Abstract
We investigated the possibility of positive selection acting on members of the putative seven-pass chemoreceptor superfamily in Caenorhabditis elegans, which comprises ≈1,300 genes encoding seven-pass G protein-coupled receptors (GPCRs). Using a maximum-likelihood approach, we conducted statistical tests for evidence of codon sites where the ratio of nonsynonymous substitutions per nonsynonymous site to synonymous substitutions per synonymous site (dN/dS) was >1. Evidence for positive selection was found only for the srz family, about which virtually nothing specific is known. We extended the annotation of the srz gene family, establishing gene models for 60 srz genes in C. elegans and 28 srz genes in Caenorhabditis briggsae. dN/dS ratios varied dramatically in different regions of the SRZ proteins, peaking in predicted extracellular regions. These regions included 23 sites where evidence of positive selection was highly significant, corresponding remarkably well with regions implicated in ligand binding in other GPCR family members. We interpret these results as indicating that the srz family is under positive selection, probably driven by ligand binding.
Keywords: positive selection, ligand binding, maximum likelihood, synonymous, nonsynonymous
Caenorhabditis elegans has ≈1,300 predicted genes that encode members of putative chemosensory receptors and together define the seven-pass receptor (SR) superfamily (1–4), which belongs to the broader class of G protein-coupled receptors (GPCRs). Based on sequence alignment and phylogenetic analysis, SR superfamily members fall into about a dozen families. These families range in size from the large srh and str families (a few hundred genes each) to the modestly sized sra and srv families (≈30 genes each). Each SR family appears to have arisen by gene duplication and divergence from a founder gene. These duplications have occurred sporadically over a long evolutionary period, giving rise to complex relationships. Near one extreme, str-5 and str-6 result from a recent duplication and differ by only two nucleotides in their coding regions. Near the other extreme, the str-1 and str-47 proteins are only 19% identical and presumably arose from an ancient duplication. Members of different SR families are even more distantly related, with the most distant pairs barely alignable.
Proteins in the SR superfamily appear to be more rapidly diverging than the average gene (2, 3, 5, 6). For example, the average ortholog pair between C. elegans and Caenorhabditis briggsae has 80% amino acid identity (5), whereas the average str pair from the same data set has 59% identity (J.H.T., unpublished data). Rapidly diverging proteins may result from relaxed selective constraints, in which changes in protein sequence are relatively well tolerated. Alternatively, they may result from selective pressure for changes in amino acid sequence (positive selection or diversifying selection). A clear signal of positive selection is an excess in the number of nonsynonymous substitutions per nonsynonymous site (dN or KA, amino acid altering changes) compared with the number of synonymous substitutions per synonymous site (dS or KS, silent changes). Because dN and dS are normalized to the number of sites, in cases of neutral drift, dN/dS approximates 1.0. Coding regions under negative selection typically have much lower dN/dS ratios [e.g., there is an average dN/dS ratio of ≈0.2 between humans and mice (7)].
In extreme cases, positive selection acts on all or most of a protein and can be detected in pairwise dN/dS ratios from entire genes (8–10). Preliminary analysis indicated that the SR superfamily is not undergoing such broad positive selection: dN/dS ratios for pairs of SR genes were in the range of 0.2–0.5 (by the method described in ref. 11), consistent with negative (purifying) selection. More commonly, however, positive selection acts only on specific parts of a protein sequence (12). For example, analysis of six human class I major-histocompatibility-complex (MHC) proteins showed a dN/dS ratio averaged across all sites of 0.5 (13), whereas sites in the antigen recognition site have a dN/dS ratio significantly >1 (12). Variations in dN/dS ratios among sites can be examined by using a maximum likelihood test (14–18). This method can identify positive selection in a subset of sites even when the dN/dS ratio averaged across all sites is <1, without a priori knowledge of the identity of the positively selected sites. These methods compare the likelihood of neutral models to selection models in fitting a set of data: Neutral models account for variable dN/dS ratios but constrain the ratios to be ≤1, whereas selection models allow for one additional class of sites whose ratio may be >1. If a likelihood ratio statistic indicates that the selection model fits the data better and the additional dN/dS ratio is >1, an empirical Bayes approach is used to predict sites subjected to positive selection (16, 18, 19). This method correctly identifies amino acids in the MHC antigen recognition site as being subject to positive selection (16, 20). Simulation studies have also demonstrated the power and accuracy of this likelihood method (21).
Materials and Methods
Gene Annotation and Genomic Locations. Using methods described in the supporting information, which is published on the PNAS web site, we identified 113 srz-related loci in the C. elegans genome, of which 78 were fully or partially predicted in WormBase (http://ws120.wormbase.org). Of these 113 loci, 14 appear to encode less than half of an SRZ protein and were not further characterized. Of the remaining 99 loci, we could derive plausible structures for 60 genes; the others have apparent functional defects, including stop codons, frameshifts, and deletions. Of the plausible gene structures, 51 encoded proteins aligned sufficiently well to analyze for codon evolution. A similar process of gene finding in C. briggsae yielded 28 probable functional genes, 33 probable defective genes, and ≈20 gene fragments. The probable functional genes included 7 previously unrecognized predicted genes and revisions of all 21 existing WormBase predictions. Locations of genes on chromosome arms or clusters were based on meiotic recombination rates (22). All gene predictions have been reported to WormBase (http://wormbase.org) and are available in the supporting information.
EST and ORFeome Sequence Tag (OST) Integration and Conflicts. There is only one EST (yk1186h12) for the entire srz family, and it is consistent with our independently derived T25E12.11 gene model. The OST project generates gene-specific cDNAs by RT-PCR with primers designed from gene models (23). Only 17 of 60 putative functional C. elegans srz gene models were successfully amplified. The remaining 43 genes are divided about equally among completely unpredicted genes, genes with one or both primers not in our gene model, and genes with both primers in our gene model but still failing to amplify, perhaps because of low message abundance. Eleven of 17 OST amplifications had a partial or complete sequence report; 7 of these were consistent with our gene models, and the other 4 had unspliced introns or other abnormalities that strongly suggested they are aberrant.
Genome Distance Determination. Pairwise alignments were used to assign each putative functional srz gene in C. elegans a closest functional relative in C. elegans (i.e., its closest paralog). The physical distance between each such pair was determined for all pairs that were on the same chromosome. These distances were compared with distances between randomly assigned pairs from the same gene set. Lists of distances were compared by a Mann–Whitney U test with two-tailed P values.
Phylogenetic Analysis. Gene trees were constructed by the maximum-likelihood method implemented by proml [from the phylip package (24)]. Multiple alignments for trees were generated by clustalx with blosum matrices and otherwise default settings (25, 26). For Fig. 1, maximum-likelihood bootstrap analysis (24) was performed on three subtrees (1,000 replicates each), because computer run time for the entire tree was prohibitive. Conservation between C. elegans and C. briggsae was assessed for 28 C. briggsae SRZ proteins and 125 C. briggsae STR/SRJ proteins aligned against all 60 predicted C. elegans SRZ proteins and all 229 predicted C. elegans STR/SRJ proteins. For the best-scoring pair alignment, the number of identities was divided by the average lengths of the query and match proteins.
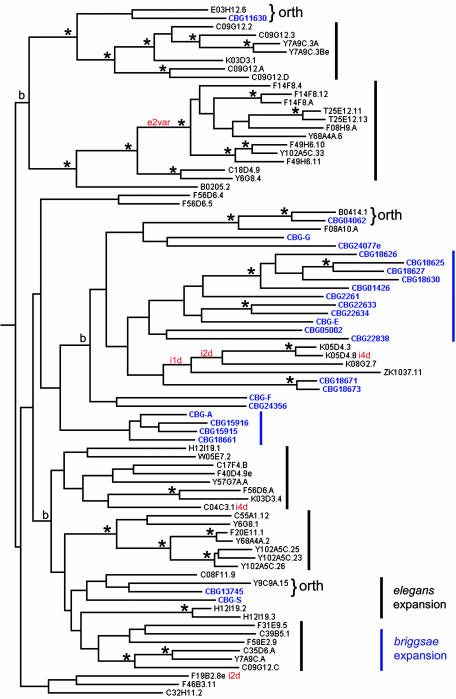
Fig. 1.
Maximum-likelihood gene tree of 82 SRZ proteins. C. elegans gene names are black; C. briggsae gene names are blue. Bootstrap analysis was performed on the subtrees marked “b.” Branch points found >95% of the time are marked with an asterisk. Probable one-to-one ortholog pairs are labeled “orth.” Bars to the right mark probable species-specific gene expansions. Red text on some branch points and sequences indicates a parsimonious interpretation of intron and exon variants in C. elegans as follows: “iNd,” loss of intron N from an ancestral four-intron structure; “e2var,” a length variant of exon 2. All genes below such a mark on the tree share the same variant.
Codon Analysis. For each set of SR genes to test, we aligned predicted translations by using clustalx (25, 26) and, from this, generated a corresponding codon alignment and tree (24). The codon alignment and tree were provided to codeml, and models 7 and 8 were run by using at least three initial dN/dS seeds. The neutral model 7 assumes a β-distribution with 10 dN/dS ratio classes constrained to lie between 0 and 1.0, whereas the selection model 8 permits 1 additional dN/dS ratio class without constraint. For cases in which the additional dN/dS ratio was >1, significance was tested by a χ2 test (with 2 degrees of freedom) on twice the negative of the log-likelihood difference between models 7 and 8, a statistic found to be conservative in simulations (21). To test whether dN/dS was significantly >1.0, we compared model 8 to a comparable model with the 11th dN/dS class fixed at 1.0. Significance was determined as above but with 1 degree of freedom. The power and accuracy of the codeml algorithm with varying tree lengths has been assessed by simulation (21). Total tree length had an optimum in the range of 1–10, although tree lengths up to 100 still allowed substantial power if the number of sequences was large. With our data set, inclusion of more sequences increases tree length, producing a tradeoff between large data sets and tree length. In addition, alignment difficulties became more pronounced with increasing divergence, a factor not yet analyzed in simulations. We made most codeml runs with a local clade of 4–12 sequences, but, in a few cases, we made codeml runs on larger sets of sequences.
Alternative SRZ Alignments. The smallest SRZ clades aligned unambiguously, but positively selected sites in more divergent SRZ clades were in segments that align with difficulty (as expected for segments under positive selection). We tested whether alignment artifacts might produce false-positive selection results. In several specific cases, we realigned regions by hand in an attempt to reduce amino acid diversity at sites identified as being under positive selection. We investigated more systematically a set of eight genes that gave a clear signal of positive selection. We aligned the proteins both by hand and with a wide range of gap penalties, including several with reduced penalties. None of these alternative alignments caused more than minor changes in codeml results. We also inspected codon usage in the C. elegans srz genes and found no substantial differences. Highly significant positive selection was also found for gene group A in C. elegans (Fig. 3), assuming equal equilibrium codon frequencies.
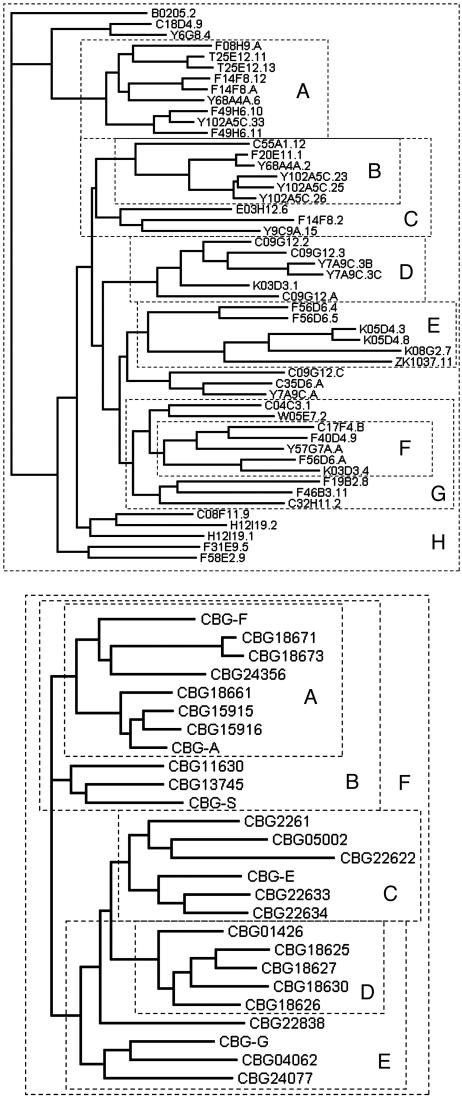
Fig. 3.
Groups analyzed for dN/dS ratio. Genes analyzed by codeml are indicated on a tree for each species. Each set is boxed by a dashed line, and the lettering identifies the results summarized in Table 2. In each species, a few proteins were excluded because they did not align robustly (or, in two cases, because they had not yet been found).
Results
The srz Gene Family. The srz gene family was recognized by H.M.R. (unpublished data) as a distant relative of better-documented SR gene families. As a step toward characterizing this gene family, we carried out a complete reannotation of all loci with the potential to encode part or all of an srz gene product. Using family-based protein homology information, we were able to derive plausible gene models for 60 srz-related genes in C. elegans. In addition, we identified 53 loci that are probably nonfunctional: 14 were gene fragments, and 39 had stop codons, frameshifts, splice site defects, or deletions that probably render them nonfunctional. A high frequency of gene fragments and apparently nonfunctional genes has also been described for other SR families (2–4). Using a similar approach, we derived gene models for 28 srz genes in C. briggsae. For unknown reasons, this related species has fewer genes than C. elegans for this gene family and for some other previously documented SR families (5). The srz loci in C. elegans also share with other SR families their unusual genomic distribution: 94 of the 113 srz loci are on chromosomes IV and V, and 109 of the 113 are on autosomal chromosome arms, which comprise 27.9% of the genome (22). The genes on chromosomes IV and V are strongly clustered in a manner related to their divergence. For example, all of the srz genes on chromosome V are on or very near the right arm, and the average genomic distance between the closest gene pairs was 380 kb (23 pairs), significantly different from 2,161 kb for randomly assigned pairs from the same set of genes (38 pairs, P < 0.0001 by a Mann–Whitney U test). We conclude that most rearrangements that duplicate or rearrange srz genes are intrachromosomal and local, such that related duplicate copies tend to stay close to each other in the genome. Similar findings have been reported in related gene families in C. elegans (2, 3) and for gene families in other organisms (27).
srz Genes Duplicate and Delete Frequently. Fig. 1 shows a maximum-likelihood tree of 82 aligned SRZ proteins from C. elegans and C. briggsae, rooted by a C. elegans srb family outgroup (not shown). For the C. elegans genes, we also analyzed the evolution of intron losses and one exon length variant; both are fully compatible with the protein tree (Fig. 1). The evolution of this family is striking in its dynamics of duplication and deletion. Only three pairs of genes could be plausibly assigned as one-to-one orthologs. Most srz genes in C. briggsae are the result of two gene amplifications absent in C. elegans, whereas those in C. elegans are the result of five amplifications absent in C. briggsae. The gene annotation and tblastn searches showed that the failure to identify one-to-one orthologs is not a result of unrecognized genes. To our knowledge, this gene family is the most dynamic one in these organisms. To permit direct comparison to another dynamic gene family, we completed the C. briggsae annotation of all members of the srj subgroup of the str family and used similar methods to construct the srj tree. For this group of 38 C. elegans proteins and 25 C. briggsae proteins, about half of the C. briggsae genes could be assigned one-to-one orthologs in C. elegans, and there was evidence of only two small gene expansions (data not shown).
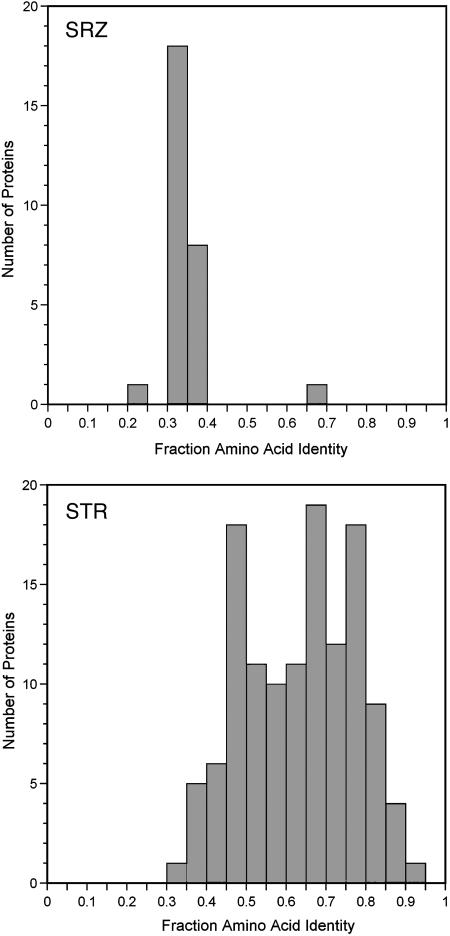
srz Genes Are Rapidly Diverging. We investigated whether the amino acid sequences of srz genes are diverging faster than those of other SR families by comparing C. briggsae family members to their best match in C. elegans. The comparison in this direction is expected to be more robust than the reverse, because the sequence and annotation in C. elegans is more complete. For comparison, we investigated the str family because it is large and the annotation in C. elegans is of high quality (2). To provide a valid data set for comparison, we manually tested or improved gene predictions for 125 C. briggsae genes in the str family. As shown in Fig. 2, SRZ proteins in C. briggsae are significantly more divergent from their closest C. elegans relative than are STR proteins. With slight quantitative differences, similar results were found from alignments of the C. elegans proteins to C. briggsae. We also carried out more limited analyses with the srh, srd, and srw SR families and found that all diverge more slowly than srz genes (data not shown). The rapid divergence in the srz family might result from more rapid gene duplication and deletion, resulting in closest relative assignments that reflect a last common ancestral gene that predated the C. elegans and C. briggsae speciation. Although this possibility is probably correct in part, results in the next section strengthen the possibility that amino acid sequences also diverge faster in the srz family.
Fig. 2.
Divergence of best-match SRZ and STR proteins. The amino acid identities for best-match protein pairs from C. briggsae to C. elegans were plotted as binned histograms. The possibly bimodal distribution for the STR genes may result from genes with one-to-one orthologs (higher identity peak) and those without (because they have an older last common ancestor).
Positive Selection in the SR Superfamily. To analyze the possibility of positive selection acting on specific codons in the SR superfamily, we used the maximum-likelihood method of Nielsen and Yang to test for variation in the dN/dS ratio among sites in sets of closely related SR genes (18, 28, 29). This method is less powerful in detecting positive selection when used with highly divergent genes (21), and it is problematic to align such cases with confidence. For these reasons, we performed most of our analyses with local clades of 4–12 genes. This approach kept genetic divergence reasonably small and kept alignments robust but permitted analysis of a substantial number of sequences. In each SR family, at least three clades were analyzed, with some intentional variation in clade size and tree length. In some cases, we also analyzed larger sets of sequences, resulting in large tree lengths but inclusion of more sequences. There was generally good correspondence between results with local clades and broader clades, perhaps as a result of a balance between tree length and amount of data available to analyze.
In the sra, srb, srd, sre, srg, srh, sri, str, sru, srv, srw, and srx gene families, we saw little or no evidence for positive selection (Table 1). In contrast to these families, with the srz family we repeatedly obtained results indicating positive selection, arguing that we should have detected a similar level of positive selection in any of the other SR families. To test this result directly, we simulated a data set by using characteristics of a set of str sequences for all features except the fraction of sites under positive selection and the dN/dS ratios, which were taken from a representative srz sequence set. We readily detected positive selection in this modeled str data set. The significance of maximum-likelihood results indicating positive selection in the srz genes could not be attributed to sampling error from multiple tests, because the results remained significant even after a conservative Bonferroni correction (30) and subsequent tests with many other srz sequences gave similar results.
Table 1. Summary of dN/dS analysis for the SR superfamily.
| SR family | No. of sequences analyzed (total tree length) | Significance |
|---|---|---|
| sra | 8 (4), 4 (9), 10 (32) | — |
| srb | 4 (7), 7 (8), 4 (10), 6 (13), 12 (20) | — |
| srd | 4 (5), 6 (6), 7 (8), 6 (10), 7 (18), 7 (19), 5 (28) | 1/7 |
| sre | 7 (13), 7 (13), 27 (143) | — |
| srg | 4 (6), 7 (7), 5 (8), 24 (61) | — |
| srh | 5 (6), 6 (6), 6 (8), 7 (8), 8 (8), 8 (15), 19 (28), 16 (31) | — |
| sri | 5 (5), 5 (8), 7 (12), 13 (18), 16 (29) | — |
| str | 6 (5), 6 (8), 5 (10), 12 (13), 40 (23), 22 (78), 27 (78) | — |
| sru | 4 (6), 7 (9), 6 (11), 8 (28) | — |
| srv | 4 (6), 5 (8), 7 (9), 12 (38) | — |
| srw | 4 (3), 5 (3), 12 (21), 9 (22), 14 (26) | 1/5 |
| srx | 8 (16), 7 (18), 8 (18), 20 (77), 6 (184) | — |
| srz | Many (see Table 2) | P < 0.0001 |
All results were statistically insignificant (marked “—”), with the exception of the srz family (see Table 2) and one group each from the srd and srw families. The significance for the srw case was marginal (P ≈ 0.01) and may result from a multiple testing artifact. The significance for the srd case was high (P < 0.0001) but was found in only one small clade of the family.
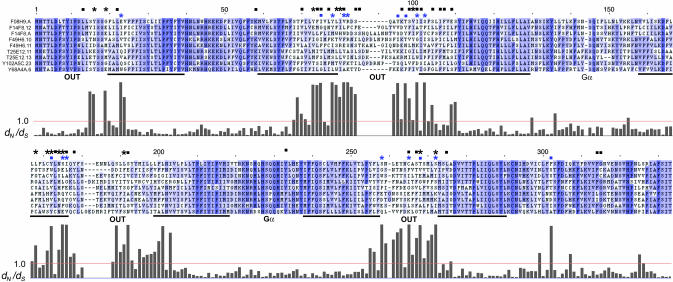
Positively Selected Sites May Define a Ligand-Binding Site. To examine the relationship of sites under positive selection to SRZ protein structure, we inspected the transmembrane (TM) topology of multiply aligned SRZ proteins (Fig. 4). As with other GPCR proteins and in keeping with the crystal structure of rhodopsin (31), there are seven TM domains, and the third TM domain appears unusually long. The N terminus is predicted to be extracellular, and the intracellular loops TM3–4 and TM5–6 are thought to form a cleft in which the cognate G protein α-subunit binds (32–34). There is a clear relationship between codon diversity and TM topology of SRZ proteins. Specifically, dN/dS ratios peak in the extracellular loops and in the extracellular-proximal part of most TM domains. Similarly, sites identified as under significant positive selection are strongly clustered with a similar pattern. With the exception of a few scattered sites, dN/dS ratios are low in intracellular loops, including the regions implicated in G protein binding. The most divergent regions are precisely those implicated in ligand binding in other GPCRs (33, 35). We predict that the sites with high dN/dS ratios form a ligand-binding domain. A similar pattern of positive selection in nociceptor GPCR genes in humans has recently been described and interpreted in a similar manner (36).
Fig. 4.
dN/dS ratios and SRZ protein structure. The results of dN/dS analysis from C. elegans on a multiple alignment of group A SRZ proteins (Fig. 3). Above the alignment, blue marks indicate results from group A: asterisks, dN/dS > 1.0 (P ≥ 0.99); squares, additional sites with P ≥ 0.95. Black asterisks and squares summarize the same values from the other seven C. elegans dN/dS analyses. Below the alignment is a histogram of the approximate posterior mean dN/dS for each ungapped column of this group A alignment, with a red reference line at dN/dS = 1.0. Nearly identical results were obtained from a similar analysis of the group A alignment by using a recent update of paml (version 3.14) that implements an empirical Bayes method for site identification (Z. Yang, personal communication). Black bars indicate positions of TM domains, and text indicates orientation with respect to the plasma membrane and regions of Gα binding, all by analogy with other GPCRs. Alignment shading indicates alignment quality, with darker blue signifying higher scores.
To determine whether the finding of positive selection was robust and to determine the extent of selection in the broader srz gene family, we conducted a systematic dN/dS analysis of many subsets of the C. elegans and C. briggsae srz gene families. The sets of genes analyzed are diagrammed on the trees in Fig. 3, and a summary of the results is shown in Table 2. All sets were substantially better fit by including a codon class with a dN/dS ratio > 1. The overlapping sets F and G in C. elegans gave relatively weak results, and the optimal dN/dS ratios chosen by codeml were relatively low, suggesting that positive selection in this part of the C. elegans gene family is somewhat weaker. Although alignment inaccuracy may remain in some of our analyses, we think this inaccuracy cannot account for the apparent positive selection. First, in several cases, local alignments were clear in regions of positive selection; an example is seen in alignment column 23 in Fig. 4. Second, we investigated the impact of alternative alignments on the dN/dS results (see Materials and Methods) and found that neither hand adjustment of alignments nor systematic variations in multiple alignment parameters changed the fundamental result of highly significant support for positive selection.
Table 2. Summary of dN/dS analysis for the srz family.
| Group | No. of genes | Tree length | Added dN/dS | No. of sites | P value | -2ΔML |
|---|---|---|---|---|---|---|
| C. briggsae | ||||||
| A | 8 | 14.0 | 5.3* | 13 | <0.0001 | 70.0 |
| B | 11 | 23.8 | 3.5* | 8 | <0.0001 | 49.4 |
| C | 6 | 36.6 | 2.9 | 8 | ∼0.0001 | 18.2 |
| D | 5 | 8.9 | 11.5* | 10 | <0.0001 | 49.4 |
| E | 9 | 37.4 | 34.1* | 11 | <0.0001 | 42.2 |
| F (all) | 26 | 82.1 | 3.1* | 6 | <0.0001 | 47.4 |
| C. elegans | ||||||
| A | 9 | 8.3 | 3.7* | 17 | <0.0001 | 95.0 |
| B | 6 | 6.5 | 2.0* | 3 | ∼0.001 | 18.2 |
| C | 9 | 17.3 | 1.7 | 9 | <0.0001 | 28.6 |
| D | 6 | 9.2 | 1.3 | 26 | <0.0001 | 23.2 |
| E | 6 | 15.7 | 2.9* | 4 | <0.0001 | 27.4 |
| F | 5 | 14.0 | 1.4 | 4 | ∼0.001 | 10.2 |
| G | 10 | 32.1 | 1.3 | 3 | ∼0.001 | 11.2 |
| H (all) | 51 | 102.7 | 2.0* | 13 | <0.0001 | 99.0 |
“Group” refers to the diagrams in Fig. 3. “Added dN/dS” is the free 11th ratio computed by model 8 in codeml. “No. of sites” is the number of sites found to be under positive selection (P ≥ 0.95). “P value” derives from the likelihood ratio test of positive selection for the group (see Materials and Methods).
Result was also significant (P < 0.0001) when compared to model 8 run with the 11th dN/dS class fixed at 1.0.
Discussion
This study indicates that srz genes are under positive selection and that the regions of highest dN/dS ratios correlate with the extracellular face of the receptors. We hypothesize that srz gene duplication and divergence is being driven by selection to recognize diverse ligands that change over time or by selection to recognize a wider range of ligands. By precedent in GPCRs, these ligands are likely to be short peptides or small organic molecules. What biological process might SRZ receptors mediate, and why are they subject to positive selection? Two processes solidly established to involve positive selection are antigen recognition and mating specificity (37–39). srz membership in the SR superfamily lends particular credence to a role in some form of chemosensation. Mate choice, sperm–egg interaction, nociception, and pathogen avoidance are specific functions that we speculate are plausible for SR proteins. Experimental evidence testing these possibilities can be obtained in C. elegans by determining srz tissue-specific expression patterns and by obtaining deletion alleles in srz genes.
Attempts to detect positive selection in other SR families were unsuccessful. We cannot rule out that some groups of genes in these families are under positive selection, although tests were negative in nearly every case with three to eight groups in each family. In the srz family, in contrast, nearly all tested groups showed significant positive selection. The distinctness of the srz family is also supported by a higher rate of evolution in the srz family when compared with other analyzed families. Comparison with C. briggsae suggests that this evolution probably happens both at the level of amino acid sequence and at the level of a high rate of gene duplication and deletion. We hypothesize that the srz family plays some unique biological role that is distinct from the other SR families, although the nature of this role remains unknown. Direct evidence for a specific function in the SR superfamily is limited to the str family, one member of which is known to mediate attractive chemotaxis toward the odorant diacetyl (40, 41). It may be that most chemoreceptor families are involved in similar processes and that the srz family is devoted to some other chemosensory process that is more likely to be subject to positive selection, such as mate choice.
Presumably because of their high rate of evolution and low expression levels, genes in the srz family pose a difficult gene prediction problem. More than half of the WormBase srz gene predictions for C. elegans and nearly all of the predictions for C. briggsae required correction. We developed an approach to comparative gene finding (see Materials and Methods) that combines use of short conserved protein motifs, construction of a position-specific score matrix (PSSM) from those motifs, and a six-frame translated genomic search to locate regions with high potential to encode query motifs. Combined local motif hits were used to aid hand annotation of splicing pattern and production of a complete gene model. Comparison to predicted srz gene structures and tblastn searches validated the sensitivity of the method. Because the method focuses on short conserved motifs, it is relatively robust to motif disruption by introns in genomic sequence. Because the method uses a PSSM, it should have better sensitivity than searches with single protein sequences, and it can efficiently cover protein diversity in gene families. The method may be generally useful in assisting gene prediction in similar divergent protein families.
Supplementary Material
Acknowledgments
We thank Emily Rocke and Mary Stewart for useful discussions. This work was supported by a University of Washington Genome Training grant and National Institutes of Health Grant RO1GM48700.
Author contributions: J.H.T., J.L.K., H.M.R., and W.J.S. designed research; J.H.T., J.L.K., H.M.R., and K.L. performed research; J.H.T., J.L.K., H.M.R., and W.J.S. analyzed data; and J.H.T. and J.L.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SR, seven-pass receptor; GPCR, G protein-coupled receptor; TM, transmembrane.
Data deposition: All gene models have been reported to the WormBase database, http://wormbase.org (release no. WS137).
References
- 1.Troemel, E. R., Chou, J. H., Dwyer, N. D., Colbert, H. A. & Bargmann, C. I. (1995) Cell 83, 207–218. [DOI] [PubMed] [Google Scholar]
- 2.Robertson, H. M. (2001) Chem. Senses 26, 151–159. [DOI] [PubMed] [Google Scholar]
- 3.Robertson, H. M. (2000) Genome Res. 10, 192–203. [DOI] [PubMed] [Google Scholar]
- 4.Robertson, H. M. (1998) Genome Res. 8, 449–463. [DOI] [PubMed] [Google Scholar]
- 5.Stein, L. D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M. R., Chen, N., Chinwalla, A., Clarke, L., Clee, C., Coghlan, A., et al. (2003) PLoS Biol. 1, E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris, T. W., Chen, N., Cunningham, F., Tello-Ruiz, M., Antoshechkin, I., Bastiani, C., Bieri, T., Blasiar, D., Bradnam, K., Chan, J., et al. (2004) Nucleic Acids Res. 32, Database issue, D411–D417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. (2002) Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 8.Swanson, W. J. & Vacquier, V. D. (1995) Proc. Natl. Acad. Sci. USA 92, 4957–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, Y. H., Ota, T. & Vacquier, V. D. (1995) Mol. Biol. Evol. 12, 231–238. [DOI] [PubMed] [Google Scholar]
- 10.Tsaur, S. C. & Wu, C. I. (1997) Mol. Biol. Evol. 14, 544–549. [DOI] [PubMed] [Google Scholar]
- 11.Nei, M. & Gojobori, M. (1986) Mol. Biol. Evol. 3, 418–426. [DOI] [PubMed] [Google Scholar]
- 12.Hughes, A. L. & Nei, M. (1988) Nature 335, 167–170. [DOI] [PubMed] [Google Scholar]
- 13.Swanson, W. J., Yang, Z., Wolfner, M. F. & Aquadro, C. F. (2001) Proc. Natl. Acad. Sci. USA 98, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang, Z., Nielsen, R., Goldman, N. & Pedersen, A. M. (2000) Genetics 155, 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang, Z. & Swanson, W. J. (2002) Mol. Biol. Evol. 19, 49–57. [DOI] [PubMed] [Google Scholar]
- 16.Yang, Z. & Bielawski, B. (2000) Trends Ecol. Evol. 15, 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang, Z. & Nielsen, R. (2002) Mol. Biol. Evol. 19, 908–917. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen, R. & Yang, Z. (1998) Genetics 148, 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, Z. & Nielsen, R. (2000) Mol. Biol. Evol. 17, 32–43. [DOI] [PubMed] [Google Scholar]
- 20.Swanson, W. J., Aquadro, C. F. & Vacquier, V. D. (2001) Mol. Biol. Evol. 18, 376–383. [DOI] [PubMed] [Google Scholar]
- 21.Anisimova, M., Bielawski, J. P. & Yang, Z. (2001) Mol. Biol. Evol. 18, 1585–1592. [DOI] [PubMed] [Google Scholar]
- 22.Barnes, T. M., Kohara, Y., Coulson, A. & Hekimi, S. (1995) Genetics 141, 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reboul, J., Vaglio, P., Rual, J. F., Lamesch, P., Martinez, M., Armstrong, C. M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. (2003) Nat. Genet. 34, 35–41. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein, J. (1989) phylip, Phylogeny Inference Package (Department of Genome Sciences, University of Washington, Seattle), Version 3.2.
- 25.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schein, M., Yang, Z., Mitchell-Olds, T. & Schmid, K. J. (2004) Mol. Biol. Evol. 21, 659–669. [DOI] [PubMed] [Google Scholar]
- 28.Yang, Z. (1997) Comput. Appl. Biosci. 13, 555–556. [DOI] [PubMed] [Google Scholar]
- 29.Yang, Z., Swanson, W. J. & Vacquier, V. D. (2000) Mol. Biol. Evol. 17, 1446–1455. [DOI] [PubMed] [Google Scholar]
- 30.Bonferroni, C. (1936) Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze 8, 3–62. [Google Scholar]
- 31.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T., Stenkamp, R. E., et al. (2000) Science 289, 739–745. [DOI] [PubMed] [Google Scholar]
- 32.Greasley, P. J., Fanelli, F., Rossier, O., Abuin, L. & Cotecchia, S. (2002) Mol. Pharmacol. 61, 1025–1032. [DOI] [PubMed] [Google Scholar]
- 33.Henry, L. K., Khare, S., Son, C., Babu, V. V., Naider, F. & Becker, J. M. (2002) Biochemistry 41, 6128–6139. [DOI] [PubMed] [Google Scholar]
- 34.Havlickova, M., Blahos, J., Brabet, I., Liu, J., Hruskova, B., Prezeau, L. & Pin, J. P. (2003) J. Biol. Chem. 278, 35063–35070. [DOI] [PubMed] [Google Scholar]
- 35.Fong, T. M. & Strader, C. D. (1994) Med. Res. Rev. 14, 387–399. [DOI] [PubMed] [Google Scholar]
- 36.Choi, S. S. & Lahn, B. T. (2003) Genome Res. 13, 2252–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes, A. L. & Nei, M. (1989) Proc. Natl. Acad. Sci. USA 86, 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes, A. L., Ota, T. & Nei, M. (1990) Mol. Biol. Evol. 7, 515–524. [DOI] [PubMed] [Google Scholar]
- 39.Swanson, W. J. & Vacquier, V. D. (2002) Nat. Rev. Genet. 3, 137–144. [DOI] [PubMed] [Google Scholar]
- 40.Troemel, E. R., Kimmel, B. E. & Bargmann, C. I. (1997) Cell 91, 161–169. [DOI] [PubMed] [Google Scholar]
- 41.Sengupta, P., Chou, J. H. & Bargmann, C. I. (1996) Cell 84, 899–909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.