Abstract
Hormones and nutrients often induce genetic programs via signaling pathways that interface with gene-specific activators. Activation of the cAMP pathway, for example, stimulates cellular gene expression by means of the PKA-mediated phosphorylation of cAMP-response element binding protein (CREB) at Ser-133. Here, we use genome-wide approaches to characterize target genes that are regulated by CREB in different cellular contexts. CREB was found to occupy ≈4,000 promoter sites in vivo, depending on the presence and methylation state of consensus cAMP response elements near the promoter. The profiles for CREB occupancy were very similar in different human tissues, and exposure to a cAMP agonist stimulated CREB phosphorylation over a majority of these sites. Only a small proportion of CREB target genes was induced by cAMP in any cell type, however, due in part to the preferential recruitment of the coactivator CREB-binding protein to those promoters. These results indicate that CREB phosphorylation alone is not a reliable predictor of target gene activation and that additional CREB regulatory partners are required for recruitment of the transcriptional apparatus to the promoter.
Keywords: cAMP, cAMP-response element binding protein-binding protein, DNA methylation
The concept of a transcription code that dictates gene expression by means of the concerted action of multiple promoter elements has served as a useful paradigm for understanding specificity in gene regulation. The ability of multiple transcription factors to recruit RNA polymerase II to the promoter by means of low affinity interactions with components of the transcriptional machinery has been documented extensively (1).
By contrast with this model, other studies suggest that some activators per se are sufficient to mediate transcriptional responses to hormonal signals depending on the occupancy of relevant sites (1). Indeed, genome-wide studies comparing binding patterns of hepatic nuclear factors in the liver and endocrine pancreas indicate that selective occupancy may often explain how different genetic programs are activated in distinct cell types (2).
The cAMP-response element binding protein (CREB) family of activators stimulates cellular gene expression after phosphorylation at a conserved serine (Ser-133 in CREB1) in response to cAMP (3). Ser-133 phosphorylation promotes target gene activation in part by means of recruitment of the coactivator paralogs CREB-binding protein (CBP)/p300 (4). Recruitment of CBP by phospho-CREB (P-CREB) appears sufficient for induction of cellular genes in response to cAMP (5, 6); in vitro transcription studies indicate that P-CREB is capable of promoting assembly of the transcriptional apparatus independent of other regulatory inputs (7).
By contrast, some reports suggest that other upstream activators in addition to CREB are required for cellular gene induction by cAMP (8). Indeed, the notion that CREB coordinates with other transcription factors is supported by recent animal studies in which CREB appeared to elicit the expression of distinct genetic programs in different tissues (9–11). Thus, depending on cellular context, CREB activity may be targeted to certain genes at the level of promoter occupancy, Ser-133 phosphorylation, or recruitment of the transcriptional apparatus. Here, we use multiple independent high-throughput techniques to examine how CREB functions in different tissues. Our studies indicate that signal sensing and transcriptional induction by CREB are separate events, the latter of which requires cooperative interactions with other upstream activators.
Methods
Bioinformatic Analysis. Genome sequences and annotations were obtained from the UCSC Genome Bioinformatics Site (http://genome.ucsc.edu). A whole genome search of full cAMP-response element (CRE) (TGACGTCA) and half CRE (TGACG/CGTCA) sites was performed on the National Center for Biotechnology Information Build 34 assembly of the human genome (hg16), and conserved CREs were chosen based on the presence of exact sequences in human/mouse/rat (hg16/mm3/rn3) multiple genome alignments. All CRE hits were mapped to promoter, exonic, intronic, and intergenic regions according to the locations of RefSeq genes. Promoters were defined as 3 kb upstream to 300 bp downstream of the annotated transcription start sites. For all CREs located in the promoter regions, a search of downstream (within 300 bp) TATA boxes was performed by using a weight matrix (12). CREs located within 50 bp of each other were considered to form clusters of CREs. Profile hidden Markov models (pHMMs) for full CRE and half CRE sites were built based on known CREB target genes and were used to search for positional conserved sites as described in ref. 13. The training set sequences and the pHMMs are available at http://natural.salk.edu/CREB.
Statistical Analysis. To determine whether a certain category of genes is enriched in a list compared with the whole population of genes, P values were computed as the upper bound of the distribution of Jackknife Fisher exact probabilities (14). This P value is a sliding-scale, conservative adjustment of the Fisher exact probability that strongly penalizes the significance of categories supported by few genes and negligibly penalizes categories supported by many genes. It therefore yields more robust results than Fisher exact scores. When determining the number of genes from a list, LocusLink numbers were used as unique identifiers.
RNA and Microarray Analysis. HEK293T and MIN6 cells, human islets, and primary hepatocytes were cultured, transfected with dominant negative A-CREB expression vector or infected with A-CREB adenovirus, and harvested for mRNA analyses (10, 13, 15). To identify fasting-inducible genes, liver RNAs were harvested from male C57BL/6 mice after 18 h of fasting or 14 h of fasting followed by 4 h of refeeding. Total RNA samples were amplified, labeled, and hybridized to Affymetrix GeneChip (Affymetrix, Santa Clara, CA) arrays by using standard protocols. Scanned images were analyzed by using dchip software (16). Lower bounds of the 90% confidence intervals of fold changes (LFC) (16) were used to identify cAMP-inducible genes. Expression data and recommended cutoffs for LFC (usually between 1 and 1.3) for each experiment are available at http://natural.salk.edu/CREB.
Chromatin Immunoprecipitation (ChIP) on Chip Analysis. Nondiscriminating and phospho (Ser-133)-specific CREB antisera are described in ref. 17; CBP antibody (A-22) was from Santa Cruz Biotechnology. Promoter arrays were manufactured as described in ref. 2, except that ≈6,000 additional spots were added. ChIP on chip assays with human hepatoctytes were performed as described in ref. 2. ChIP assays on HEK293T cells were performed (13) and then subjected to the same protocol for amplification and array hybridization as above. For data analysis, an improved error model using intergenic regions located in “gene deserts” was used as a background distribution. ChIP-positive probes were identified based on the following cutoff: confidence level P value ≤0.001 and binding ratio ≥2.
Results
Identification of CRE-Containing Genes. CREB regulates cellular gene expression by binding to a conserved CRE that occurs either as a palindrome (TGACGTCA) or half site (CGTCA/TGACG) (3). A comprehensive scan of the human genome revealed 10,447 full CREs and 740,390 half CREs, which we mapped to promoters (regions from 3 kb upstream to 300 bp downstream of the annotated transcription start sites), as well as exonic, intronic, and intergenic regions based on RefSeq annotations (18). Because functional sites on the genome are often maintained through evolution, we also evaluated the conservation of CREs between rodent and human sequences by using mouse/rat/human multiple genome alignments. Compared with other regions in the genome, promoter-associated CREs occur two to three times more frequently and are more highly conserved across species (Fig. 1 a and b). By contrast with nonconserved sites, most conserved promoter CREs are located within 200 nucleotides of the transcription start site, where they are most likely to be functional (3) (Fig. 1c).
Fig. 1.
Identification of CREB target genes through bioinformatic analysis. (a) Frequency of CREs in promoter, intergenic, intronic, and exonic regions of the human genome, expressed as number of sites per megabase of DNA. Relative occurrence of full-site (TGACGTCA) and half-site (TGACG/CGTCA) CREs in each category is shown. (b) Percent conservation of full and half CRE sites in orthologous sequences from human, rat, and mouse genomes. (c) Relative distribution of promoter-associated CRE sites that are conserved or not conserved between species as a function of distance from the transcription start site. (d) Identification of CRE-containing genes in the human genome by using three independent methods (conserved CRE, CRE model + position, and CRE cluster). Number of genes identified by each method and overlap between the three methods is shown. Selected Gene Ontology categories enriched in predicted CREB target genes are shown.
Based on the idea that conserved promoter-proximal CREs are likely to be CREB-occupied, we used three independent algorithms to identify CREB target genes in silico. An initial search for CRE sites that were conserved between human and rodent orthologs yielded 3,025 human genes. As a complementary approach, we also built statistical models (profile hidden Markov models) that allow flexibility in the CRE sequence while selecting for positional conservation of the site (1,045 genes). Finally, looking for clusters of CREs that occur at promoter regions, because multiple copies of a promoter element are often indicative of function, we identified 1,024 genes. The union of these sets, containing 4,084 putative CREB target genes, is referred to hereafter as the CRE_All list (see Fig. 1d; Tables 2 and 3, which are published as supporting information on the PNAS web site; and the searchable database at http://natural.salk.edu/CREB).
We compared the CRE_All list against well documented CREB targets and found 64 of 82 published genes (3) (77%, P = 2 × 10–11), suggesting that the in silico methods we used yielded biologically relevant sites. Because TATA boxes down-stream of CREs are required for robust transcriptional induction by cAMP (13), we searched the CRE_All list for consensus TATA sequences (12) located within 300 bp downstream of the CRE. About one-third of CRE-containing genes (1,518) also contained TATA boxes (this list is referred to as CRE_TATA).
To determine the functional roles of the putative CREB target genes in the CRE_All list, we selected Gene Ontology categories (19) that are highly enriched in CRE-containing genes relative to all human genes (14) (Table 1). Transcription factors (332/866 or 38%) accounted for one of the most significant sets of CREB target genes (Table 4, which is published as supporting information on the PNAS web site), followed by genes involved in metabolic control, cell cycle regulation, and regulated secretion (Tables 1 and 4).
Table 1. Functional grouping of putative CREB target genes.
| Category | CREB targets/all genes | P value |
|---|---|---|
| Transcription factor activity | 332/866 | 2.2 × 10-19 |
| Metabolism | 1,903/7,902 | 3.9 × 10-7 |
| Cell cycle | 237/726 | 1.6 × 10-6 |
| Secretory pathway | 55/147 | 6.0 × 10-4 |
Role of CRE Methylation. Although the number of putative CREB target genes (4,084) in the CRE_All list is fewer than the estimated number of CREB dimers per cell (20,000) (17), the total number of CREs in the human genome (750,837) far exceeds that estimate. The ability of CpG methylation at the CRE to inhibit CREB binding (20) prompted us to test whether CRE methylation might restrict CREB occupancy to functionally relevant sites.
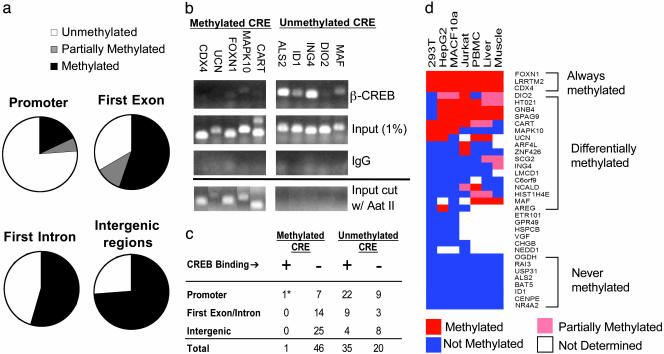
CRE methylation frequency in HEK293T cells was highest (>70%) at intergenic regions, where CREB is presumably nonfunctional, and lowest over promoters (20%) (Fig. 2a). CREB did not occupy CREs that were methylated in vivo by ChIP assay of 47 methylated CRE-containing genes (Fig. 2 b and c). By contrast, most unmethylated CREs were occupied by CREB at the promoter in HEK293T cells (22/31; Fig. 2 b and c), suggesting that CREB binding in the genome is indeed restricted by means of a DNA methylation-dependent mechanism.
Fig. 2.
CRE methylation blocks CREB binding to nonfunctional sites. (a) CRE methylation frequency on promoters and exonic, intronic, or intergenic regions, as determined by endonuclease assay with methylation-sensitive restriction enzyme Aat II. (b) Effect of CRE methylation on CREB occupancy in HEK293T cells by ChIP assay. (Upper) Relative binding of CREB to unmethylated or methylated CREs in five different promoters for each group by using anti-CREB antiserum. Input levels of DNA (1%) for each gene are indicated; nonspecific IgG control is shown. (Lower) PCR analysis of genomic fragments showing that methylated CREs are resistant to Aat II digestion, whereas unmethylated CREs are completely digested. (c) Summary of CRE methylation and CREB-binding patterns in promoters, intergenic, or intronic/exonic regions of the genome. * marks one CREB-occupied CRE (LRRTM2 gene) that was partially methylated in HEK293T cells. (d) Relative methylation state (color-coded) of 34 genes across seven cell contexts. Genes are clustered based on methylation profiles.
Because CRE methylation has been proposed to silence CREB target genes in a tissue-specific manner (21, 22), we examined the methylation status of 34 CRE-containing genes across seven cell types. A full panel of methylation profiles was observed: some CREs are always unmethylated, some are uniformly methylated, and others are methylated only in certain tissue types (Fig. 2d). For example, the CRE on the somatostatin promoter is methylated in HEK293T cells and consequently is not occupied by CREB (Table 5, which is published as supporting information on the PNAS web site). Similarly, expression of the homeobox gene CDX4 development is confined to early development (23), and the CDX4 CRE is correspondingly methylated in all tissues except for embryonic stem cells (data not shown).
Global Analysis of CREB Occupancy. Having seen that CREB binding is likely confined to unmethylated CREs at promoters, we identified cellular genes that are occupied by CREB in vivo by using a promoter microarray (Hu19K) that contains genomic PCR products covering ≈1 kb around the transcription start sites of 16,000 human genes, as well as 623 intergenic probes specifically designed against gene-sparse regions. Such gene deserts are thought to be unoccupied by transcriptional regulators, permitting unbiased normalization and more rigorous characterization of the significance of CREB binding (Fig. 5a, which is published as supporting information on the PNAS web site).
Using CREB:DNA complexes obtained from HEK293T cells by ChIP to screen the Hu19K array (P ≤ 0.001, binding ratio ≥2), we found ≈3,000 CREB-positive promoters, accounting for nearly 20% of protein coding genes (2,811/16,361 or 17%; see Table 6, which is published as supporting information on the PNAS web site, and http://natural.salk.edu/CREB). By contrast, only 3 of 151 coding regions and 1 of 623 intergenic regions were CREB-positive in this assay, indicating that CREB binding is restricted to functionally important CREs (Fig. 5 a and b). CREB also appears to occupy a significant fraction of noncoding RNA genes, as well (33/160 or 21%; Fig. 5b).
To assess the sensitivity and specificity of the ChIP-chip method, we performed manual ChIP assays on CRE-containing promoters identified in the CRE_All list. Of 14 promoters not occupied by CREB by manual ChIP assay, only one scored as a positive in the ChIP-chip study, suggesting that this method is highly specific (2). The number of CREB-occupied promoters by ChIP-chip assay likely represents a conservative estimate of total CREB target genes; of 28 CREB-occupied promoters identified by manual ChIP assay, 15 (54%) were also positive by ChIP-chip analysis. Indeed, a blunting of the sensitivity and enrichment ratios in ChIP-chip assays when compared with site-specific querying of binding has been noted previously (ref. 24 and D.T.O., unpublished work). Extrapolating from the estimated sensitivity of this method, the actual number of genes occupied by CREB in HEK293T cells may be >5,000.
Having identified a large set of promoters that are occupied by CREB in HEK293T cells, we examined whether these genes were accurately predicted by bioinformatic analyses (Fig. 1d). Although each of the three search methods showed significant improvement over a simple CRE consensus site algorithm, the CRE_All list provided an optimal balance between predictive power for CRE occupancy and sensitivity (Fig. 5c). Indeed, genes in the CRE_All list were far more likely to score significant P values by ChIP-chip assay than genes without detectable CREs (Fig. 3a).
Fig. 3.
cAMP stimulates Ser-133 phosphorylation uniformly over CREB-occupied genes. (a) Comparison of CREB occupancy and presence of CREs in human promoters. For each promoter category (those with predicted CRE, those without predicted CRE, and all genes), the distribution of confidence levels (P values) from ChIP-chip results is shown. Graph shows percentage of promoters within each category at specific P-value range. P values were computed from three independent CREB ChIP experiments with HEK293T cells. (b) Analysis of P-CREB levels over human promoters by ChIP-chip assay of HEK293T cells at 0, 1, and 4 h after exposure to FSK. Randomly selected subsets of ChIP-positive CREB target genes from cAMP responsive and nonresponsive genes were used to compare P-CREB profiles. (c) Western blot assay showing levels of total CREB and P-CREB in HEK293T cells after FSK treatment for times indicated.
Having seen that CREB occupies a large number of promoters, we performed gene profiling experiments on HEK293T cells to determine whether binding of CREB to these genes is sufficient for target gene activation by the cAMP agonist forskolin (FSK). To eliminate CREB-independent effects of FSK, we used a dominant negative form of CREB called A-CREB, which selectively disrupts target gene activation by CREB (25). Most genes that were both up-regulated by FSK treatment and repressed by A-CREB in HEK283T cells contain functional CRE and TATA box motifs (Table 7, which is published as supporting information on the PNAS web site). Compared with the estimated number of CREB-occupied genes in HEK293T cells (>5,000), however, only a small fraction (100 or <2%) actually responded to FSK, indicating that the majority of CREB-occupied genes are not induced by cAMP.
Genome-Wide Analysis of CREB Phosphorylation. To test whether the inability of most CREB-occupied promoters to respond to a cAMP agonist reflects a selective block in Ser-133 phosphorylation of CREB at these sites, we performed promoter occupancy assays with P-CREB:DNA complexes from control or FSK-treated HEK293T cells by using a P-CREB-specific anti-serum (17) (Fig. 3 b and c). Nearly all CREB-positive promoters show similar kinetics of Ser-133 phosphorylation in response to cAMP; levels of P-CREB were low under resting conditions (0 h) but increased sharply within 1 h after exposure to FSK (Fig. 3 b and c). We also noted comparable changes in P-CREB levels on cAMP inducible vs. noninducible CREB target genes by manual ChIP assay, arguing against differential Ser-133 phosphorylation as a predominant mechanism by which gene subsets are selectively induced in response to cAMP (Fig. 6, which is published as supporting information on the PNAS web site).
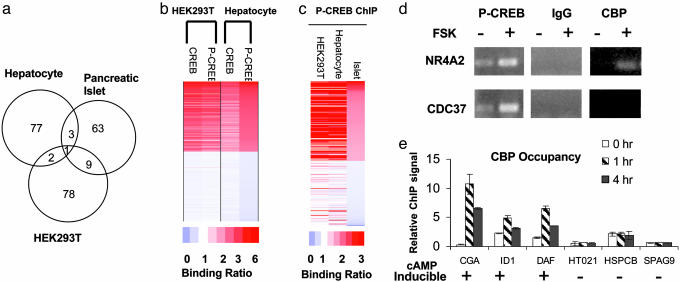
The ability of CREB to carry out distinct functions in different tissues led us to examine the importance of cellular context for target gene activation. Exposure to FSK reliably induced ≈100 genes in HEK293T cells as well as in primary cultures of mouse hepatocytes or human islets by gene profiling assays (see Fig. 4a and the database at http://natural.salk.edu/CREB), but the sets of cAMP responsive genes in each case were almost completely distinct. About half of the cAMP responsive genes from each cell type contain consensus CRE sites, and CRE_TATA genes exhibited a much higher tendency to be cAMP responsive than CRE_NoTATA genes, confirming the importance of a TATA box for induction (13) (Table 7; see also Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 4.
cAMP stimulates distinct profiles of CREB target genes in different tissues. (a) Venn diagram showing overlap between CREB target genes in human islets, hepatocytes, and HEK293T cells. (b) ChIP-chip assay of promoters occupied by CREB and P-CREB in human hepatocytes compared with HEK293T cells. Top 1,000 and bottom 1,000 scoring genes from hepatocyte P-CREB ChIP-chip assays were plotted. Relative binding ratio observed for each gene is color-coded. (c) Comparison of P-CREB binding profiles from ChIP-chip assays of HEK293T cells, human pancreatic islets, and human hepatocytes. Top 200 and bottom 200 scoring genes from islet P-CREB ChIP-chip assays were plotted. (d) ChIP assay of HEK293T cells showing effects of FSK on recruitment of CBP to cAMP inducible (NR4A2) vs. noninducible (CDC37) genes. Levels of P-CREB on each promoter are indicated. (e) Quantitative PCR assay showing relative levels of CBP over cAMP inducible and noninducible promoters after 0, 1, or4hofFSK treatment by ChIP.
In keeping with the role of CREB in promoting islet cell proliferation and viability (10, 11), exposure of cultured human islets to cAMP stimulated a number of growth factor genes and antiapoptotic factors. By contrast, exposure of primary hepatocytes to cAMP induced genes involved in fasting glucose and lipid metabolism. And treatment of HEK293T cells with FSK stimulated genes that were again distinct from either liver or pancreatic islets (Table 7).
CREB Occupancy and Phosphorylation in Different Tissues. To determine whether the tissue-specific activation of CREB targets is due to differences in CREB occupancy, we performed promoter location experiments using CREB:DNA complexes collected from each tissue by ChIP (see the database at http://natural.salk.edu/CREB). Most of the CREB-occupied genes in hepatocytes were also bound by CREB in HEK293T cells (1,795/2,144 or 84%), and virtually none of the ChIP-chip-negative genes (using a CREB binding P > 0.5 as a cutoff) in hepatocytes was positive in HEK293T cells (11/3,090 or 0.36%) (Fig. 4b), arguing against differential occupancy in explaining the tissue-specific regulation of CREB target genes. Although overall hybridization signals from ChIP assays of human pancreatic islets were weak, the limited number of CREB target genes we were able to identify showed extensive overlap with genes in HEK293T cells and primary hepatocytes (Fig. 4c). Indeed, the patterns of CREB phosphorylation over target promoters were also highly similar among human hepatocytes, pancreatic islets, and HEK293T cells, arguing against a role for the cAMP/PKA pathway in discriminating between different genetic programs (Fig. 4 b and c).
Recruitment of CBP to CREB Target Genes. Ser-133 phosphorylation of CREB in response to cAMP is thought to be sufficient for CBP/p300 recruitment to the promoter and for target gene activation (5, 6). The ability of cAMP agonists to trigger Ser-133 phosphorylation over a majority of CREB-positive promoters in liver, islet, and HEK293T cells led us to explore the involvement of CBP/p300 in discriminating between cAMP inducible vs. noninducible genes. Confirming the ChIP-chip studies, FSK induced comparable levels of P-CREB on the cAMP-responsive NR4A2 and unresponsive CDC37 genes by manual ChIP assay of HEK293T cells (Fig. 4d). However, exposure to FSK selectively induced recruitment of CBP to the NR4A2 promoter but not to the CDC37 promoter, indicating that Ser-133 phosphorylation may not be sufficient in all cases to trigger CREB:CBP complex formation. We noted similar differences in CBP recruitment between a number of cAMP inducible (CGA, ID1, DAF) vs. noninducible (HT021, HSPCB, SPAG9) genes, despite the fact that levels of P-CREB over both sets of promoters were comparable by ChIP assay (Fig. 4e). Taken together, these results suggest that the ability of cAMP to activate selective CREB target gene subsets is reflected at the level of CBP occupancy and that additional CREB regulatory partners are required for CBP recruitment to the promoter.
Discussion
Like other second messenger pathways, cAMP stimulates distinct genetic programs in different tissues. Our results reveal that these tissue-specific profiles in gene activation are not explained by differences in CREB occupancy or Ser-133 phosphorylation but reflect the selective recruitment of CBP and perhaps other cofactors to relevant promoters.
CREB was found to regulate ≈4,000 target genes in the human genome, and a majority of these are occupied in vivo (see the database at http://natural.salk.edu/CREB). Goodman and colleagues (26) have also identified a large number of CREB-occupied loci in the rat genome; a majority of CREB-binding sites in that study were similarly detected near expected transcription start sites. In a separate study using a human chromosome 22 tiling array, Euskerchin et al. (27) found that CREB occupancy was widely disseminated with only modest sequence-specific occupancy over conserved CRE sites. The reasons for this discrepancy are unclear; but the use of an IgG ChIP sample rather than genomic DNA as a control channel in that study may explain, in part, differences with our observations.
P-CREB has been shown to stimulate target gene expression by associating with a number of coactivators, including CBP/p300 (28, 29), TORC (30, 31), and TAFII4 (32, 33). However, our results suggest that the interaction of P-CREB with these proteins is too weak for cellular gene activation per se and that additional CREB regulatory partners are required for stable recruitment of such cofactors to the promoter. Indeed, bioinformatic analysis reveals that CRE elements often cosegregate with sites for other activators (Table 8, which is published as supporting information on the PNAS web site). Identifying these regulatory partners may permit the characterization of transcription codes that reliably predict which CREB target genes are induced by cAMP in a given cell type.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM RO1-037828 (to M.M.) and DK068655 (to R.A.Y.).
Author contributions: X.Z., D.T.O., S.-H.K., R.A.Y., and M.R.M. designed research; X.Z., D.T.O., S.-H.K., M.D.C., G.C., R.J., E.H., E.J., and S.K. performed research; X.Z., D.T.O., M.D.C., G.C., J.L.B., H.C., R.J., J.R.E., B.E., J.B.H., T.U., and R.A.Y. contributed new reagents/analytic tools; X.Z., D.T.O., S.-H.K., M.D.C., G.C., J.L.B., H.C., B.E., J.B.H., T.U., R.A.Y., and M.R.M. analyzed data; and X.Z., D.T.O., and M.R.M. wrote the paper.
Abbreviations: CRE, cAMP-response element; CREB, CRE binding protein; CBP, CREB-binding protein; P-CREB; phospho-CREB; FSK, forskolin; ChIP, chromatin immunoprecipitation.
Data deposition: The gene expression data reported in this paper have been deposited in the Gene Expression Omnibus database (accession no. GSE2060).
References
- 1.Kadonaga, J. T. (2004) Cell 116, 247–257. [DOI] [PubMed] [Google Scholar]
- 2.Odom, D. T., Zizlsperger, N., Gordon, D. B., Bell, G. W., Rinaldi, N. J., Murray, H. L., Volkert, T. L., Schreiber, J., Rolfe, P. A., Gifford, D. K., et al. (2004) Science 303, 1378–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayr, B. & Montminy, M. (2001) Nat. Rev. Mol. Cell Biol. 2, 599–609. [DOI] [PubMed] [Google Scholar]
- 4.Goodman, R. H. & Smolik, S. (2000) Genes Dev. 14, 1553–1577. [PubMed] [Google Scholar]
- 5.Cardinaux, J. R., Notis, J. C., Zhang, Q., Vo, N., Craig, J. C., Fass, D. M., Brennan, R. G. & Goodman, R. H. (2000) Mol. Cell. Biol. 20, 1546–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du, K., Asahara, H., Jhala, U., Wagner, B. & Montminy, M. (2000) Mol. Cell. Biol. 20, 4320–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asahara, H., Santoso, B., Du, K., Cole, P. & Montminy, M. (2001) Mol. Cell. Biol. 21, 7892–7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson, L. M., Kerppola, T. K., Vendrell, M., Luk, D., Smeyne, R. J., Bocchiaro, C., Morgan, J. I. & Curran, T. (1995) Neuron 14, 241–252. [DOI] [PubMed] [Google Scholar]
- 9.Herzig, S., Long, F., Jhala, U., Hedrick, S., Quinn, R., Bauer, A., Schutz, G., Yoon, C., Puisgever, P., Spiegelman, B. & Montminy, M. (2001) Nature 413, 179–183. [DOI] [PubMed] [Google Scholar]
- 10.Jhala, U. S., Canettieri, G., Screaton, R. A., Kulkarni, R. N., Krajewski, S., Reed, J., Walker, J., Lin, X., White, M. & Montminy, M. (2003) Genes Dev. 17, 1575–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inada, A., Hamamoto, Y., Tsuura, Y., Miyazaki, J., Toyokuni, S., Ihara, Y., Nagai, K., Yamada, Y., Bonner-Weir, S. & Seino, Y. (2004) Mol. Cell. Biol. 24, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bucher, P. (1990) J. Mol. Biol. 212, 563–578. [DOI] [PubMed] [Google Scholar]
- 13.Conkright, M. D., Guzmán, E., Flechner, L., Su, A. I., Hogenesch, J. & Montminy, M. (2003) Mol. Cell 11, 1101–1108. [DOI] [PubMed] [Google Scholar]
- 14.Hosack, D. A., Dennis, G., Jr., Sherman, B. T., Lane, H. C. & Lempicki, R. A. (2003) Genome Biol. 4, R70.1–R70.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koo, S. H., Satoh, H., Herzig, S., Lee, C. H., Hedrick, S., Kulkarni, R., Evans, R. M., Olefsky, J. & Montminy, M. (2004) Nat. Med. 10, 530–534. [DOI] [PubMed] [Google Scholar]
- 16.Li, C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagiwara, M., Brindle, P., Harootunian, A., Armstrong, R., Rivier, J., Vale, W., Tsien, R. & Montminy, M. R. (1993) Mol. Cell. Biol. 13, 4852–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruitt, K. D. & Maglott, D. R. (2001) Nucleic Acids Res. 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iguchi-Ariga, S. M. & Schaffner, W. (1989) Genes Dev. 3, 612–619. [DOI] [PubMed] [Google Scholar]
- 21.Iannello, R. C., Gould, J. A., Young, J. C., Giudice, A., Medcalf, R. & Kola, I. (2000) J. Biol. Chem. 275, 19603–19608. [DOI] [PubMed] [Google Scholar]
- 22.Mancini, D. N., Singh, S. M., Archer, T. K. & Rodenhiser, D. I. (1999) Oncogene 18, 4108–4119. [DOI] [PubMed] [Google Scholar]
- 23.Gamer, L. W. & Wright, C. V. (1993) Mech. Dev. 43, 71–81. [DOI] [PubMed] [Google Scholar]
- 24.Friedman, J. R., Larris, B., Le, P. P., Peiris, T. H., Arsenlis, A., Schug, J., Tobias, J. W., Kaestner, K. H. & Greenbaum, L. E. (2004) Proc. Natl. Acad. Sci. USA 101, 12986–12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahn, S., Olive, M., Aggarwal, S., Krylov, D., Ginty, D. & Vinson, C. (1998) Mol. Cell. Biol. 18, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Impey, S., McCorkle, S. R., Cha-Molstad, H., Dwyer, J. M., Yochum, G. S., Boss, J. M., McWeeney, S., Dunn, J. J., Mandel, G. & Goodman, R. H. (2004) Cell 119, 1041–1054. [DOI] [PubMed] [Google Scholar]
- 27.Euskirchen, G., Royce, T. E., Bertone, P., Martone, R., Rinn, J. L., Nelson, F. K., Sayward, F., Luscombe, N. M., Miller, P., Gerstein, M., et al. (2004) Mol. Cell. Biol. 24, 3804–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwok, R., Lundblad, J., Chrivia, J., Richards, J., Bachinger, H., Brennan, R., Roberts, S., Green, M. & Goodman, R. (1994) Nature 370, 223–226. [DOI] [PubMed] [Google Scholar]
- 29.Arias, J., Alberts, A., Brindle, P., Claret, F., Smeal, T., Karin, M., Feramisco, J. & Montminy, M. (1994) Nature 370, 226–228. [DOI] [PubMed] [Google Scholar]
- 30.Conkright, M. D., Canettieri, G., Screaton, R., Guzman, E., Miraglia, L., Hogenesch, J. B. & Montminy, M. (2003) Mol. Cell 12, 413–423. [DOI] [PubMed] [Google Scholar]
- 31.Iourgenko, V., Zhang, W., Mickanin, C., Daly, I., Jiang, C., Hexham, J. M., Orth, A. P., Miraglia, L., Meltzer, J., Garza, D., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 12147–12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreri, K., Gill, G. & Montminy, M. (1994) Proc. Natl. Acad. Sci. USA 91, 1210–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felinski, E. A. & Quinn, P. G. (1999) J. Biol. Chem. 274, 11672–11678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.