Abstract
The polyglutamine-containing neurodegenerative protein ataxin 3 (AT3) has deubiquitylating activity and binds ubiquitin chains with a preference for chains of four or more ubiquitins. Here we characterize the deubiquitylating activity of AT3 in vitro and show it trims/edits K48-linked ubiquitin chains. AT3 also edits polyubiquitylated 125I-lysozyme and decreases its degradation by proteasomes. Cellular studies show that endogenous AT3 colocalizes with aggresomes and preaggresome particles of the misfolded cystic fibrosis transmembrane regulator (CFTR) mutant CFTRΔF508 and associates with histone deacetylase 6 and dynein, proteins required for aggresome formation and transport of misfolded protein. Small interfering RNA knockdown of AT3 greatly reduces aggresomes formed by CFTRΔF508, demonstrating a critical role of AT3 in this process. Wild-type AT3 restores aggresome formation; however, AT3 with mutations in the active site or ubiquitin interacting motifs cannot restore aggresome formation in AT3 knockdown cells. These same mutations decrease the association of AT3 and dynein. These data indicate that the deubiquitylating activity of AT3 and its ubiquitin interacting motifs as well play essential roles in CFTRΔF508 aggresome formation.
Keywords: proteasome, protein aggregation, spinocerebellar ataxia type 3/Machado–Joseph disease, deubiquitinization, ubiquitin-interacting motif
Spinocerebellar ataxia type 3/Machado–Joseph disease is a dominantly inherited neurodegenerative disease caused by expansion of a polyglutamine domain in the protein ataxin 3 (AT3) (1). Expansion of the glutamine domain in AT3, as well as in eight other members of the polyglutamine neurodegenerative disease family, increases protein misfolding, resulting in aggregation and formation of nuclear and cytoplasmic inclusions (2–6). Inclusions of polyglutamine proteins are ubiquitylated and contain proteasomes, suggesting an attempt by the ubiquitin proteasome pathway (UPP) to degrade the misfolded protein (7–11). Misfolded aggregated protein is not only a characteristic of polyglutamine diseases but also a common feature of many neurodegenerative diseases, including Alzheimer's and Parkinson's diseases (12).
Cells deal with misfolded proteins in several ways, including chaperone-mediated refolding and proteasome-dependent degradation. In situations where proteasomes are compromised or overwhelmed, misfolded protein is transported to a perinuclear location near the microtubule-organizing center (MTOC) to form aggresomes (13, 14). Although formation of aggresomes seems to be important for cellular homeostasis, little is known about the proteins responsible for recognizing, processing, and transporting misfolded proteins. Other than microtubules, the only proteins known to be involved in this critical regulatory process are the microtubule motor, dynein/dynactin, and the microtubule-associated deacetylase histone deacetylase 6 (HDAC6) (13–16).
Like other polyglutamine neurodegenerative proteins, AT3 is prone to misfold and aggregate and therefore is a target of the UPP; however, recent data suggest that AT3 may also function in the UPP. We recently showed that AT3 is a deubiquitylating enzyme (DUB) (17), and we and others showed that AT3 binds chains of four or more ubiquitins through its ubiquitin interacting motifs (UIMs) (17–20). Mutating its active site cysteine inhibits DUB activity, whereas mutating conserved residues in the UIMs prevents ubiquitin binding. Although DUBs constitute one of the largest families of proteins in the UPP, relatively little specific information is available on the myriad of functions thought to be regulated by them (21, 22).
Here we characterize some of the DUB properties of AT3 in vitro and show that cellular AT3 regulates aggresome formation by the highly unstable cystic fibrosis transmembrane regulator (CFTR) mutant CFTRΔF508. AT3 localizes to aggresomes and preaggresome particles and interacts with dynein and HDAC6, and its DUB activity and UIMs are required for CFTRΔF508 aggresome formation.
Materials and Methods
Plasmids and Proteins. Maltose-binding protein (MBP)/AT3 bacterial expression constructs were described previously (17). The myc-tagged AT3 eukaryotic expression construct was described previously (23). myc-AT3-C14A and myc-AT3-L229/249A were generated by site-directed mutagenesis as described (17). FLAG-tagged HDAC6 and GFP-CFTRΔF508 cDNAs were provided by Stuart Schreiber (Harvard University, Cambridge, MA; ref. 24) and Ron Kopito (Stanford University, Stanford, CA; ref. 13).
DUB Assays. Ubiquitin chain cleavage. For time course experiments, 250 ng of polyubiquitin chains of heterogeneous lengths (Ub 2-7; Affiniti, Nottingham, U.K.) was incubated with MBP or the indicated amounts of MBP-AT3 fusion proteins for various times up to 48 h at 37°C in assay buffer (50 mM Hepes and 0.5 mM EDTA at pH 7.5 containing 0.1 mg/ml ovalbumin and 1 mM DTT). The reaction was stopped by adding SDS sample buffer and proteins separated by SDS/PAGE and was analyzed by Western blot by using FK1 polyubiquitin antibody (Affiniti). The relative intensities of the bands were determined by densitometry (ImageQuant, Amersham Pharmacia Biosciences). Some experiments were carried out at 37°C for 8 h with approximately equimolar concentrations of MBP-AT3 and Ub 2-7 chains. MBP controls were included in all DUB assays.
Ubiquitylated 125I-lysozyme. Ubiquitylation and deubiquitylation of 125I-lysozyme was carried out as described (17).
Ubiquitin-AMC. Protease assays using the fluorogenic peptide substrate ubiquitin-AMC (Boston Biochem, Cambridge, MA) were carried out for 15 min as described (17).
Proteasome Degradation Assays. Ubiquitylated 125I-lysozyme was prepared as described (17) and then incubated with 5 nM purified 26S proteasome (Affiniti) for 30 min at 37°C in the presence or absence of 25, 150, or 250 nM MBP-AT3 fusion proteins. 125I-lysozyme was precipitated on ice with 100 μl of 10% trichloroacetic acid (TCA), and the number of TCA-soluble protein fragments in the supernatant was quantitated by using a gamma counter. The effect of AT3 on the peptidase activity of the purified proteasomes was measured by incubating 5 nM 26S proteasomes with the fluorogenic peptide substrates (100 μM) Suc-LLVY-AMC (chymotryptic-like) or Boc-LLR-AMC (tryptic-like) in the presence or absence of 0.25 μM MBP-AT3 fusion proteins for 1 h at 37°C. AMC fluorescence was measured by using a Bio-Tek Instruments (Winooski, VT) FLX800 fluorimeter.
Immunofluorescence. COS 7 cells were fixed with 4% paraformaldehyde in PBS and permeabilized with 0.1% Triton X-100. After blocking with 3% BSA in PBS, cells were incubated with anti-AT3 (2) or anti-FLAG M2 (Sigma) for 1.5 h at room temperature, and then secondary antibody was conjugated with Alexa Fluor 568.
Small Interfering RNA (siRNA) Knockdown of Endogenous AT3 and Quantitation of Aggresomes. A 72-bp siRNA with antisense (5′-TGGCAGAAGGAGTTAC-3′), loop (5′-TTGATATCCG-3′), and sense (5′-GTAACTCCTCCTTCTGCCA-3′) sequences was inserted into the pRNAin-H1.2/Neo GenScript siRNA expression vector (GenScript, Piscataway, NJ). Scrambled AT3 siRNA was also generated: antisense (5′-GGAACAAGGTGAGTGAGTCT-3′), loop (5′-TTGATATCCG-3′), and sense (5′-GACTCACTCACCTTGTTCCT-3′). pRNAin-H1.2-AT3 was transfected into COS 7 cells with FuGene 6 (Roche Diagnostics), followed 48 h later by a second transfection. siRNA-resistant forms of wild-type AT3, AT3-C14A, and AT3-L229/249A were generated by introducing silent mutations in the siRNA-targeting region (A150G, A153C, A156C, and T159A) of myc-tagged AT3 constructs. After AT3 knockdown, cells were transfected with GFP-CFTRΔF508 and treated 24 h later with 10 μM MG132. For each condition in each experiment, 200 transfected cells were scored for the presence of aggresomes under blinded conditions 2 h after MG132 treatment; data represent the average of three independent experiments.
Cotransfections and Immunoprecipitations. Cells were transfected with FuGENE 6 according to the manufacturer's protocol. Immunoprecipitations were performed essentially as described (25). Briefly, cleared cell lysates were incubated with anti-FLAG M2 (Sigma), antidynein intermediate chain (Chemicon), or anti-AT3 (2) for 6 h followed by overnight incubation with Dynabeads Protein G (Dynal, Oslo). The beads were washed, and bound proteins were separated by SDS/PAGE and analyzed by immunoblotting with anti-myc or anti-AT3.
Results
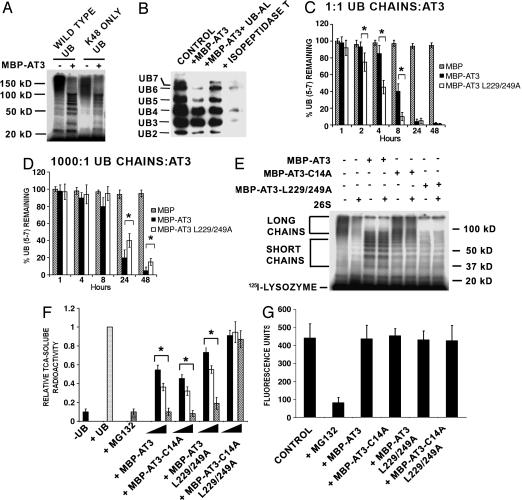
AT3 Trims/dits K48-Linked Ubiquitin Chains. Our previous work (17) indicated that AT3 has DUB activity and, similarly to the proteasome, binds to chains of four or more ubiquitins; this suggests that AT3 may interact with and/or process proteins targeted for proteasome degradation. The proteasome preferentially recognizes and degrades proteins tagged with ubiquitin chains linked through lysine 48 (K48). To determine whether AT3 cleaves K48-linked ubiquitin chains, 125I-lysozyme was ubiquitylated by using wild-type ubiquitin or mutant ubiquitin lacking all but K48 (K48 Only in Fig. 1A). AT3 removes K48-linked ubiquitins from 125I-lysozyme, similar to its removal of wild-type ubiquitin chains; high-molecular-weight ubiquitin chains are preferentially cleaved/edited, but lower-molecular-weight chains are spared (Fig. 1 A). AT3 also cleaves unanchored K48-linked polyubiquitin chains containing five to seven ubiquitins from a heterogeneous population of ubiquitin chains containing two to seven ubiquitins (Fig. 1B). It is difficult to follow the cleavage of ubiquitin chains with fewer than five ubiquitins, because cleavage of long chains results in the buildup of shorter chains, as well as inefficient cleavage of these short chains even after 48 h with a 10-fold excess of AT3 (data not shown). Chain cleavage by AT3 is blocked by ubiquitin aldehyde, a specific ubiquitin protease inhibitor, and, in contrast to AT3, isopeptidase T does not trim ubiquitin chains but rather disassembles them completely (Fig. 1B).
Fig. 1.
AT3 deubiquitylating activity. (A) AT3 trims anchored K48-linked ubiquitin (UB) chains. Wild-type ubiquitin or mutated ubiquitin lacking all but one lysine (K48 ONLY UB) was used to ubiquitylate 125I-lysozyme, which was incubated with MBP-AT3. Similar to wild-type ubiquitin, K48 ubiquitin forms high-molecular-weight polyubiquitin chains that are cleaved to shorter chains by AT3. kD, kilodalton. (B) AT3 trims unanchored K48-linked ubiquitin chains. Polyubiquitin immunoblot (FK1 antibody) showing that MBP-AT3 preferentially cleaves free polyubiquitin chains containing five to seven ubiquitins (lane 2; 1:1 ratio AT3:chains) compared with isopeptidase T (lane 4), which completely disassembles chains. Ubiquitin aldehyde inhibits AT3 DUB activity (lane 3). (C and D) UIMs of AT3 modulate ubiquitin chain cleavage. Quantitation of cleavage of free polyubiquitin chains containing five to seven ubiquitins. At equimolar concentrations of polyubiquitin chains and AT3 (0.25 μM), MBP-AT3-L229-249A (the UIM mutant) significantly increases cleavage of chains of five to seven ubiquitins than is MBP-AT3. At a ratio of 1,000:1 (ubiquitin chains to 0.25 nM AT3), MBP-AT3 containing functional UIMs cleaves chains slightly better than does MBP-AT3-L229-249A. Data represent mean ± SEM of three experiments. *, P < 0.05. (E and F) AT3 inhibits degradation of ubiquitylated 125I-lysozyme by proteasomes. (E) Ubiquitylated 125I-lysozyme is degraded by 26S proteasomes, corresponding to a loss of high-molecular-weight polyubiquitylated 125I-lysozyme in the presence of 26S proteasome (compare lanes 1 and 2). MBP/AT3 trims long polyubiquitin chains and blocks degradation of polyubiquitylated 125I-lysozyme by 26S proteasomes (lanes 3 and 4), whereas MBP-AT3-L229-249A removes both short and long ubiquitin chains and inhibits degradation (lanes 7 and 8). MBP-AT3-C14A does not trim ubiquitin chains but blocks degradation of polyubiquitylated 125I-lysozyme (lanes 5 and 6). (F) Quantitation of 125I-lysozyme degradation. 26S proteasomes degrade (TCA-soluble radioactivity) polyubiquitylated 125I-lysozyme (+UB) but not nonubiquitylated 125I-lysozyme (–UB). The proteasome inhibitor MG132 (10 μM) inhibits degradation of polyubiquitylated 125I-lysozyme. Different concentrations (25, 150, or 250 nM, represented by filled, open, and stippled bars, respectively) of MBP-AT3, MBP-AT3-L229-249A, or MBP-AT3-C14A, but not AT3-C14A-L229/249A, decrease the degradation of polyubiquitylated 125I-lysozyme incubated with 26S proteasomes. Data represent mean ± SEM from four experiments. *, P < 0.05 compared with polyubiquitylated 125I-lysozyme (+UB). (G) AT3 does not affect proteolytic activity of the 26S proteasome. MBP-AT3 fusion proteins (250 nM) do not affect chymotryptic-like (or tryptic-like; data not shown) activity of 26S proteasomes against a fluorogenic peptide substrate.
UIMs in AT3 may contribute to its editing functions; therefore, we examined the effects of the UIMs on the cleavage of chains containing five to seven ubiquitins. At equimolar concentrations of AT3 and ubiquitin chains, AT3-L229/249A [a UIM mutant that does not bind chains (17)] cleaves ubiquitin chains faster than does wild-type AT3 (Fig. 1C). However, at a ratio of 1:1,000 (AT3 to chains), wild-type AT3 containing functional UIMs cleaves chains more efficiently than does AT3-L229/249A (Fig. 1D). This suggests that UIMs may aid in recruiting ubiquitin chains at low concentrations of AT3, whereas at higher concentrations, UIMs hinder chain cleavage.
AT3 Inhibits Degradation of Polyubiquitylated 125I-Lysozyme by the Proteasome. To determine whether AT3 has an effect on proteasome degradation, ubiquitylated 125I-lysozyme was incubated with purified 26S proteasomes in the presence or absence of AT3 (Fig. 1E). Proteasomes degrade polyubiquitylated 125I-lysozyme in vitro as indicated by the loss of high-molecular-weight ubiquitylated 125I-lysozyme (lane 2, Fig. 1E) as well as by the increase in TCA-soluble 125I-lysozyme fragments (+UB in Fig. 1F); degradation is inhibited by the proteasome inhibitor MG132 (Fig. 1F). AT3 trims/edits ubiquitin chains conjugated to the 125I-lysozyme, and edited ubiquitylated 125I-lysozyme is not effectively degraded by proteasomes (Fig. 1 E and F). Reduced degradation may be due to AT3's removal of ubiquitins from the 125I-lysozyme below the threshold required for proteasome recognition. Alternatively, the UIMs of AT3 may bind ubiquitin chains and physically prevent access by the proteasome. To address this question, we incubated ubiquitylated 125I-lysozyme with AT3-C14A (the active site mutant), AT3-L229/249A (the UIM mutant), or AT3-C14A-L229/249A (the UIM and active site mutant) in the presence or absence of proteasomes. If AT3 DUB activity is responsible for blocking degradation, then mutating the active site should reverse this effect. AT3-C14A (the active site mutant) does not trim ubiquitin chains on 125I-lysozyme but still blocks its degradation (Fig. 1 E and F). AT3 with both the active site and UIMs mutated, however, does not inhibit 125I-lysozyme degradation (Fig. 1F), suggesting that the UIMs are primarily responsible for inhibiting degradation. In the absence of functional UIMs, AT3 removes both long and short ubiquitin chains (see also ref. 17) and decreases degradation of 125I-lysozyme, presumably by removing the ubiquitin degradation signal (Fig. 1 E and F). MBP-AT3 fusion proteins do not bind to 26S proteasomes (data not shown), nor do they affect the proteolytic activity of purified 26S proteasomes when assayed by using peptide fluorogenic substrates, indicating that the effects of AT3 are not made through binding the proteasomes or inhibiting the core proteasome (Fig. 1G).
These data suggest that UIMs of AT3 bind ubiquitylated 125I-lysozyme and prevent access to the proteasome, thereby inhibiting degradation. In the absence of functional UIM domains, AT3 may also inhibit degradation by trimming ubiquitin chains below the signal required for recognition by the proteasome.
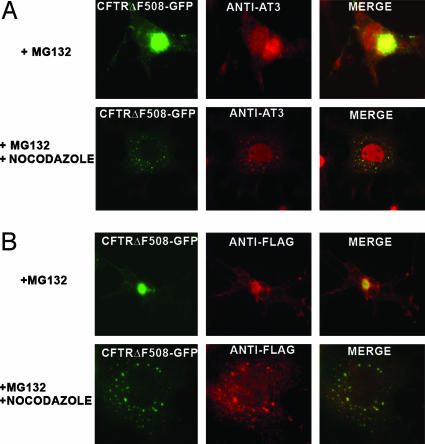
Endogenous AT3 Associates with Aggresomes Formed by CFTRΔF508. Many proteins with short half-lives, as well as misfolded proteins, are ubiquitylated and degraded by proteasomes. Inhibiting proteasomes causes accumulation of proteins targeted for degradation; these proteins are transported on microtubules to the MTOC and form large perinuclear aggregates/aggresomes (13, 14). Initially we wanted to determine whether endogenous AT3 associates with aggresomes formed by CFTRΔF508, a deletion mutant of the CFTR, which is used as a model for studying protein aggregation and aggresome formation (13). Cells transfected with CFTRΔF508-GFP and treated with the proteasome inhibitor MG132 develop large perinuclear aggresomes (Fig. 2A). Endogenous AT3 colocalizes with CFTRΔF508 aggresomes (Fig. 2 A), suggesting that AT3 either associates with aggresomes once they are formed or functions in the pathway responsible for aggresome formation. To determine whether AT3 associates with the CFTR before moving to the MTOC, misfolded CFTRΔF508 was blocked from reaching the MTOC by destabilizing microtubules with nocodazole. As reported (26), destabilizing microtubules results in accumulation of CFTRΔF508 in widely dispersed cytoplasmic aggregates/preaggresome particles. Endogenous AT3 also colocalizes with these cytoplasmic particles (Fig. 2 A). Recent data indicate that HDAC6 is necessary for aggresome formation (15). HDAC6 binds polyubiquitylated proteins and interacts with the dynein motor complex, facilitating transport of misfolded proteins to the MTOC. We confirmed that HDAC6 associates with both aggresomes and preaggresome particles formed by CFTRΔF508-GFP (Fig. 2B). Therefore, both AT3 and HDAC6 colocalize with preaggresome particles.
Fig. 2.
Endogenous AT3 is present in aggresomes formed by CFTRΔF508. (A) Endogenous AT3 colocalizes with aggresomes formed by CFTRΔF508-GFP in COS 7 cells after proteasome inhibition with MG132 as well as preaggresome particles after nocodazole disruption of microtubules. (B) FLAG-tagged HDAC6 colocalizes with aggresomes and preaggresome particles of CFTRΔF508-GFP.
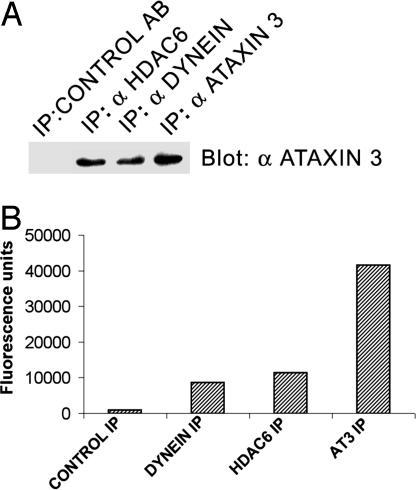
Endogenous AT3 Associates with Components of the Complex That Transports Misfolded Proteins to the MTOC. To determine whether AT3 associates with HDAC6 and/or dynein, immunoprecipitations were performed. Endogenous AT3 coimmunoprecipitates with both dynein and HDAC6, and these immunoprecipitates contain ubiquitin protease activity (Fig. 3). These data are consistent with the function of AT3 with HDAC6 and dynein in the pathway responsible for trafficking aggregated proteins to form aggresomes.
Fig. 3.
Endogenous AT3 associates with dynein and HDAC6. (A) Endogenous AT3 coimmunoprecipitates with endogenous dynein and transfected FLAG-tagged HDAC6. (B) Immunoprecipitated (IP) dynein, HDAC6, and AT3 all contain ubiquitin protease activity against the fluorogenic substrate ubiquitin-AMC. Control antibody does not immunoprecipitate AT3 or ubiquitin protease activity.
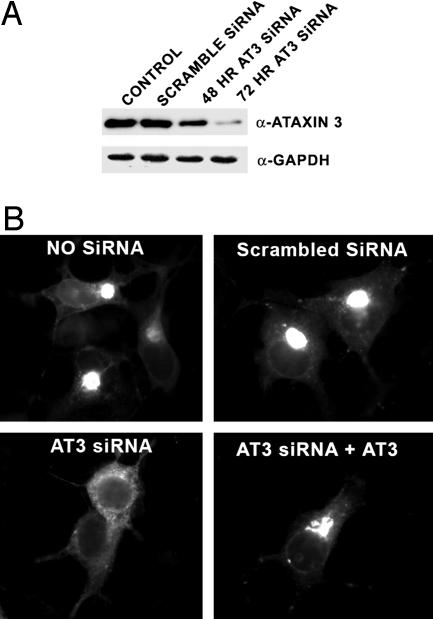
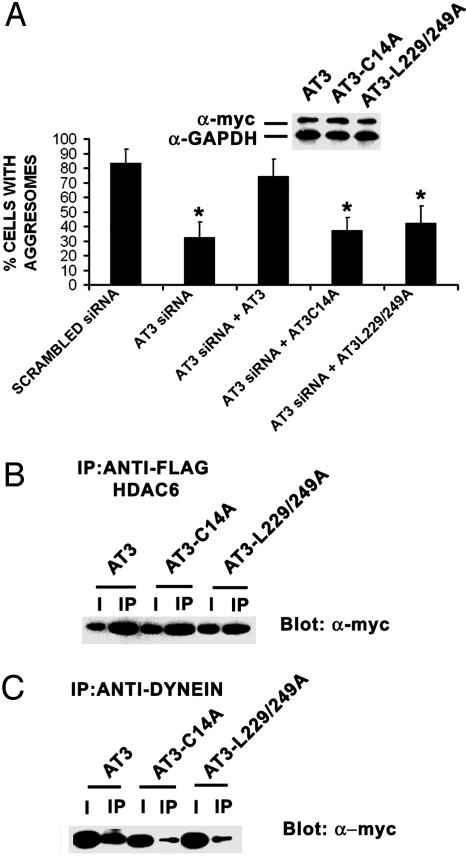
The Deubiquitylating Activity and UIMs of AT3 Are Required for CFTRΔF508 Aggresome Formation and Regulate Association with Dynein. Because AT3 interacts with proteins involved in forming aggresomes, we examined the potential importance of AT3 in aggresome formation by reducing AT3 levels using siRNA. After 72 h, siRNA very effectively decreases AT3, compared with untransfected cells and cells transfected with scrambled AT3 siRNA (Fig. 4A). AT3 siRNA also dramatically decreases CFTRΔF508-GFP aggresomes after MG132 treatment, compared with cells transfected with scrambled AT3 siRNA or untransfected controls (Figs. 4B and 5A). Transfecting knockdown cells with AT3 that is not a target for siRNA restores aggresome formation, indicating that AT3 is required for CFTRΔF508-GFP aggresome formation (Figs. 4B and 5A). To gain insight into whether the DUB activity or ubiquitin-binding properties of AT3 are involved in aggresome formation, the ability of AT3-C14A (the active site mutant) or AT3-L229/249A (the UIM mutant) to restore CFTRΔF508 aggresome formation was determined. Transfecting AT3 knockdown cells with AT3-C14A or AT3-L229/249A that is not a target for siRNA does not restore aggresome formation (Fig. 5A), indicating that the DUB activity and ubiquitin-binding properties of AT3 play a critical role in aggresome formation. To help define potential functions of AT3 in aggresome formation, the effect of mutating the active site or UIMs on interactions with HDAC6 and dynein was determined. Coimmunoprecipitation assays showed that both AT3-C14A and AT3-L229/249A associate with HDAC6 (Fig. 5B); however, neither associates efficiently with dynein (Fig. 5C).
Fig. 4.
siRNA knockdown of AT3 decreases aggresomes formed by CFTRΔF508. (A) An siRNA vector targeting AT3 was transfected into COS 7 cells and cells either collected after 48 h or transfected again and collected 24 h later and blotted for AT3 or GAPDH as a control. AT3 siRNA greatly reduces AT3 after 72 h and has no apparent effect on cell morphology or viability. (B) CFTRΔF508-GFP aggresomes are dramatically decreased in AT3 knockdown cells compared with untransfected cells or cells transfected with scrambled AT3 siRNA. Cotransfecting CFTRΔF508-GFP with an siRNA-resistant AT3 restores aggresome formation in AT3 knockdown cells.
Fig. 5.
The deubiquitylating activity and UIMs of AT3 regulate formation of aggresomes and association with dynein. (A) AT3 knockdown cells were cotransfected with CFTRΔF508-GFP and one of the following: pcDNA3 vector, myc-AT3, myc-AT3-C14A, or myc-AT3-L229/249A. myc-AT3 restores CFTRΔF508 aggresome formation, whereas neither myc-AT3-C14A nor myc-AT3-L229/249A restores formation of aggresomes. (Inset) Transfected myc-AT3 constructs are not targets of siRNA and show similar expression. Data represent mean ± SEM of three independent experiments with at least 600 cells counted for each condition. *, P < 0.05 compared with scrambled siRNA or AT3 siRNA plus AT3. (B) FLAG-tagged HDAC6 coimmunoprecipitates myc-AT3, myc-AT3-C14A, and myc-AT3-L229/249A from COS 7 cells. (C) Dynein intermediate chain coimmunoprecipitates myc-AT3 but considerably less myc-AT3-C14A or myc-AT3-L229/249A. I, 10% input; IP, immunoprecipitate.
These data indicate that both the DUB activity and the UIMs of AT3 are required for aggresome formation and suggest that these functions may be needed for a complex containing AT3 to associate with dynein before transport to the MTOC.
Discussion
The functions of DUBs range from editing ubiquitin chains to recycling ubiquitin by complete disassembly of polyubiquitin chains. The large number of DUBs suggests multiple cellular roles and high substrate specificity; however, few DUBs have been characterized (22, 27). We recently identified AT3 as a protein with DUB activity (17). Here we show that AT3 edits K48-linked ubiquitylated 125I-lysozyme as well as unanchored K48-linked ubiquitin chains. In time course experiments at high concentrations of AT3, its UIMs slow the cleavage of free ubiquitin chains of five to seven ubiquitins, whereas at lower concentrations of AT3, the UIMs modestly increase the efficiency of chain cleavage, possibly by recruiting ubiquitin chains. Although we show that AT3 with UIM mutations cleaves shorter ubiquitin chains on 125I-lysozyme (Fig. 1E and ref. 17), short unanchored ubiquitin chains are poor substrates for AT3 DUB activity (data not shown), suggesting that AT3 may function primarily in editing ubiquitin chains anchored to proteins and not in disassembling free ubiquitin chains. We also show that AT3 decreases degradation of ubiquitylated 125I-lysozyme by purified proteasomes in vitro. AT3 trims the longer ubiquitin chains and likely remains bound to shorter chains, which blocks degradation by preventing access to proteasomes. This implies that AT3 may trim ubiquitin chains to no fewer than four ubiquitins, because AT3 UIMs bind chains of four or more ubiquitins (17–20). When UIMs are mutated, AT3 removes even the short ubiquitin chains from 125I-lysozyme, which is probably responsible for decreasing degradation by the proteasome. AT3 did not bind to proteasomes in our assay (data not shown) and did not alter degradation of a peptide proteasome substrate; therefore, AT3 decreases proteasome degradation by altering the substrate and not through its effects on the proteasome.
The ability of AT3 to partially deubiquitylate and prevent degradation of a protein while possibly maintaining its degradation signal by binding to the remaining ubiquitin chain is an interesting characteristic for a DUB. DUBs such as USP25 and USP37 also contain UIMs, and USP44, USP45, USP49, and USP51 contain zinc-finger ubiquitin-binding domains (28). Therefore, ubiquitin-binding domains may play a general role in deubiquitylation and protein degradation by binding ubiquitylated proteins and possibly regulating the specificity/selectivity of chain cleavage.
The UPP is the major pathway responsible for degrading short-lived and misfolded proteins. In situations where the proteasome cannot effectively deal with excess misfolded proteins, some of these proteins are sequestered into aggresomes at the MTOC (13). Indeed, transport of misfolded proteins to form aggresomes seems to be a critical process for cellular homeostasis (13, 15). We show that endogenous AT3 colocalizes with aggresomes and preaggresome particles of CFTRΔF508, and that it associates with components of the complex that transports misfolded proteins to the MTOC. More importantly, our data indicate that transport of misfolded CFTRΔF508 requires endogenous AT3 and that rescue of aggresome formation in AT3 knockdown cells requires both the DUB activity and the UIMs of AT3. Dynein is required to transport CFTRΔF508 to the MTOC (26), and we provide evidence that the DUB activity and the UIMs of AT3 are necessary for AT3 and/or a complex containing AT3 to interact with dynein.
Our results are consistent with a model in which AT3 is recruited to bind and trim ubiquitin chains on misfolded ubiquitylated proteins, shielding them from the proteasome before transport to the MTOC for aggresome formation. Alternatively, AT3 may use its deubiquitylating activity to stabilize proteins involved in the trafficking of misfolded proteins. As a microtubule motor protein, dynein has multiple cellular functions, including vesicle trafficking and fast retrograde axonal transport (29–31). Endogenous AT3 interacts with dynein, and transfection studies show that this interaction depends on both the DUB activity and UIMs of AT3 (Fig. 5). This raises the possibility that AT3 may regulate other dynein-dependent processes, which is particularly interesting given that disruption of dynein-mediated transport is implicated in neurodegenerative diseases (32, 33).
Protein aggregates are hallmarks of many neurodegenerative diseases including polyglutamine diseases (12). Expansion of the polyglutamine domain of AT3 is thought to destabilize the protein structure, resulting in aggregation (34, 35). AT3 seems to function in the UPP; therefore, a potentially dangerous situation is set up wherein pathological AT3, a protein that misfolds and aggregates, is exposed to misfolded aggregateprone proteins as part of its normal function. It is intriguing that wild-type AT3 is present in inclusions and aggregates of non-polyglutamine-containing proteins, including Marinesco bodies (36) and aggregated protein inclusions in amyotrophic lateral sclerosis (37). The presence of AT3 in these pathophysiological aggregates is consistent with the data presented here, showing that AT3 plays an essential role in formation of CFTRΔF508 inclusions/aggresomes and is present in the inclusions.
Polyglutamine expansion may affect the interactions of AT3 with other proteins and/or affect its DUB activity. Although preliminary data suggest that wild-type and pathological AT3 have very similar DUB activity (B.G.B. and R.N.P., unpublished observations), pathological AT3 cleaves short ubiquitin chains on 125I-lysozyme more efficiently than does wild-type AT3 (17). Additionally, differences in substrate specificity could result from pathological AT3 having altered protein interactions. For instance, AT3 interacts with p97/VCP/Cdc48p in a polyglutamine-dependent manner, with pathological AT3 binding more effectively than wild-type AT3 (38). Valosin-containing protein (VCP) is involved in extracting ubiquitylated misfolded proteins from the endoplasmic reticulum for subsequent degradation by proteasomes (39). Therefore, pathological AT3 could be preferentially involved in VCP-dependent processes. The recent observation that AT3 is a DUB (17, 40), in addition to the present study, which shows that AT3 DUB activity regulates association with dynein and is required for CFTRΔF508 aggresome formation, provides insights for studies investigating both the physiological and potential pathological functions of AT3.
Acknowledgments
We thank Nancy Bonini and Hank Paulson for helpful comments on the manuscript, Ron Kopito for GFP-CFTRΔF508 cDNA, and Stuart Schreiber for FLAG-tagged HDAC6 cDNA. This work was supported by National Institutes of Health Grant NS42625 (to R.N.P.).
Author contributions: B.G.B. and R.N.P. designed research; B.G.B. performed research; B.G.B. analyzed data; and B.G.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AT3, ataxin 3; UPP, ubiquitin proteasome pathway; MTOC, microtubule-organizing center; HDAC6, histone deacetylase 6; DUB, deubiquitylating enzyme; UIM, ubiquitin interacting motif; CFTR, cystic fibrosis transmembrane regulator; MBP, maltose-binding protein; siRNA, small interfering RNA; TCA, trichloroacetic acid.
References
- 1.Takiyama, Y., Nishizawa, M., Tanaka, H., Kawashima, S., Sakamoto, H., Karube, Y., Shimazaki, H., Soutome, M., Endo, K., Ohta, S., et al. (1993) Nat. Genet. 4, 300–304. [DOI] [PubMed] [Google Scholar]
- 2.Paulson, H. L., Perez, M. K., Trottier, Y., Trojanowski, J. Q., Subramony, S. H., Das, S. S., Vig, P., Mandel, J. L., Fischbeck, K. H. & Pittman, R. N. (1997) Neuron 19, 333–344. [DOI] [PubMed] [Google Scholar]
- 3.DiFiglia, M., Sapp, E., Chase, K. O., Davies, S. W., Bates, G. P., Vonsattel, J. P. & Aronin, N. (1997) Science 277, 1990–1993. [DOI] [PubMed] [Google Scholar]
- 4.Skinner, P. J., Koshy, B. T., Cummings, C. J., Klement, I. A., Helin, K., Servadio, A., Zoghbi, H. Y. & Orr, H. T. (1997) Nature 389, 971–974. [DOI] [PubMed] [Google Scholar]
- 5.Li, M., Miwa, S., Kobayashi, Y., Merry, D. E., Yamamoto, M., Tanaka, F., Doyu, M., Hashizume, Y., Fischbeck, K. H., et al. (1998) Ann. Neurol. 44, 249–254. [DOI] [PubMed] [Google Scholar]
- 6.Holmberg, M., Duyckaerts, C., Durr, A., Cancel, G., Gourfinkel-An, I., Damier, P., Faucheux, B., Trottier, Y., Hirsch, E. C., Agid, Y., et al. (1998) Hum. Mol. Genet. 7, 913–918. [DOI] [PubMed] [Google Scholar]
- 7.Chai, Y., Koppenhafer, S. L., Shoesmith, S. J., Perez, M. K. & Paulson, H. L. (1999) Hum. Mol. Genet. 8, 673–682. [DOI] [PubMed] [Google Scholar]
- 8.Abel, A., Walcott, J., Woods, J., Duda, J. & Merry, D. E. (2001) Hum. Mol. Genet. 10, 107–116. [DOI] [PubMed] [Google Scholar]
- 9.Cummings, C. J., Mancini, M. A., Antalffy, B., DeFranco, D. B., Orr, H. T. & Zoghbi, H. Y. (1998) Nat. Genet. 19, 148–154. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt, T., Lindenberg, K. S., Krebs, A., Schols, L., Laccone, F., Herms, J., Rechsteiner, M., Riess, O. & Landwehrmeyer, G. B. (2002) Ann. Neurol. 51, 302–310. [DOI] [PubMed] [Google Scholar]
- 11.Stenoien, D. L., Cummings, C. J., Adams, H. P., Mancini, M. G., Patel, K., DeMartino, G. N., Marcelli, M., Weigel, N. L. & Mancini, M. A. (1999) Hum. Mol. Genet. 8, 731–741. [DOI] [PubMed] [Google Scholar]
- 12.Ross, C. A. & Poirier, M. A. (2004) Nat. Med. 10, S10–S17. [DOI] [PubMed] [Google Scholar]
- 13.Johnston, J. A., Ward, C. L. & Kopito, R. R. (1998) J. Cell Biol. 143, 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Mata, R., Bebok, Z., Sorscher, E. J. & Sztul, E. S. (1999) J. Cell Biol. 146, 1239–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawaguchi, Y., Kovacs, J. J., McLaurin, A., Vance, J. M., Ito, A. & Yao, T. P. (2003) Cell 115, 727–738. [DOI] [PubMed] [Google Scholar]
- 16.Kopito, R. R. (2000) Trends Cell Biol. 10, 524–530. [DOI] [PubMed] [Google Scholar]
- 17.Burnett, B., Li, F. & Pittman, R. N. (2003) Hum. Mol. Genet. 12, 3195–3205. [DOI] [PubMed] [Google Scholar]
- 18.Donaldson, K. M., Li, W., Ching, K. A., Batalov, S., Tsai, C. C. & Joazeiro, C. A. (2003) Proc. Natl. Acad. Sci. USA 100, 8892–8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chai, Y., Berke, S. S., Cohen, R. E. & Paulson, H. L. (2004) J. Biol. Chem. 279, 3605–3611. [DOI] [PubMed] [Google Scholar]
- 20.Doss-Pepe, E. W., Stenroos, E. S., Johnson, W. G. & Madura, K. (2003) Mol. Cell. Biol. 23, 6469–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkinson, K. D. (2000) Semin. Cell Dev. Biol. 11, 141–148. [DOI] [PubMed] [Google Scholar]
- 22.Wing, S. S. (2003) Int. J. Biochem. Cell Biol. 35, 590–605. [DOI] [PubMed] [Google Scholar]
- 23.Perez, M. K., Paulson, H. L., Pendse, S. J., Saionz, S. J., Bonini, N. M. & Pittman, R. N. (1998) J. Cell Biol. 143, 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grozinger, C. M., Hassig, C. A. & Schreiber, S. L. (1999) Proc. Natl. Acad. Sci. USA 96, 4868–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, F., Macfarlan, T., Pittman, R. N. & Chakravarti, D. (2002) J. Biol. Chem. 277, 45004–45012. [DOI] [PubMed] [Google Scholar]
- 26.Johnston, J. A., Illing, M. E. & Kopito, R. R. (2002) Cell Motil. Cytoskeleton 53, 26–38. [DOI] [PubMed] [Google Scholar]
- 27.Guterman, A. & Glickman, M. H. (2004) Curr. Protein Pept. Sci. 5, 201–211. [DOI] [PubMed] [Google Scholar]
- 28.Quesada, V., Diaz-Perales, A., Gutierrez-Fernandez, A., Garabaya, C., Cal, S. & Lopez-Otin, C. (2004) Biochem. Biophys. Res. Commun. 314, 54–62. [DOI] [PubMed] [Google Scholar]
- 29.Martin, M., Iyadurai, S. J., Gassman, A., Gindhart, J. G., Jr., Hays, T. S. & Saxton, W. M. (1999) Mol. Biol. Cell 10, 3717–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirokawa, N., Sato-Yoshitake, R., Yoshida, T. & Kawashima, T. (1990) J. Cell Biol. 111, 1027–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paschal, B. M. & Vallee, R. B. (1987) Nature 330, 181–183. [DOI] [PubMed] [Google Scholar]
- 32.LaMonte, B. H., Wallace, K. E., Holloway, B. A., Shelly, S. S., Ascano, J., Tokito, M., Van Winkle, T., Howland, D. S. & Holzbaur, E. L. (2002) Neuron 34, 715–727. [DOI] [PubMed] [Google Scholar]
- 33.Hafezparast, M., Klocke, R., Ruhrberg, C., Marquardt, A., Ahmad-Annuar, A., Bowen, S., Lalli, G., Witherden, A. S., Hummerich, H., Nicholson, S., et al. (2003) Science 300, 808–812. [DOI] [PubMed] [Google Scholar]
- 34.Perutz, M. F., Johnson, T., Suzuki, M. & Finch, J. T. (1994) Proc. Natl. Acad. Sci. USA 91, 5355–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bevivino, A. E. & Loll, P. J. (2001) Proc. Natl. Acad. Sci. USA 98, 11955–11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujigasaki, H., Uchihara, T., Koyano, S., Iwabuchi, K., Yagishita, S., Makifuchi, T., Nakamura, A., Ishida, K., Toru, S., Hirai, S., et al. (2000) Exp. Neurol. 165, 248–256. [DOI] [PubMed] [Google Scholar]
- 37.Seilhean, D., Takahashi, J., El Hachimi, K. H., Fujigasaki, H., Lebre, A. S., Biancalana, V., Durr, A., Salachas, F., Hogenhuis, J., de The, H., et al. (2004) Acta Neuropathol. 108, 81–87. [DOI] [PubMed] [Google Scholar]
- 38.Hirabayashi, M., Inoue, K., Tanaka, K., Nakadate, K., Ohsawa, Y., Kamei, Y., Popiel, A. H., Sinohara, A., Iwamatsu, A., Kimura, Y., et al. (2001) Cell Death Differ. 8, 977–984. [DOI] [PubMed] [Google Scholar]
- 39.Jarosch, E., Taxis, C., Volkwein, C., Bordallo, J., Finley, D., Wolf, D. H. & Sommer, T. (2002) Nat. Cell Biol. 4, 134–139. [DOI] [PubMed] [Google Scholar]
- 40.Scheel, H., Tomiuk, S. & Hofmann, K. (2003) Hum. Mol. Genet. 12, 2845–2852. [DOI] [PubMed] [Google Scholar]