of cells of the dorsal retina to undergo morphological differentiation.
Fig. 5.
Markers of dorsal and ventral retinal compartments are expressed normally in Tbx2b-SP morphants. (A and B) EphrinB2A is expressed normally in the lens, and a stripe of cells is expressed in the dorsal retina at 24 hpf (A). Expression is unaffected in Tbx2b-SP morphants. (B) msx-c is also expressed in a stripe of cells in the dorsal retina in both control (C) and Tbx2b-SP-injected embryos (D). tbx5.1 is expressed from 24 hpf to ≈32 hpf in a small region of the dorsal retina. (E and F) Expression at 32 hpf is unaffected in control or Tbx2b-SP-injected morphants. (G) The ventral marker pax2a is expressed at 24 hpf in the optic stalk, an extension between the forebrain and ventral retina. (H) Expression is unaffected in Tbx2b-SP morphants. (I and J) vax2, a ventral retina marker is also unaffected in control or Tbx2b-SP-injected embryos. Dorsal is up in all images.
Discussion
Here, we describe the observation that the molecular regulation of neuronal differentiation and morphogenesis during zebrafish retinal development differs between the dorsal and ventral retinal compartments. Previous work (3, 31, 32) has identified factors that function in the formation of the ventral retina and these include RA and the pandora/Spt6 transcription elongation factor. Recent studies (5, 6) have also begun to elucidate the molecular mechanisms underlying retinal neuron differentiation and morphogenesis, implicating the Brahma chromatin-remodeling complex in this process. What is unique about the current study is that functional interference to Tbx2b results in a block to neuronal differentiation and morphogenesis only in the dorsal retina. This finding suggests that not only are there specific pathways involved in the overall formation of the dorsal and ventral retinal compartments but also that the cellular mechanisms that direct individual retinal cells to form specific neuronal subtypes within these compartments differ between them.
What is the normal developmental function of Tbx2b in the retina and how is this achieved? Perturbations such as modulation of Sonic hedgehog, BMP4, or RA levels all result in distinct changes in the distribution of gene products normally restricted to the dorsal or ventral retinal compartments concomitant with severe morphological disruptions in gross retinal structure (33–35). Tbx2b functional interference results in no obvious differences in the expression of these compartmentally restricted gene products, thereby placing it genetically downstream of these overall pattern-generating mechanisms within the developing retina (Fig. 6). In the mouse, Tbx2 distribution is also restricted to the dorsal retina and a knockout line has recently been reported in which cardiac defects were characterized (36). It will be interesting to examine the retinas of these embryos to determine whether Tbx2 plays a similar role in asymmetrically regulating retinal neuron differentiation in mammals. In other systems, Tbx2 orthologues have been shown to function as transcriptional repressors (37, 38). Whereas this function remains to be biochemically demonstrated in zebrafish, if consistent, it suggests that Tbx2b indirectly facilitates neuronal differentiation by repressing an intermediate factor that, when active, prevents neuronal differentiation in the dorsal retina. The identity of this factor is currently unknown.
Fig. 6.
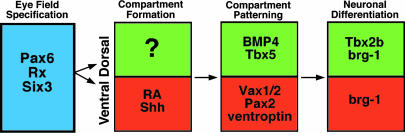
A model of D/V axis formation within the eye. Shown is a summary of eye development and D/V axis formation. See text for details.
Several questions arise with regard to the Tbx2b morphant phenotype. Aspects of RGC formation in the dorsal retina of both Tbx2b morphants are normal (Fig. 4), yet Tbx2b mRNA is strongly detected within this cell layer (Fig. 1). Why are RGCs unaffected in Tbx2b morphants? Because several T-box factors are expressed in similar or overlapping regions within the retina, it is possible that one of these can compensate for the loss of Tbx2b function, rescuing, in part, normal RGC formation but not that of other retinal neurons. It is also possible that, whereas the dorsal RGCs express several markers for neuronal differentiation (HuC/D, Islet, and zn5), these markers might be indicative of early events in that differentiation, and that later morphogenetic events are perturbed. Indeed, the morphology of RGCs in the dorsal retina appears abnormal when viewed histologically, and only small regions of inner plexiform layer are evident. RGCs are the first cells to differentiate in the retina, so perhaps, in Tbx2b morphants, these cells begin the process of differentiation but are impaired at later stages (18). It will be interesting to examine retinotectal projections, and thereby axogenesis, to determine the extent to which dorsal RGCs undergo morphogenesis and axonal pathfinding to the tectum.
Most of the markers available for individual cell types in the retina recognize cell-specific differentiation products (39). Beyond RGCs, cells of the dorsal retina in Tbx2b morphants do not express any of these markers assayed. This fact, however, does not address whether these cells are specified to specific outer retinal lineages. That they are postmitotic implies that they are specified and unable to undergo differentiation and morphogenesis. Without an understanding of what gene products are expressed in a given cell during each step from specification to early and late phases of neuronal differentiation and on to terminal morphogenesis, however, we are only able to conclusively indicate that they remain morphologically undifferentiated. We do not know at which point along the developmental pathway from postmitotic cell to morphologically discrete retinal neuron the cells of the dorsal retina are stalled in these morphants, a subject that warrants further investigation.
Fig. 6 provides a summary and model of D/V axis formation within the developing eye. Ignored in this model are the cell-fate determination events underway in both compartments directing retinal progenitor cells to distinct neuronal fates, likely occurring in conjunction with the patterning and differentiation events depicted. Eye field specification involves a combination of factors, including Pax6, Rx, and Six3. Dorsal and ventral compartments form within this eye field. In the prospective ventral compartment, RA is necessary for retina formation, whereas Sonic hedgehog facilitates optic stalk formation, a ventral derivative. No corresponding factors within the dorsal compartment have yet been identified as necessary for retina formation. The dorsal and ventral compartments are patterned, possibly through the combined functions of signaling factors (BMP4), signaling antagonists (ventroptin), and transcriptional regulators (Tbx5, Vax1/2, and Pax2). It is important to note that while it has been shown that several of these factors are sufficient to alter gene expression and morphogenesis of dorsal and ventral retinal compartments when overexpressed, the necessity in these processes has not yet been adequately demonstrated for most of them. Once committed, compartmentally restricted cells undergo terminal differentiation and morphogenesis. Tbx2b participates asymmetrically in regulating these events, acting mainly in the dorsal compartment, whereas young/brg-1 functions within both.
Supplementary Material
Acknowledgments
We thank T. Darland, B. Link, G. D. Ween, C. Chien, and C. Cepko for helpful discussions and comments on this work. This work was supported by National Eye Institute Grants EY15064 (to J.M.G.) and EY00811 (to J.E.D.).
Abbreviations: RGC, retinal ganglion cell; D/V, dorsal/ventral; RA, retinoic acid; BMP, bone morphogenetic protein; MASO, morpholino antisense oligonucleotide.
References
- 1.McLaughlin, T., Hindges, R. & O'Leary, D. D. (2003) Curr. Opin. Neurobiol. 13, 57–69. [DOI] [PubMed] [Google Scholar]
- 2.Peters, M. A. (2002) Curr. Opin. Neurobiol. 12, 43–48. [DOI] [PubMed] [Google Scholar]
- 3.Marsh-Armstrong, N., McCaffery, P., Gilbert, W., Dowling, J. E. & Drager, U. C. (1994) Proc. Natl. Acad. Sci. USA 91, 7286–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyatt, G. A., Schmitt, E. A., Marsh-Armstrong, N. R. & Dowling, J. E. (1992) Proc. Natl. Acad. Sci. USA 89, 8293–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Link, B. A., Fadool, J. M., Malicki, J. & Dowling, J. E. (2000) Development (Cambridge, U.K.) 127, 2177–2188. [DOI] [PubMed] [Google Scholar]
- 6.Gregg, R. G., Willer, G. B., Fadool, J. M., Dowling, J. E. & Link, B. A. (2003) Proc. Natl. Acad. Sci. USA 100, 6535–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dheen, T., Sleptsova-Friedrich, I., Xu, Y., Clark, M., Lehrach, H., Gong, Z. & Korzh, V. (1999) Development (Cambridge, U.K.) 126, 2703–2713. [DOI] [PubMed] [Google Scholar]
- 8.Ruvinsky, I., Oates, A. C., Silver, L. M. & Ho, R. K. (2000) Dev. Genes Evol. 210, 82–91. [DOI] [PubMed] [Google Scholar]
- 9.Ahn, D. G., Ruvinsky, I., Oates, A. C., Silver, L. M. & Ho, R. K. (2000) Mech. Dev. 95, 253–258. [DOI] [PubMed] [Google Scholar]
- 10.Smith, J. (1999) Trends Genet. 15, 154–158. [DOI] [PubMed] [Google Scholar]
- 11.Packham, E. A. & Brook, J. D. (2003) Hum. Mol. Genet. 12, R37–R44. [DOI] [PubMed] [Google Scholar]
- 12.Westerfield, M. (1995) The Zebrafish Book (Univ. of Oregon Press, Eugene).
- 13.Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. & Schilling, T. F. (1995) Dev. Dyn. 203, 253–310. [DOI] [PubMed] [Google Scholar]
- 14.Gross, J. M., Perkins, B. D., Amsterdam, A., Egana, A., Darland, T., Matsui, J. I., Sciascia, S., Hopkins, S. & Dowling, J. E. (2005) Genetics, 10.1534/genetics.104.039727. [DOI] [PMC free article] [PubMed]
- 15.Jowett, T. & Lettice, L. (1994) Trends Genet. 10, 73–74. [DOI] [PubMed] [Google Scholar]
- 16.Nasevicius, A. & Ekker, S. C. (2000) Nat. Genet. 26, 216–220. [DOI] [PubMed] [Google Scholar]
- 17.Draper, B. W., Morcos, P. A. & Kimmel, C. B. (2001) Genesis 30, 154–156. [DOI] [PubMed] [Google Scholar]
- 18.Schmitt, E. A. & Dowling, J. E. (1996) J. Comp. Neurol. 371, 222–234. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt, E. A. & Dowling, J. E. (1999) J. Comp. Neurol. 404, 515–536. [PubMed] [Google Scholar]
- 20.Kay, J. N., Roeser, T., Mumm, J. S., Godinho, L., Mrejeru, A., Wong, R. O. & Baier, H. (2004) Development (Cambridge, U.K.) 131, 1331–1342. [DOI] [PubMed] [Google Scholar]
- 21.Marusich, M. F., Furneaux, H. M., Henion, P. D. & Weston, J. A. (1994) J. Neurobiol. 25, 143–155. [DOI] [PubMed] [Google Scholar]
- 22.Korzh, V., Edlund, T. & Thor, S. (1993) Development (Cambridge, U.K.) 118, 417–425. [DOI] [PubMed] [Google Scholar]
- 23.Shkumatava, A., Fischer, S., Muller, F., Strahle, U. & Neumann, C. J. (2004) Development (Cambridge, U.K.) 131, 3849–3858. [DOI] [PubMed] [Google Scholar]
- 24.Turner, D. L., Snyder, E. Y. & Cepko, C. L. (1990) Neuron 4, 833–845. [DOI] [PubMed] [Google Scholar]
- 25.Chan, J., Mably, J. D., Serluca, F. C., Chen, J. N., Goldstein, N. B., Thomas, M. C., Cleary, J. A., Brennan, C., Fishman, M. C. & Roberts, T. M. (2001) Dev. Biol. 234, 470–482. [DOI] [PubMed] [Google Scholar]
- 26.Ekker, M., Akimenko, M. A., Bremiller, R. & Westerfield, M. (1992) Neuron 9, 27–35. [DOI] [PubMed] [Google Scholar]
- 27.Begemann, G. & Ingham, P. W. (2000) Mech. Dev. 90, 299–304. [DOI] [PubMed] [Google Scholar]
- 28.Hammerschmidt, M., Kramer, C., Nowak, M., Herzog, W. & Wittbrodt, J. (2003) Dev. Dyn. 227, 128–133. [DOI] [PubMed] [Google Scholar]
- 29.Krauss, S., Johansen, T., Korzh, V. & Fjose, A. (1991) Development (Cambridge, U.K.) 113, 1193–1206. [DOI] [PubMed] [Google Scholar]
- 30.Take-uchi, M., Clarke, J. D. & Wilson, S. W. (2003) Development (Cambridge, U.K.) 130, 955–968. [DOI] [PubMed] [Google Scholar]
- 31.Malicki, J., Neuhauss, S. C., Schier, A. F., Solnica-Krezel, L., Stemple, D. L., Stainier, D. Y., Abdelilah, S., Zwartkruis, F., Rangini, Z. & Driever, W. (1996) Development (Cambridge, U.K.) 123, 263–273. [DOI] [PubMed] [Google Scholar]
- 32.Keegan, B. R., Feldman, J. L., Lee, D. H., Koos, D. S., Ho, R. K., Stainier, D. Y. & Yelon, D. (2002) Development (Cambridge, U.K.) 129, 1623–1632. [DOI] [PubMed] [Google Scholar]
- 33.Macdonald, R., Barth, K. A., Xu, Q., Holder, N., Mikkola, I. & Wilson, S. W. (1995) Development (Cambridge, U.K.) 121, 3267–3278. [DOI] [PubMed] [Google Scholar]
- 34.Koshiba-Takeuchi, K., Takeuchi, J. K., Matsumoto, K., Momose, T., Uno, K., Hoepker, V., Ogura, K., Takahashi, N., Nakamura, H., Yasuda, K. & Ogura, T. (2000) Science 287, 134–137. [DOI] [PubMed] [Google Scholar]
- 35.Hyatt, G. A., Schmitt, E. A., Marsh-Armstrong, N., McCaffery, P., Drager, U. C. & Dowling, J. E. (1996) Development (Cambridge, U.K.) 122, 195–204. [DOI] [PubMed] [Google Scholar]
- 36.Harrelson, Z., Kelly, R. G., Goldin, S. N., Gibson-Brown, J. J., Bollag, R. J., Silver, L. M. & Papaioannou, V. E. (2004) Development (Cambridge, U.K.) 131, 5041–5052. [DOI] [PubMed] [Google Scholar]
- 37.Carreira, S., Dexter, T. J., Yavuzer, U., Easty, D. J. & Goding, C. R. (1998) Mol. Cell. Biol. 18, 5099–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He, M., Wen, L., Campbell, C. E., Wu, J. Y. & Rao, Y. (1999) Proc. Natl. Acad. Sci. USA 96, 10212–10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avanesov, A. & Malicki, J. (2004) Methods Cell Biol. 76, 333–384. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.