Abstract
Although eradicated from nature more than two decades ago, the threat of smallpox has reemerged because of concerns over its use as a biological weapon. We present the structure of the poxvirus L1 protein, a molecule that is conserved throughout the poxvirus family and is nearly identical in vaccinia virus and in variola virus, which causes smallpox. L1 is a myristoylated envelope protein that is a potent target for neutralizing antibodies and an important component of current experimental vaccines. The L1 structure reveals a hydrophobic cavity located adjacent to its N terminus. The cavity would be capable of shielding the myristate moiety, which is essential for virion assembly. The structure of L1 is a step in the elucidation of molecular mechanisms common to all poxviruses that may stimulate the design of safer vaccines and new antipoxvirus drugs.
Keywords: myristoylation, virion protein, x-ray structure
Poxviruses are a family of large DNA viruses, the most infamous of which is variola virus, the cause of smallpox. After the worldwide eradication of smallpox more than two decades ago by immunization with the closely related vaccinia virus, routine immunization against smallpox ceased (1). With the threat of an intentional release of variola virus and the emergence of monkeypox virus that also infects humans, the development of antiviral drugs and safer vaccines has been urged (2). By better understanding mechanisms involved in poxvirus biology, therapeutics and vaccines can be designed to act against the whole family of poxviruses (3).
Poxviruses have ≈200 genes and a complex life cycle. After entry into the host cell, the virus reproduces in the cytoplasm, encoding its own replication machinery and transcription factors. Maturing vaccinia virions undergo a transformation from spherical immature particles to the brick-shaped intracellular mature virions (IMVs) that have lipid membranes and are the first infectious form of the virus. Most IMVs are released directly by lysis of the host cell. Some IMVs acquire additional membranes and are then transported out through the host cell membrane. They can either remain attached to the outside of the cell or detach from the cell as an extracellular enveloped virus (EEV). Disruption of the EEV exterior membrane, either by mechanical force or by host complement factors, can also release the IMV form (4). Vaccine development is targeting proteins from each form (5, 6), although because of the fragile nature of the EEV outermost membrane, the IMV form of the virus is thought to play the major role in host-to-host transmission (7).
L1 (also called L1R) is a myristoylated transmembrane protein of 250 residues that is expressed on the surface of the IMV form of the virus. It is an essential protein because vaccinia viruses with the L1 gene deleted are not capable of maturation (8). Although L1 has a C-terminal hydrophobic segment embedded in the viral membrane, the 185-residue, disulfide-bonded ectodomain is located in the cytoplasm before lysis of the host cell. In the otherwise reducing environment of the cytoplasm, the virus encodes an enzymatic pathway that oxidizes the L1 cysteines to disulfide bonds. Disruption of this pathway prevents the maturation of viral particles (9). Virally expressed L1 has a myristate that is attached covalently to the N-terminal Gly residue. Myristoylation of viral proteins occurs in many viral families and is involved in virus assembly and viral infection (10). L1 is also involved in infection, because antibodies to L1 are able to block host cell invasion by the IMV form of the virus (11, 12). L1 has been used in multigene DNA vaccines that are able to block lethal viral challenge in mice and nonhuman primates (6). A multicomponent protein vaccine containing L1 has also been able to block vaccinia virus infection in mice (5).
Materials and Methods
Cloning and Expression. The first 185 residues of L1 were PCR amplified by using the primers gggaattctaaggaggatattcatatgggtgccgcagcaagcatacag and ggggatcctcactgaactcctgtaccagcaacttg and were cloned into the expression vector, pLM1, by using EcoRI and BamHI. For T7 promoter-driven expression of the protein, the plasmid was transformed into BL21(DE3)RIL cells (Stratagene). When the OD600 reached 0.6, protein expression was induced with 1 mM isopropyl β-d-thiogalactoside (IPTG). The cell pellets were resuspended in lysis buffer (100 mM Tris·HCl, pH 8.0/150 mM NaCl/1% Triton X-100/1% sodium deoxycholate/5 mM MgCl2) with the addition of lysozyme to 0.02 mg/ml, DNase to 0.02 mg/ml, and 1 mM phenylmethylsulfonylfluoride. The cells were lysed by cycles of freezing and thawing, followed by homogenization. “Inclusion body protein” was washed several times with lysis buffer, followed by several washes with detergent-free buffer (100 mM Tris·HCl, pH 8.0/150 mM NaCl). The inclusion bodies were dissolved in a denaturing buffer containing 100 mM Tris·HCl, 6 M guanidine-HCl, and 2 mM DTT. The selenomethionine (SeMet)-derivatized protein was produced in BL21(DE3)RIL-X cells (Stratagene) in SelenoMethionine media (Molecular Dimensions, Apopka, FL) supplemented with 50 mg/liter l-selenomethionine (Sigma). When the OD600 reached 0.6, the cells were induced for 6 h with 1 mM IPTG.
Refolding and Purification. Inclusion body L1 was refolded by rapid dilution into 100 mM Tris·HCl/400 mM l-arginine-HCl/5 mM reduced l-glutathione-HCl/0.5 mM oxidized l-glutathione-HCl. After allowing 24 h for refolding, the buffer was dialyzed for 48 h against buffer containing 10 mM Tris·HCl (pH 8.0). The protein was concentrated in Centricon-80 concentrators (Millipore) and purified at least twice by size exclusion chromatography. The purified protein was dialyzed overnight against 10 mM Tris·HCl (pH 8.0). The purity and integrity of the protein was verified by using N-terminal sequencing and mass spectrometry. The refolding and purification of SeMet L1 was the same as for native L1.
Crystallization. Native L1 crystals were obtained in hanging drops by the vapor diffusion method against a mother liquor of 100 mM Hepes (pH 8.0) and 23–25% polyethylene glycol (PEG) 3350. SeMet derivatives were obtained by multiple rounds of streak seeding with either native crystals or SeMet crystal clusters into hanging drops in a buffer of 100 mM Hepes (pH 8.0) and 19–22% PEG-3350. The crystals were frozen in a buffer of 100 mM Hepes, 35% PEG 3350, and 20% glycerol.
Data Collection and Processing. Data (240°) from a SeMet-substituted crystal were collected at the high-energy remote wavelength for selenium on the 19-ID (Structural Biology Center-Collaborative Access Team) beamline at the Advanced Photon Source (APS). Native data (180°) was collected on the 22-ID (Southeast Regional Collaborative Access Team) beamline at the APS at Argonne National Laboratory (Table 1). All data were processed and scaled with HKL2000 (13).
Table 1. Crystallographic statistics.
| Data set | Native | SeMet |
|---|---|---|
| Resolution, Å | 50-1.51 (1.56-1.51) | 50-2.86 (2.96-2.86) |
| X-ray source | 22-ID (SER-CAT) | 19-ID (SBC-CAT) |
| Energy, eV | 12,398 | 12,690 |
| Wavelength, Å | 1.00 | 0.97702 |
| Completeness, % | 99.1 (100) | 99.8 (98.5) |
| Average redundancy | 3.7 (3.7) | 4.1 (3.5) |
| Rsym, % | 6.4 (50.0) | 4.9 (9.0) |
| I/σI | 12.3 (3.6) | 26.1 (13.5) |
| Observed/unique reflections | 197,807/52,912 | 62,894/15,507 |
| Dimensions of C2 cell | ||
| a, Å | 108.7 | 109.6 |
| b, Å | 56.6 | 56.9 |
| c, Å | 72.6 | 73.3 |
| β, ° | 129.3 | 129.7 |
| Figure of merit | ||
| solve | 0.38 | |
| resolve | 0.74 | |
| rms deviations | ||
| Bond lengths, Å | 0.076 | |
| Bond angles, ° | 2.1 | |
| Dihedral angles, ° | 22.6 | |
| Improper angles, ° | 1.15 | |
| Ramachandran | ||
| Favored, % | 93.5 | |
| Allowed, % | 6.5 | |
| Mean B value (Å2) for protein | ||
| Main chain | 23.5 | |
| Side chain | 25.2 | |
| No. of protein atoms (non-hydrogen) | 2,612 | |
| No. of waters | 329 | |
| Mean B value of all atoms | 26.1 | |
| No. of reflections refined/no. free | 52,902/2,646 | |
| Rwork/Rfree, % | 23.9/24.6 |
Values in parentheses are for the highest resolution bin. SBC-CAT, Structural Biology Center-Collaborative Access Team; SER-CAT, Southeast Regional Collaborative Access Team.
Phasing and Refinement. Selenium sites and initial phases were obtained by using solve (14) in a single wavelength anomalous dispersion experiment. resolve improved the maps and automatically built a partial model of glycines and alanines that had interpretable helices (15, 16). The model was improved by iterative cycles of rebuilding in o (17), followed by refinement with cns (18). Asparagine, histidine, and glutamine rotamers were verified by using molprobity (19). Figures were generated by using pymol (20). The hydrophobic surface was calculated by using the protscale plug-in and the electrostatic potential surface calculated with the Adaptive Poisson-Boltzmann Solver (apbs) plug-in (21). The buried surface area calculation was performed by using cns and areaimol (22). Model geometry was verified by using procheck (23). Secondary structure assignment was performed by using dssp (24). Sequences were aligned by using multalin (25). The volume of the cavity was calculated by using a combination of the programs mapman, mama, and flood (26, 27).
Results and Discussion
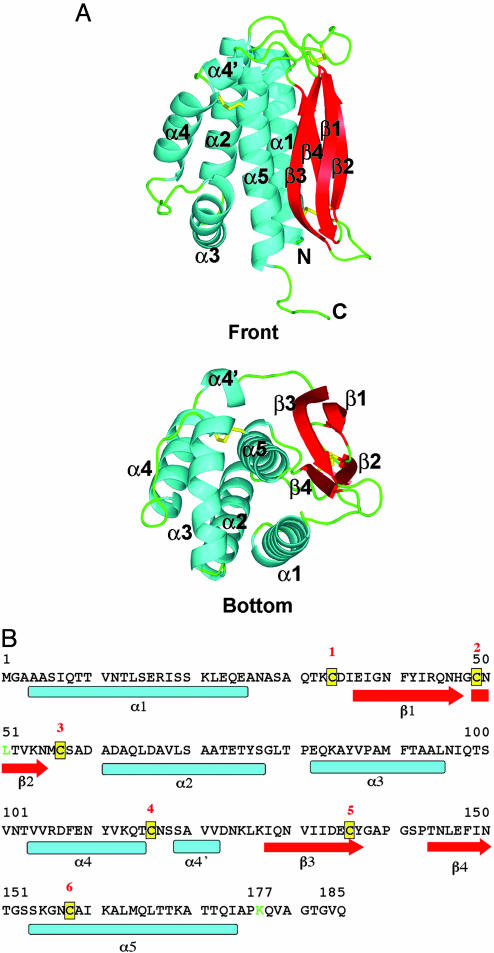
The Structure of the Poxvirus L1 Protein. We have determined the structure of the L1 ectodomain to 1.51-Å resolution by x-ray crystallography. The L1 structure exhibits a fold composed of a bundle of α-helices packed against a pair of two-stranded β-sheets (Fig. 1A). Loops connecting the α-helices to the β-sheets are found only at the top of the molecule. One β-sheet is made up of β1-β3, and the other β-sheet is β2-β4. Between the two sheets, a short loop connects the β1 strand to β2, and a longer loop connects β3 to β4 at the bottom. Thus, β-strands in opposing β-sheets are covalently linked through the polypeptide chain. The β-sheet arrangement is further stabilized by a disulfide bond between β2 and β3, tying the strand-loop-strand arrangements of β1-β2 and of β3-β4 to each other (Fig. 1 A). The juxtaposition of the long N-terminal (α1) and C-terminal (α5) helices bring the N and C termini close together at the bottom of the molecule. The collection of α-helices (α2, α3, α4, and α4′) in the middle of the sequence has several loops, a kink, and a small single-turn helix (α4′) (Fig. 1 A and B). A view of the “bottom” of the molecule reveals that the last 60 residues, containing helix α5 and the β-strands β3 and β4, are at the middle of the molecule (Fig. 1 A Lower). A search of the dali server (28) did not yield any similar structures.
Fig. 1.
The structure of L1. (A) Two views of L1 as ribbon diagrams. The front view (Upper) shows the location of helices (blue) on one side of the molecule, and on the other side, parallel β-sheets (red) formed by nonconsecutive β-strands, connected by loops (green) and the three disulfide bonds (yellow). The bottom view (Lower) shows β3, β4, and α5 packed near the center of the molecule. Helices (α1-α5), β-strands (β1-β4), the N terminus (N), and the C terminus (C) are labeled. (B) Secondary structure elements in L1. Vaccinia virus and variola virus sequences differ at a single residue; Lys-177 in vaccinia virus is an Arg in variola virus (green). This single change is in the linker region between the L1 ectodomain and its transmembrane region. The monkeypox virus sequence also has the Lys to Arg change and a second difference at residue 51, a Leu to Ile change (green). The sequence locations of helices (blue), β-strands (red), and the six cysteines (yellow) are shown. Numerals label the six cysteines (1 to 6) that form disulfide bonds in the pattern, 1–3, 2–5, and 4–6. The L1 transmembrane segment and residues inside the virus (residues 186–250 in vaccinia virus) are not shown.
The sequence of L1 is nearly identical in the vaccinia, variola, and monkeypox viruses. Comparing the ectodomains (residues 1–185) between vaccinia virus and variola virus, there is a single difference at Lys-177 in vaccinia virus, which is conservatively changed to Arg in variola virus (Fig. 1B). This change is located after the C-terminal helix in the linker region that connects the ectodomain and the transmembrane segment. This region is ordered in one molecule of the asymmetric unit, presumably stabilized by crystal packing. The monkeypox virus sequence has a second conservative change of an Ile in place of Leu-51. With just two conservative changes, the L1 structures from variola virus and monkeypox virus will be identical to the vaccinia virus structure.
Conserved Disulfide Bonds of L1. L1 contains six cysteines that are conserved throughout the family of poxviruses. The electron density shows that three bonds are formed between Cys-34 and Cys-57, Cys-49 and Cys-136, and Cys-116 and Cys-158. Numbered in order of appearance in the sequence, this pattern is 1–3, 2–5, and 4–6. Cys-34 and Cys-57 are located in loops, whereas Cys-49 and Cys-136 are both in strands. Cys-116, which is located in the region between α4 and α4′, is disulfide bonded to Cys-158 in helix α5. The disulfide bonds in L1 are formed in the cytoplasm by the virus-encoded glutaredoxin, G4 (9), but from the differing environments surrounding the bonds, G4 does not appear to recognize a specific structural motif. Some conformationally sensitive antibodies to L1 require intact disulfide bonds (11). For example, the neutralizing monoclonal antibody, 2D5, recognizes an epitope that has been mapped to include Asp-35 (12); this epitope is adjacent to Cys-34 and is likely a discontinuous epitope held together by the disulfide bond between Cys-34 and Cys-57. In the model, only 13 Å2, or 12%, of the side chain of Cys-158 is accessible to the solvent, the most of any L1 cysteine. Cys-34 has only 5 Å2 of its side chain exposed to solvent, and the side chains of the other four cysteines are not solvent accessible. The disulfide bonds that are made by G4 during virion maturation in the relatively reducing environment of the cytoplasm may be kept in an oxidized form by the relative solvent inaccessibility of the bonds.
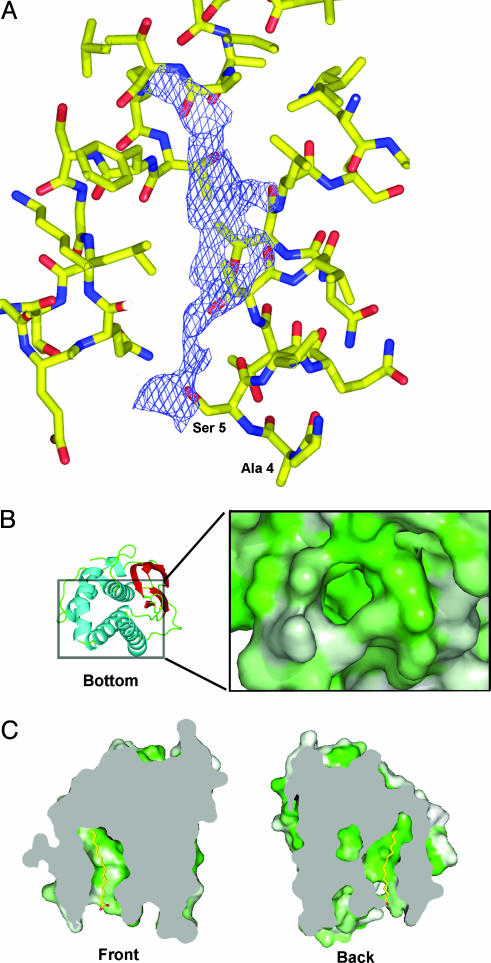
The L1 Hydrophobic Cavity and Myristoylation. The structure reveals a large hydrophobic cavity at the base of the tent-like helices with a volume of ≈536 Å3. We see electron density in the cavities of both the native and selenomethionine crystals that cannot be explained by the L1 amino acid sequence and does not appear to be water molecules (Fig. 2A). The molecule(s) in the cavity likely derive from the refolding or crystallization process, because L1 was refolded from bacterially produced, denatured protein. The cavity is lined with the 16 hydrophobic amino acids: Ile-7, Thr-10, Val-11, Leu-14, Ala-72, Thr-75, Tyr-76, Leu-79, Val-87, Met-90, Phe-91, Val-104, Phe-108, Leu-163, Leu-166, and Ala-170. Fig. 2B shows a surface representation of the cavity in L1, colored by hydrophobicity, and viewed from a point just outside the cavity opening.
Fig. 2.
Extra electron density and the cavity in L1. (A) The unidentified electron density inside the L1 cavity is shown (purple) from a 2Fo-Fc composite omit electron density map contoured at 1 σ. Residues neighboring the cavity are shown in stick representation. Ala-4 and Ser-5 of helix α1 are labeled to show the proximity of the density to the N terminus. (B) A ribbon diagram of the bottom of L1 (Left) shows the location of the hydrophobic cavity. The rectangle (Left) represents the region that is expanded (Right). A molecular surface representation of the cavity, without the unknown density (Right), is colored by degree of hydrophobicity on a green (most hydrophobic) to white (least hydrophobic) scale. (C) Myristate is modeled in the hydrophobic cavity in stick representation. Two sections through L1 (gray planes) reveal the extent of the hydrophobic cavity as viewed from the front (Left) and the back (Right) of L1.
Because of its proximity to the N terminus, this hydrophobic pocket may be able to shield the myristate moiety, which is present on L1 in infected cells. L1 becomes myristoylated in the cell on the N-terminal Gly-2, and this residue is conserved throughout all poxvirus L1 sequences. Preventing myristoylation by mutating Gly-2 to Ala inhibits the incorporation of L1 in immature virions and blocks the maturation of virus (8). We have modeled a myristate in the hydrophobic cavity based on the location of the unknown electron density (Fig. 2C). In addition to the residues that line the hydrophobic cavity, Ile-174 is positioned at the outer ridge of the cavity and may act as a gate for the cavity. Other proteins, such as the tyrosine kinase, Abl, use a myristate moiety to regulate function and localization to the membrane (29). In both picornavirus and polyomavirus, myristoylation mediates protein–protein interactions that are required for capsid assembly (10). The myristylprotein, VP4, of poliovirus packs inside the capsid but can become exposed in a reversible manner that has been proposed to play a role in cell entry (30). It would be interesting to test how poxviruses are affected by myristate analogues or by compounds that target the hydrophobic cavity.
Because L1 contains a transmembrane region, the myristate may exit the pocket to interact with other membranes and retract into the pocket through conformational changes, possibly regulated by the state of its disulfide bonds. Three other myristylproteins have been identified in vaccinia virus. Despite the presence of this hydrophobic moiety, two are known to be soluble in the cytoplasm (31), suggesting that they have a mechanism of shielding their myristate moieties. In reoviruses, the μ1 myristylprotein contains a shielded myristate. The removal of the σ3 protein and the proteolytic cleavage of μ1 results in the exposure of the μ1 myristate and its insertion into the host membrane (32). The L1 myristate may also become exposed through a proteolytic mechanism. In such a mechanism, the disulfide bonds of L1 may be important for its stability. A comparison of the L1 cavity and the μ1 cavity reveals that they are of similar size and are both capable of accommodating a whole myristate molecule, although the cavities in the two proteins are formed in structurally different ways.
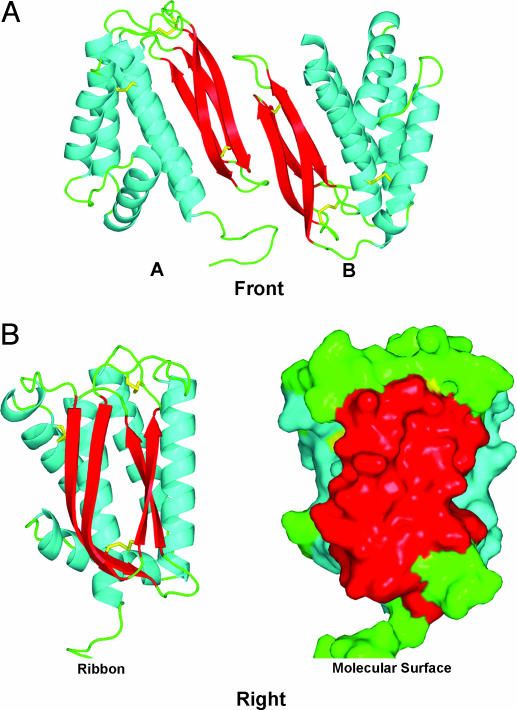
The Two Molecules of the L1 Asymmetric Unit. Although L1 is a monomer by size exclusion chromatography, it packs into the crystal with two molecules in the asymmetric unit. As shown in Fig. 3, the molecules are packed with contacts between the β-sheet regions that bury 1,027 Å2 of solvent-accessible surface area. Because L1 is not a dimer in solution, this contact surface may be a binding site for other protein(s). The neutralizing monoclonal antibody, 2D5, binds near or on the L1 β-sheets because its epitope has been mapped to include Asp-35, located two residues from the start of the β1 strand (12). Mutations of this surface can be tested for their effect on the virus life cycle.
Fig. 3.
Interaction of the two L1 molecules in the asymmetric unit. (A) The two molecules pack through interactions at the edge of the β-sheets. (B) View of the β-sheet side of L1. A ribbon representation of L1 shows the β-sheet region (Left) that mediates packing in the asymmetric unit. (Right) The same view is depicted as a molecular surface representation of the helices (blue), β-strands (red), loops (green), and disulfide bonds (yellow).
Between the two molecules of the asymmetric unit, the electron density is excellent for the entire L1 model except for the first two residues at the N terminus. Molecule A of the asymmetric unit has electron density for residues 4–185, except for a portion of the loop between α4 and β3 from residue 118 to 125 that is well ordered in molecule B. Molecule B has density for residues 5–172, missing part of the last turn of α5 and the linker region that follows. The large unidentified electron density is seen in molecule A, perhaps stabilized by the packing of the C terminus (Ile-174) at the cavity opening. Molecule B contains a smaller density located deeper in the cavity, away from the opening. Because of the difference in size of the densities, the unknown molecule is likely to be polyethylene glycol from the crystallization solution, because a molecule retained from refolding would be expected to be present in both molecules of the asymmetric unit. There is disorder in the loop from residue 95 to 101 in molecule B but good density for the same loop in molecule A. The last 13 residues of molecule B could not be modeled. The root mean square (rms) difference between all α-carbon atoms in the two molecules of the asymmetric unit is 0.77 Å. Overall, there is excellent density to support all but the first two residues of the model of L1, validating the use of this 1.51 Å model in drug design.
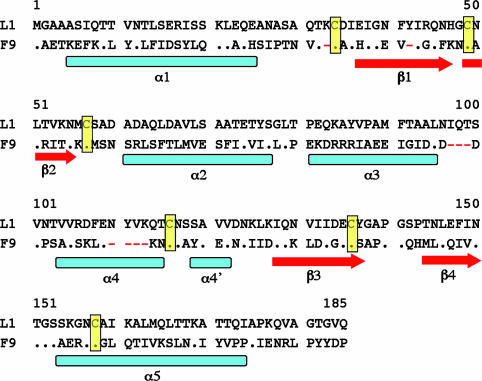
Comparison Between L1 and F9. The F9 protein of vaccinia virus has 24% sequence identity with L1 and is highly conserved among poxviruses, although its function is unknown. Despite their low sequence identity, we modeled F9 on the x-ray structure of L1 because most of the predicted secondary structural elements are the same as those determined for L1 and the six cysteines, as well as the spacing between them, are conserved (Fig. 4). F9 likely has a similar fold and pattern of disulfide bond formation but does not have an N-terminal glycine for myristoylation. Compared with L1, F9 has one residue missing in the loop between the α1 helix and the β1 strand (Fig. 4). There is one residue missing in β1. The biggest difference between L1 and F9 appears to be in the region around the α3 and α4 helices. F9 also appears to be missing at least one turn of the α4 helix, although the end of α4 appears to be anchored by a disulfide bond between Cys-107 and Cys-149. The α5 helix of F9 appears to be shorter by one turn and is likely terminated by consecutive prolines. This shorter helix and the absence of a residue equivalent to Ile-174 of L1 reduce the depth of the hydrophobic cavity in F9 compared with L1. Changes in the residues lining the hydrophobic cavity, for example, the larger Phe-70 in F9 replacing Ala-72 in L1, may occlude the back of the cavity and further reduce its depth. Finally, the sequence similarity ends after the position of the final helix, α5.
Fig. 4.
Sequence alignment of the ectodomains of vaccinia virus L1 and F9. Identical residues are represented as dots. The locations of helices and β-strands based on the L1 structure are shown. The six cysteines forming the disulfide bonds are highlighted in yellow. Here, gaps are represented as red dashes; four gaps have been introduced in the F9 sequence by the alignment. There is a single residue gap in the loop between α1 and β2 and another that shortens β1. The loop between α3 and α4 is shorter by three residues in F9. There is a gap of four residues in α4 of F9 that shortens it by one turn of helix.
Concluding Remarks. L1 plays critical roles in both morphogenesis and infection of poxviruses. With the structure of L1, new experiments can now address the biological role of L1 on a molecular level. The model allows the mapping of antibody epitopes that may lead to additional target surfaces on L1 for drug design. It would be interesting to find neutralizing antibodies with epitopes that do not map to the surface, suggesting either a conformational change or proteolytic cleavage of L1 on the virion surface. Based on the structure, therapeutics can be designed to bind the putative protein interaction surface or the hydrophobic pocket. The structure forms a basis for the design of drugs that directly interact with L1 to prevent poxvirus infection.
Acknowledgments
We thank the members of the Structural Biology Section for helpful discussions, Mark Garfield and Carl Hammer for help with mass spectrometry and N-terminal sequencing, and the staff of the Structural Biology Center–Collaborative Access Team and the Southeast Regional Collaborative Access Team for assistance with data collection. Data were collected at the Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source beamlines was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-Eng-38.
Abbreviations: IMV, intracellular mature virion; SeMet, selenomethionine.
Data deposition: The atomic coordinates and structure factors of L1 have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1YPY).
References
- 1.Fields, B. N., Knipe, D. M. & Howley, P. M. (1996) Fields Virology (Lippincott-Raven, Philadelphia).
- 2.Harrison, S. C., Alberts, B., Ehrenfeld, E., Enquist, L., Fineberg, H., McKnight, S. L., Moss, B., O`Donnell, M., Ploegh, H., Schmid, S. L., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 11178–11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henderson, D. A., Inglesby, T. V., Bartlett, J. G., Ascher, M. S., Eitzen, E., Jahrling, P. B., Hauer, J., Layton, M., McDade, J., Osterholm, M. T., et al. (1999) J. Am. Med. Assoc. 281, 2127–2137. [DOI] [PubMed] [Google Scholar]
- 4.Lustig, S., Fogg, C., Whitbeck, J. C. & Moss, B. (2004) Virology 328, 30–35. [DOI] [PubMed] [Google Scholar]
- 5.Fogg, C., Lustig, S., Whitbeck, J. C., Eisenberg, R. J., Cohen, G. H. & Moss, B. (2004) J. Virol. 78, 10230–10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper, J. W., Thompson, E., Wilhelmsen, C., Zimmerman, M., Ichou, M. A., Steffen, S. E., Schmaljohn, C. S., Schmaljohn, A. L. & Jahrling, P. B. (2004) J. Virol. 78, 4433–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooper, J. W., Custer, D. M. & Thompson, E. (2003) Virology 306, 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravanello, M. P. & Hruby, D. E. (1994) J. Virol. 68, 6401–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senkevich, T. G., White, C. L., Koonin, E. V. & Moss, B. (2002) Proc. Natl. Acad. Sci. USA 99, 6667–6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurer-Stroh, S. & Eisenhaber, F. (2004) Trends Microbiol. 12, 178–185. [DOI] [PubMed] [Google Scholar]
- 11.Wolffe, E. J., Vijaya, S. & Moss, B. (1995) Virology 211, 53–63. [DOI] [PubMed] [Google Scholar]
- 12.Ichihashi, Y. & Oie, M. (1996) Virology 220, 491–494. [DOI] [PubMed] [Google Scholar]
- 13.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 14.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terwilliger, T. C. (2000) Acta Crystallogr. D 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terwilliger, T. C. (2003) Acta Crystallogr. D 59, 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 18.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 19.Lovell, S. C., Davis, I. W., Arendall, W. B., III, de Bakker, P. I., Word, J. M., Prisant, M. G., Richardson, J. S. & Richardson, D. C. (2003) Proteins 50, 437–450. [DOI] [PubMed] [Google Scholar]
- 20.DeLano, W. L. (2002) The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, CA).
- 21.Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. (2001) Proc. Natl. Acad. Sci. USA 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 23.Laskowski, R. A., McArthur, M. W., Moss, D. S. & Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 24.Kabsch, W. & Sander, C. (1983) Biopolymers 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 25.Corpet, F. (1988) Nucleic Acids Res. 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleywegt, G. J. & Jones, T. A. (1999) Acta Crystallogr. D 55, 941–944. [DOI] [PubMed] [Google Scholar]
- 27.Kleywegt, G. J. (1994) Acta Crystallogr. D 50, 178–185. [DOI] [PubMed] [Google Scholar]
- 28.Holm, L. & Sander, C. (1996) Science 273, 595–603. [DOI] [PubMed] [Google Scholar]
- 29.Hantschel, O., Nagar, B., Guettler, S., Kretzschmar, J., Dorey, K., Kuriyan, J. & Superti-Furga, G. (2003) Cell 112, 845–857. [DOI] [PubMed] [Google Scholar]
- 30.Hogle, J. M. (2002) Annu. Rev. Microbiol. 56, 677–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, K. H., Grosenbach, D. W., Franke, C. A. & Hruby, D. E. (1997) J. Virol. 71, 5218–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liemann, S., Chandran, K., Baker, T. S., Nibert, M. L. & Harrison, S. C. (2002) Cell 108, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]