Abstract
Using a maskless photolithography method, we produced DNA oligonucleotide microarrays with probe sequences tiled throughout the genome of the plant Arabidopsis thaliana. RNA expression was determined for the complete nuclear, mitochondrial, and chloroplast genomes by tiling 5 million 36-mer probes. These probes were hybridized to labeled mRNA isolated from liquid grown T87 cells, an undifferentiated Arabidopsis cell culture line. Transcripts were detected from at least 60% of the nearly 26,330 annotated genes, which included 151 predicted genes that were not identified previously by a similar genome-wide hybridization study on four different cell lines. In comparison with previously published results with 25-mer tiling arrays produced by chromium masking-based photolithography technique, 36-mer oligonucleotide probes were found to be more useful in identifying intron–exon boundaries. Using two-dimensional HPLC tandem mass spectrometry, a small-scale proteomic analysis was performed with the same cells. A large amount of strongly hybridizing RNA was found in regions “antisense” to known genes. Similarity of antisense activities between the 25-mer and 36-mer data sets suggests that it is a reproducible and inherent property of the experiments. Transcription activities were also detected for many of the intergenic regions and the small RNAs, including tRNA, small nuclear RNA, small nucleolar RNA, and microRNA. Expression of tRNAs correlates with genome-wide amino acid usage.
Keywords: higher plant, transcriptome, maskless array synthesizer
The flowering plant Arabidopsis thaliana is an important genetic model organism for investigating the basic mechanisms involved in the growth and development of multicellular eukaryotes. In addition, it was the first plant for which a complete genome sequence was determined (1). The availability of a complete genome sequence opens many avenues for new experiments probing different cellular functions on a genome-wide scale (2). However, designing such large-scale experiments is only possible when a complete map of all transcribed regions on the genome is available. Typical computationally derived preliminary annotations of the new sequences are error-prone, especially for shorter genes and for genes with multiple introns. Therefore, experimentally determined annotation is considered to be the most definitive proof for transcription of predicted genes and for the correct identification of coding regions.
Several experimental methods have been developed in recent years to carry out this difficult task. They include cloning and cataloging of expressed sequence tags (ESTs) and full-length cDNAs, serial analysis of gene expression (3) and “sequence tagged site”-based genetic mapping (4). However, none of the above-mentioned methods comprehensively probe the complete genome sequences for proof of RNA expression. Moreover, these methods are not sufficiently scalable to test multiple tissue types or experimental conditions in a cost-effective manner. Genome-wide tiling arrays can overcome many of the shortcomings of the previous approaches by comprehensively probing transcription in all regions of the genome. This technology has been used successfully on different organisms (5–12). A recent study on A. thaliana reported measuring transcriptional activities of four different cell lines by using 25-mer-based tiling arrays that were developed with a chromium masking-based photolithography technique (11).
In this study, we used a modified optical synthesis technique to develop a genome-wide tiling array for the complete genome of A. thaliana. This procedure used a maskless array synthesizer (5–7) that created high-density oligonucleotide arrays rapidly without the need to invest resources in creating expensive and unchangeable chromium masks. For this experiment, the maskless array synthesizer was used to create a whole-genome tiling array containing 5 million different 36-mer oligonucleotide probes that covered the Arabidopsis genome from end to end, with 10-bp gaps on both strands, for all nuclear, mitochondrial, and chloroplast chromosomes. The arrays were hybridized with mRNA isolated from an undifferentiated, rapidly dividing, A. thaliana cell line (T87; ref. 13) for which preliminary evidence had suggested that a large number of genes were being expressed. The results of this study are reported here.
Materials and Methods
Design of the Arrays. The version of the sequence released in January 2002 at GenBank (www.ncbi.nlm.nih.gov) with accession entries NC_003076, NC_003075, NC_003074, NC_003071, NC_003070, NC_000932, and NC_001284 was used to design the arrays. Based on the chromosomal sequences, 13 high-density arrays were constructed, each with ≈400,000 different 36-mer oligonucleotide probes. Probes were selected uniformly from both strands of the chromosomal sequences with gaps of 10 base pairs between two consecutive probes covering the entire genome. Additional details about the probe selection algorithm are provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Hybridization Experiment. The selected oligonucleotides were synthesized on 13 glass-based arrays by using an in situ maskless photolithographic synthesis device and hybridized with mRNA extracted from the established T87 A. thaliana cultured cell line.
Sample labeling. Total RNA was extracted from the T87 line (13) of liquid grown tissue culture cells of A. thaliana, converted to double-stranded cDNA by using a SuperScript Choice System (GIBCO/BRL), and mRNA was labeled by using an oligo(dT) primer containing the T7 RNA polymerase promoter (5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-T24-3′). Briefly, 10 μg of total RNA was incubated with 1× first strand buffer/10 mM DTT/500 μM dNTPs/5 pM primer for 60 min at room temperature. Second-strand synthesis was accomplished by incubation with 200 μM dNTPs/0.07 units/μl DNA ligase/0.27 units/μl DNA polymerase I/0.013 units/μl RNase, 1× second-strand buffer/10 units T4 DNA polymerase for 2 h. Double-stranded cDNA was purified by using phenol-chloroform extraction and Eppendorf Phase-Lock Gel tubes, and ethanol was precipitated, washed with 80% ethanol, and resuspended in 3 μl of water. In vitro transcription was used to produce biotin-labeled cRNA from the cDNA by using the Ambion (Austin, TX) MEGAscript T7 kit. Briefly, 1 μg of double-stranded cDNA was incubated with 7.5 mM ATP and GTP/5.6 mM UTP and CTP/1.9 mM bio-11-CTP and bio-16-UTP (Sigma-Aldrich) in 1× transcription buffer and 1× T7 enzyme mix for 5 h at 37°C. Before hybridization, cRNA was fragmented to an average size of 50–200 bp by incubation in 100 mM potassium acetate/30 mM magnesium acetate/40 mM Tris-acetate for 35 min at 94°C. For quality control at all steps, including input RNA quality, first- and second-strand cDNA synthesis, in vitro transcription, and fragmentation, assay performance was monitored by running small sample aliquots on the Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA). Hybridization and washing. High-density 36-mer tiling arrays were hybridized with 12 μg of cRNA in 300 μl, in the presence of 50 mM Mes/0.5 M NaCl/10 mM EDTA/0.005% (vol/vol) Tween 20 for 16 h at 45°C. Before application, samples were heated to 95°C for 5 min, 45°C for 5 min, and then centrifuged at 10,000 × g for 5 min. Hybridization was performed in a hybridization oven with continuous mixing. After hybridization, arrays were washed in nonstringent (NS) buffer [6× standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA); 0.01% Tween 20] for 5 min at room temperature, followed by washing in stringent buffer (100 mM Mes, 0.01 M NaCl, and 0.01% Tween 20) for 30 min at 45°C. After washing, arrays were stained with streptavidin-Cy3 conjugate (Amersham Pharmacia) for 25 min at room temperature, followed by a 5 min wash in NS buffer, a 30-sec rinse with final rinse buffer, and a blow dry step by using high-pressure grade 5 argon.
Scanning and data analysis. Arrays were scanned on an Axon 4000B scanner (Axon Instruments, Union City, CA), and features were extracted by using nimblescan software (NimbleGen Systems, Madison, WI). The raw data from this measurement has been deposited to the National Center for Biotechnology Information GEO database in the MIAME format (with the following entry numbers: series, GSE 2247; samples, GSM 41324–41336; platforms, GPL 1840–1842 and 1844–1853). In the subsequent analysis, raw data from all probes were mapped back onto the latest released version of the genome (version 5 was released on Feb. 19, 2004). Probe signals were normalized by dividing them by the median intensity of all probes of the respective array and logarithms (base 2) of the normalized numbers were considered. Reproducibility of the data. Reproducibility of the array platform was verified in ref. 12, where multiple arrays were hybridized with cDNA samples taken from identical and independent labeling reactions. This experiment produced replicates with r2 correlation between 0.90 and 0.95, indicating a high level of reproducibility of the experiments. The effect of potential variation across individual Arabidopsis cDNA samples was further reduced by pooling reverse transcription products of several separate labeling reactions (data not shown).
Signals on the Genes. Probes matching unique locations on the genome were considered for all subsequent analysis. For each protein-coding gene annotated in the V5 GenBank release of A. thaliana, the average expression level was computed by taking the arithmetic mean of log-normalized signals on all probes that were at least half within the exon regions of the gene. Of 26,330 annotated protein-coding genes in the V5 release, 245 (0.9%) genes were excluded from this study because they did not have any representative probes for their exons. This result happened because either those genes were located on repeat regions of the genome or the segments containing them were not present in the earlier release of the chromosome, based on which the arrays were designed.
To decide whether a gene was expressed, a threshold level representing the “background signal” was calculated for the whole genome based on the average expression levels in the promoter regions of genes (11). Because promoters are known to be transcriptionally inactive, it is reasoned that the majority of signal on them would be hybridization noise and not real signal. Only genes verified by full-length cDNAs, for which promoter regions were known for certain, were considered in this calculation, and the average signal was calculated based on probes on both strands between 150 and 500 bases upstream of these genes. In V2 release, 4,671 genes were explicitly marked as verified by full-length cDNAs, among which 4,658 had more than one probe in the promoter regions. A hybridization signal that was above the average signal levels exhibited by 85% of the promoters was 0.734 on a log (base 2)-normalized scale. A more stringent level, i.e., >90% of the promoter averages, was found to be 0.936.
RNA Analysis. A list of known small RNAs and their locations on the genome was obtained from the latest annotation (Feb. 19, 2004) files at the GenBank web site. For microRNAs (miRNAs), premiRNA sequences were obtained from the database at the Sanger Institute (www.sanger.ac.uk). As of Jan. 5, 2005, this database contained 112 miRNA sequences for A. thaliana. For all of the small RNAs mentioned above, average intensity was computed in the following manner. Signals on all probes within the reported length of the small RNA and an additional 250 bases on both 5′ and 3′ ends were considered, and the arithmetic mean of log-normalized signals on these probes was computed. In addition, plots of signals along these genes were manually inspected to verify that the activity was not due to other factors such as nearby protein-coding regions. The detailed plots of signals on all probes along these genes are available upon request.
Proteomic Analysis of T87 Soluble Protein By Using Two-Dimensional HPLC-ESI-Tandem MS Analysis of Tryptic Peptides. T87 cells were grown in liquid culture and collected by vacuum filtration over a sintered glass frit. Ice-cold grinding buffer (290 mM sucrose/250 mM Tris·HCl, pH 7.6/25 mM EDTA/0.5% polyvinyl pyrolidine/1 mM DTT/1 mM PMSF/10 μg/ml leupeptin/1 μg/ml pepstatin/1 μg/ml E64/1 μM Bestatin/100 μM 1,10-phenanthroline) was added to cells at a ratio of 3 ml of grinding buffer per gram of wet cells. The cells were then disrupted by using a Polytron homogenizer (Brinkmann) and filtered through two layers of Miracloth (Calbiochem). The lysate was fractionated by centrifugation in two steps: first for 10 min at 5,000 × g to remove organelles and second for1hat 100,000 × g to pellet membranes and to leave soluble proteins in the supernatant. The supernatant was removed and incubated for 30 min at 25°C after the addition of DTT to 5 mM. Iodoacetamide was added to a concentration of 50 mM and further incubated for 30 min at 25°C in the dark. The acetamidylated protein was split into two batches, one was exchanged into 8 M urea/50 mM ammonium bicarbonate, pH 7.5 by gel filtration chromatography (10 ml of D-salt column 43243, Pierce) and the other was precipitated by the addition of acetone to 80%, and the protein pellet was collected by centrifugation after a 14-h incubation at –20°C. The acetone precipitate was dissolved in 8 M urea/50 mM ammonium bicarbonate pH 7.5/1 mM DTT, and the concentrations of both batches were estimated by the method of Warburg and Christian (14). Aliquots containing ≈1 mg of protein were removed for trypsinolysis after dilution to 1 M urea, with 50 mM ammonium bicarbonate, pH 7.5/1 mM DTT. Proteolysis was initiated by the addition of 20 μg of sequencing grade trypsin (Promega) and incubated for 14 h at 37°C, after which the reaction was terminated by addition of formic acid to 1%. Each digest was then subjected to a strong cation exchange chromatography on a Mono-S column (1 ml) with solvent delivered (2 ml/min) by an FPLC (Amersham Pharmacia) system at 4°C. Fractionation was accomplished by salt gradient elution by using buffers A and B (A is 25% acetonitrile and 5 mM sodium phosphate, pH 3.0 in water, and B is 25% acetonitrile, 5 mM sodium phosphate (pH 3.0), 500 mM KCl in water, and gradient: 0–70% B over 20 min, 70–100% B in 2 min, a 10-min wash in 100% B, followed by reequilibration to 0% B) with 0.5-ml fractions collected. Fractions were assembled into 10 pools of approximately equal content as judged by 280 nm UV traces. Peptides from each of these pools were purified by solid phase extraction (Spec-PT-C18, Varian) before analysis by C18-reverse phase μESI tandem MS on a Q-TOF2 (Micromass, Manchester, U.K.) mass spectrometer with a previously described modified LC electrospray source (15). Briefly, chromatographic separation of peptides before MS was accomplished by using columns that were made with fused silica tubing (OD at 365 μm and inner diameter at 100 μm) with pulled tips (1 μm of orifice), packed with Zorbax Eclipse XDB-C18, 5 μm, 300 Å pore-size media (Agilent, Palo Alto, CA) to 12 cm (16). An Agilent 1100 series HPLC was used for sample application (1 μl/min) and delivery of a 0.1% (vol/vol) formic acid/water to 70% (vol/vol) acetonitrile with a 0.1% (vol/vol) formic acid gradient (150–200 nl/min) and flow rates achieved by splitting. Voltage was applied upstream of the column by using a platinum wire electrode introduced through a PEEK T junction. Tandem mass spectra (50–2,200 m/z) were collected over four channels from MS spectral features observed between 400 and 2,000 m/z; redundancy was limited by dynamic exclusion, and charge-dependent collision energy profiles were empirically predetermined. Tandem MS data were converted to pkl file format by using masslynx 3.5 (Micromass), and the resulting pkl files from each pool from both the gel-filtered and acetone-precipitated batches were concatenated. The combined pkl files were used to search A. thaliana protein amino acid sequences (GenBank V5) by using mascot (Matrix Science, London) with N-terminal acetylation, methionine oxidation, and serine and threonine phosphorylation as variable modifications, and cysteine acetamidylation as a fixed modification. Output from mascot was further processed to remove nonunique peptides from the analysis; proteins with Mowse scores ≥52 were included as statistically significant.
RT-PCR Experiment. RT-PCR were performed on 947 genes selected as part of a large protein expression study through the University of Wisconsin Center for Eukaryotic Structural Genomics (17). They were chosen based on several criteria, including the lack of homopolymer stretches, size, and, most importantly, the absence of any currently known structural information for that protein and its homologues. Oligonucleotide primer pairs (42-mer) were designed based on the predicted start and stop sites and were used to amplify cDNA out of a T87 cell line derived mRNA population that had been converted into ssDNA by using reverse transcriptase. An RT-PCR-positive result meant that an ethidium bromide stained band corresponding to the predicted size for that gene was observed after amplification. Furthermore, all of the RT-PCR-positive cDNA products were verified by sequence confirmation (data not shown).
Results and Discussion
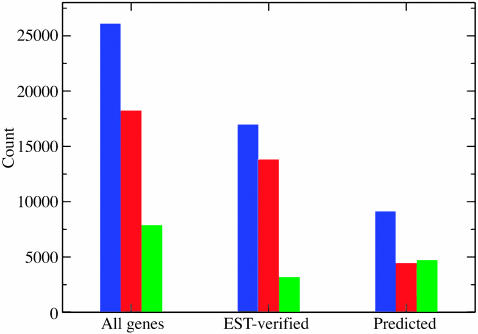
After mapping and proper normalization of the raw data, the expression levels of the annotated genes were computed (Data Sets 1 and 2, which are published as supporting information on the PNAS web site). The latest GenBank annotation, version 5, (released Feb. 19, 2004) of the A. thaliana genome reported a total of 30,873 genes from all of the nuclear, mitochondrial, and chloroplastic chromosomes. This list included 26,330 protein-coding genes (17,016 nuclear EST/cDNA-verified, 9,192 nuclear predicted/hypothetical and 122 from mitochondria/chloroplast), 3,786 pseudogenes, 673 tRNAs, 12 rRNAs, 15 small nuclear RNAs, and 57 small nucleolar RNAs. We found transcriptional activities for 18,244 (70%) of the protein-coding genes to be above a threshold level of 0.734, and 16,054 (61%) of the protein-coding genes to be above a threshold level of 0.936. These threshold levels were determined based on expression levels at the promoter regions (see Materials and Methods for details). Splitting all nuclear protein-coding genes into two categories, EST-verified and predicted, 13,820 of 17,016 (81%) of the EST/cDNA supported genes were expressed and 4,338 of 9,192 (47%) of the hypothetical genes were expressed by using the lower threshold value (0.734) (Fig. 1). When the higher threshold value (0.936) was used, 12,556 of 17,016 (74%) of the EST-verified genes (Fig. 7, which is published as supporting information on the PNAS web site) and 3,417 of 9,192 (37%) of the hypothetical genes were expressed.
Fig. 1.
Relative number of genes whose expression into mRNA was detectable (threshold > 0.734) by the 36-mer tiling experiment reported here (blue, total; red, detected; green, undetected). The left set of bars shows the relative count from all genes reported in GenBank V5 release, the middle set represents the count from those genes that were previously verified by ESTs or cDNAs, and the right set shows the subset of genes that were predicted.
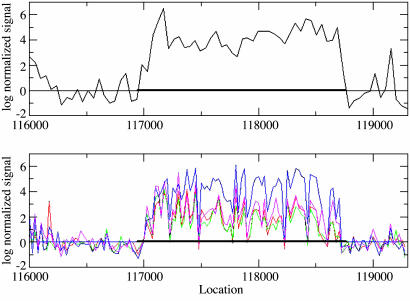
Similar whole-genome microarray measurements were recently reported for A. thaliana by using a 25-mer oligonucleotide array with mRNA extracted from four different cell types, namely root cells, flower cells, suspended cell culture, and cold-treated light-grown cells (11). A comparison of our 36-mer tiling results with the previous 25-mer data set has provided a useful benchmark for further investigations of tiling technology and confirmation that unexpected transcriptional activity in intergenic and antisense regions is a reproducible and potentially important biological phenomenon. Among the expressed genes detected by this study (threshold > 0.936), 207 (56 EST/cDNA verified and 151 predicted/hypothetical) were not detected in any of the four cell types measured by the previous experiment (Data Set 2). A comparison of two data sets suggests that the longer oligonucleotides used in this study may provide a better resolution of intron–exon boundaries (Fig. 2). For longer exons, 36-mer data were more likely to show uninterrupted high signals for the entire exon. In the 25-mer data set, signals on such long exons fell to <0 more often and, therefore, such long exons could be misidentified as multiple exons.
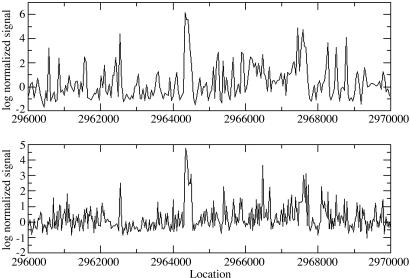
Fig. 2.
Hybridization intensities for the gene At1g01300, encoding an aspartyl protease family protein previously verified by EST and cDNA, are shown here. (Upper) This study. (Lower) Previous study (11). Red, flower; green, root; blue, suspended cell; magenta, cells in the presence of light. The gene is located on Watson strand of chromosome 1 and has only one exon marked by solid black line near y = 0. For long exons, this study obtained signals above cutoff for all or most probes, whereas the signals measured by the previous study more often fell below the cutoff. This result could be due to the difference in choices of probe lengths or to other differences in hybridization protocol between the two experiments.
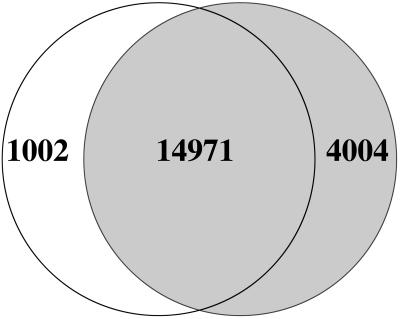
Because the T87 cell line of this study is biologically closest to the suspended cell line of the previous work (11), a direct comparison was made between these two data sets. Only the nuclear genes were considered for this comparison because the previous study did not measure mitochondrial or chloroplast transcription. Using the more stringent cutoff criterion, the current and previous study detected 15,973 and 18,975 nuclear genes, respectively (Fig. 3). The higher number of genes detected by the previous study could be due to either false identification or differences in RNA representation by differential transcription. The second factor is not insignificant as we discuss below. Approximately 75% of the 100 highest-expressed genes in either data set were within the top 1,000 in the other one. However, even among the top 100 of each study, a small fraction was undetected by the other one. For example, 9 of the 100 most expressed genes of the previous study were undetected by this work (At2g43535, At5g03545, At2g38860, At2g38210, At3g16430, At3g48580 At2g39310, At3g16450, and At3g09260). Similarly, three of the 100 most expressed genes in the current study were undetected by the previous measurement (At3g29700, At1g47400, and At2g04320). Data Set 2 shows the relative numerical ranks of all genes in both data sets based on their expression levels.
Fig. 3.
Overlap among sets of genes expressed in T87 cell line (white circle) and suspended cell line of the previous study (gray circle) (11).
To determine whether there is a correlation between mRNA level and protein abundance, a proteomic analysis was performed on total soluble protein extracted from the T87 cell cultures by using two-dimensional HPLC-tandem MS. This technique provides a listing of the most abundant proteins in the sample because when a small sampling of the entire population is performed, the most abundant proteins are the most likely to be identified. By focusing only on those several thousand tryptic peptides that provided robust and easily matched fragmentation patterns with the ≈26,500 annotated proteins, we identified 127 highly abundant proteins from the same cell line from which mRNA levels were measured. Comparison with gene expression data showed that the genes coding for these proteins had very strong transcription levels, with 96 being in the category of genes with the top 10% expression levels, 121 in the top 30% category, 126 in the top 50% category, and only one with an anomalously low mRNA measurement. Visual inspection of hybridization patterns for the top 96 genes confirmed their strong activity (Fig. 8). Their signal-to-noise ratios were so high that the intron–exon boundaries were clearly visible for these genes.
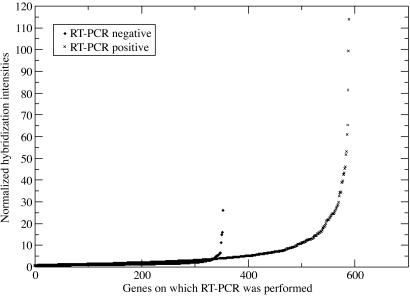
Extensive verifications of the results from this large-scale tiling array-based study were performed, both by RT-PCR and additional “gene-only” microarray measurements, as discussed in Materials and Methods (see Table 1, which is published as supporting information on the PNAS web site). RT-PCR data were available for a subset of the genome containing 947 (3.5%) genes, which were chosen as part of a large protein expression study through the University of Wisconsin Center for Eukaryotic Structural Genomics (17). The RT-PCR cloning effort amplified 594 full-length cDNAs of 947 genes (Data Set 3, which is published as supporting information on the PNAS web site). Fig. 4 shows that mRNA transcripts confirmed by RT-PCR have stronger hybridization intensities than the mRNA transcripts not detected by the RT-PCR method. Of the 594 RT-PCR positive genes, only 18 were not detectable by microarray hybridization. However, of the 353 RT-PCR negative genes, 130 genes had positive hybridization signals. Because the RT-PCR experiments were designed to produce full-length cDNAs, the choice of primers was limited exactly to the predicted start and stop sites. Therefore, it is possible that some of the 130 ORF-based primers chosen for these 130 ORFs were at fault, either because they had secondary structures that impaired their ability in a PCR reaction or they spanned an incorrectly predicted intron/exon boundary.
Fig. 4.
Plot of hybridization intensities (T87 cell line) for the 594 RT-PCR positive (X) and 353 RT-PCR negative (♦) genes. Strong hybridization intensities observed for some RT-PCR negative genes (near gene no. 350) were artifacts due to ORF-based primer choices. Seventeen of 20 RT-PCR negative genes in this region were later confirmed positive by a measurement that chose different sets of primers.
To test whether RT-PCR negative results for the strongest hybridizing genes were actually due to the limitation in the ORF-based primer choice, we reanalyzed 20 of these genes by using new primers that spanned a 300- to 700-bp region of each of the selected mRNAs. These primers were chosen so that they lacked secondary structure as well as primer pair interaction. In addition, the product of these PCR reactions was designed to span an intron, thus allowing distinction between cDNA amplification and amplification from possible contaminating genomic DNA. The results of these experiments showed RT-PCR bands of the correct predicted sizes for 17 of the 20 strongest hybridizing genes that were RT-PCR negative in Fig. 4. These results indicate that our explanation for the RT-PCR negative genes in the original University of Wisconsin Center for Eukaryotic Structural Genomics experiment is probably correct.
Data Set 3 includes the list of all 947 genes with their RT-PCR results, average expression level, and the number of probes. Gene-only arrays comprised of 60-mers and 25-mers chosen at the 3′ end of the 30,000 genes together with mismatch oligonucleotides were also used to study RNA expression levels in T87 cells. These results together were compared with the tiling arrays and RT-PCR observations and are presented in Data Set 1.
In addition to regions of the genome with protein-coding genes, a large amount of strongly hybridizing RNA is found in regions that are annotated as either (i) antisense sequences corresponding to known genes or (ii) regions between known genes, and (iii) to regions producing known small RNA products such as tRNA and miRNA. Among all protein-coding genes, antisense activities were observed for 12,090 genes (cutoff = 0.936). In this calculation, an antisense signal of a gene was estimated as the mean of log-normalized signals on all probes antisense to its exons. Such large-scale antisense activities were reported by a previous genome-wide study on Arabidopsis (11), and a comparison of the two data sets shows that the antisense expression is a reproducible feature of these experiments. Interestingly, for many genes, the average activity for the antisense strand was much stronger than the sense strand. Thus, 7,911 genes showed stronger antisense activity than sense activity. This finding is an unexpected phenomenon, and the similarity of antisense activities between the 25-mer and 36-mer data sets suggests that it is a reproducible and inherent property of the experiments. For further confirmation of antisense activities, three genes (At4g01985, At5g46730, and At5g49440) were chosen based on their high ratios (≈10-fold) of raw antisense/sense hybridization levels. RT-PCR measurement by using strand-specific cDNA pools for each gene confirmed antisense activities for two of them (At4g01985 and At5g49440).
Additional transcriptional activity was observed in several short- and long-expressed blocks of the intergenic region. In Data Sets 4 and 5, which are published as supporting information on the PNAS web site, a full list of such regions of strong intergenic activity is provided, and, in Fig. 5, one example of this phenomenon found in both data sets is shown. Some of the expressed blocks could be identified with short protein-coding genes. A detailed analysis for all expressed intergenic regions that could potentially code for proteins of length >50 aa are presented in Data Set 6, which is published as supporting information on the PNAS web site. The following issues were considered in this analysis: (i) whether they were based on more than two probes, (ii) whether they contained a translation initiation codon, (iii) whether they had a polyadenylation code within 100 bp from the 3′ end, (iv) whether they matched Rice, Brassica, or another region of the Arabidopsis genome, and (v) whether they were confirmed by serial analysis of gene expression measurement (3, 18).
Fig. 5.
Hybridization signals in a region on chromosome 1 (Crick strand) are shown. Upper and Lower present data from this study and the suspended cell line of previous study (11), respectively. The figure illustrates close correlation in hybridization patterns between two measurements, even though they are conducted with different platforms and probe lengths. The large peak between bases 296,400–296,500 is a putative expressed intergenic region identified in this study.
Because the whole-genome hybridization data showed strong activity at many regions of the genome other than the protein-coding genes, it was investigated whether the small RNAs, such as tRNA, small nuclear RNA, small nucleolar RNA, rRNA, and miRNA, showed hybridizations above the background noise. Some of these RNAs (such as miRNAs) are derived from precursors containing polyA tails (19), and this analysis was intended to verify their hybridization activities. Even for rRNAs that were generally thought not to derive from polyadenylated precursors, other whole-genome tiling array experiments recently noticed such activity. More careful studies subsequently concluded that they were in fact labeled by these protocols, perhaps as a degradation signal for deficient processing (20). This study measured strong signals (up to 8 times array median level) in both sense and antisense strand for all four rRNAs. Fifty-one of 57 small nucleolar RNAs showed average lognormalized expression levels >1.0, and 17 of 112 miRNAs had average signals above the cutoff at 0.936 (Fig. 9, which is published as supporting information on the PNAS web site). Only the small nuclear RNAs had lower signals, with all 15 of them having signal <1.0. Among the 3,786 pseudogenes annotated on the latest version of the GenBank release, 508 had average expression levels above the cutoff 0.734, and 402 had average expression levels above the more stringent cutoff of 0.936. One hundred eighty-two of the 402 pseudogenes were also expressed in at least two of four cell lines measured by the previous study (11), and 92 were expressed in all four cell lines.
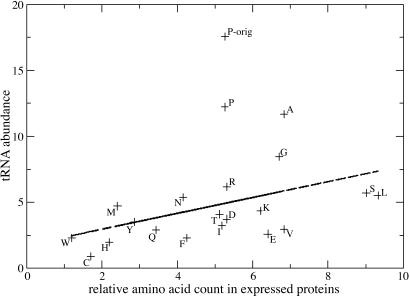
Analysis of tRNA expression has been difficult because most tRNAs with strong hybridization signal appear to be in close vicinity to some expressed protein-coding genes either in the same strand or in the opposite strand. However, when only the hybridization values within the tRNA genes, including 250 bases on both sides, were selected and these values were plotted against the use of that specific group of tRNAs' amino acid usage (added over all codons) within the entire Arabidopsis genome, an approximately linear relationship with correlation coefficient 0.45 was observed (Fig. 6). This result suggests that the tRNA hybridization values are relevant to tRNA function in protein biosynthesis.
Fig. 6.
tRNA abundance vs. amino acid usage. One outlier (P-orig) in the graph for proline was caused by 25 proline identical tRNAs located closely on chromosome 1 that most likely measure the same signal many times due to cross-hybridization. They were excluded from the analysis.
This 36-mer data set with mRNA from tissue culture cells, together with the previously published 25-mer data set, are available to the research community on request in a user-friendly format and can provide a benchmark for future work on investigating the biological significance of extractable RNA. All raw data are also deposited at the NCBI GEO database in the MIAME format under the following entries (Series, GSE 2247; samples, GSM 41324–41336; platforms, GPL 1840–1842 and 1844–1853).
In conclusion, we have described the results of measuring genome-wide transcriptional activities with high-density oligonucleotide arrays containing 36-mers that tiled across the entire Arabidopsis genome. Although this study used mRNAs from the T87 cell line, the set of arrays designed were versatile and could be used to study other tissues and growth conditions from the same plant. Our observation that the small RNAs could be identified by such genome-wide tiling studies opens new possibilities for identifying them in other organisms.
Supplementary Material
Acknowledgments
This work was supported by grants to V.S. from the National Aeronautics and Space Administration Center for Nanotechnology, the National Aeornautics and Space Administration Fundamental Biology Program, and the Computing, Information, and Communication Technology Program (contract (NAS2-99092).
Author contributions: V.S., M.P.S., and W.T. designed research; V.S., M.P.S., W.T., H.S., D.C.N., A.H., C.N., D.R., S.B., E.U., Q.Z., R.L.W., and C.S.N. performed research; V.S., M.P.S., W.T., S.L., B.G.F., G.N.P., J.L.M., and M.R.S. contributed new reagents/analytic tools; V.S., M.P.S., W.T., and H.S. analyzed data; V.S., M.P.S., and M.R.S. wrote the paper; and D.C.N. proofread the manuscript.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: miRNA, microRNA.
References
- 1.The Arabidopsis Genome Initiative (2000) Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 2.Grunenfelder, B. & Winzeler, E. A. (2002) Nat. Rev. Genet. 3, 653–661. [DOI] [PubMed] [Google Scholar]
- 3.Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 4.Hudson, T. J., Stein, L. D., Gerety, S. S., Ma, J., Castle, A. B., Silva, J., Slonim, D. K., Baptista, R., Kruglyak, L., Xu, S. H., et al. (1995) Science 270, 1945–1954. [DOI] [PubMed] [Google Scholar]
- 5.Singh-Gasson, S., Green, R. D., Yue, Y., Nelson, C., Blattner, F., Sussman, M. R. & Cerrina, F. (1999) Nat. Biotechnol. 17, 974–978. [DOI] [PubMed] [Google Scholar]
- 6.Nuwaysir, E. F., Huang, W., Albert, T. J., Singh, J., Nuwaysir, K., Pitas, A., Richmond, T., Gorski, T., Berg, J. P., Ballin, J., et al. (2002) Genome Res. 12, 1749–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stolc, V., Gauhar, Z., Mason, C., Halasz, G., van Batenburg, M. F., Rifkin, S. A., Hua, S., Herreman, T., Tongprasit, W., Barbano, P. E., et al. (2004) Science 306, 655–660. [DOI] [PubMed] [Google Scholar]
- 8.Kapranov, P., Cawley, S. E., Drenkow, J., Bekiranov, S., Strausberg, R. L., Fodor, S. P. & Gingeras, T. R. (2002) Science 296, 916–919. [DOI] [PubMed] [Google Scholar]
- 9.Shoemaker, D. D., Schadt, E. E., Armour, C. D., He, Y. D., Garrett-Engele, P., McDonagh, P. D., Loerch, P. M., Leonardson, A., Lum, P. Y., Cavet G., et al. (2001) Nature 409, 922–927. [DOI] [PubMed] [Google Scholar]
- 10.Rinn, J. L., Euskirchen, G., Bertone, P., Martone, R., Luscombe, N. M., Hartman, S., Harrison, P. M., Nelson, F. K., Miller, P., Gerstein, M., et al. (2003) Genes Dev. 17, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada, K, Lim, J., Dale, J. M., Chen, H., Shinn, P., Palm, C. J., Southwick, A. M., Wu, H. C., Kim, C., Nguyen, M., et al. (2003) Science 302, 842–846. [DOI] [PubMed] [Google Scholar]
- 12.Bertone, P., Stolc, V., Royce, T. E., Rozowsky, J. S., Urban, A. E., Zhu, X., Rinn, J. L., Tongprasit, W., Samanta, M., Weissman, S., et al. (2004) Science 306, 2242–2246. [DOI] [PubMed] [Google Scholar]
- 13.Axelos, M., Curie, C., Mazzalini, L., Bardet, C. & Lescure, B. (1992) Plant Physiol. Biochem. 30, 123–128. [Google Scholar]
- 14.Warburg, O. & Christian, C. (1941) Biochem. Z. 310, 384–421. [Google Scholar]
- 15.Hegeman, A. D., Harms, A. C., Bunner, A. E., Harper, J. F. & Sussman, M. R. (2003) J. Am. Soc. Mass Spec. 15, 647–653. [DOI] [PubMed] [Google Scholar]
- 16.Gatlin, C. L., Kleemann, G. R., Hays, L. G., Link, A. J. & Yates, J. R. (1998) Anal. Biochem. 263, 93–101. [DOI] [PubMed] [Google Scholar]
- 17.Thao, S., Zhao, Q., Kimball, T., Steffen, E., Blommel, P. G., Riters, M., Newman, C. S., Fox, B. G. & Wrobel, R. L. (2005) J. Struct. Funct. Genomics, in press. [DOI] [PubMed]
- 18.Robinson, S. J., Cram, D. J., Lewis, C. T. & Parkin, I. A. P. (2004) Plant Physiol. 136, 3223–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurihara, Y. & Watanabe, Y. (2004) Proc. Natl. Acad. Sci. USA 101, 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuai, L., Fang, F., Butler, J. S. & Sherman, F. (2004) Proc. Natl. Acad. Sci. USA 101, 8581–8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.