Abstract
Pelizaeus–Merzbacher disease (PMD) is a dysmyelinating disease caused by mutations, deletions, or duplications of the proteolipid protein (PLP) gene. Mutant forms of PLP are retained in the endoplasmic reticulum (ER), and the resulting accumulation of mutant protein is thought to be a direct cause of oligodendrocyte cell death, which is the primary clinical feature of PMD. The molecular mechanisms underlying the toxicity of mutant PLP are however currently unknown. We report here that PMD-linked mutations of PLP are associated with the accelerated assembly of the protein into stable homooligomers that resemble mature, native PLP. Thus although WT PLP forms stable oligomers after an extended maturation period, most likely at the cell surface, mutant forms of PLP rapidly assemble into such oligomers at the ER. Using PLP mutants associated with diseases of varying severity, we show that the formation of stable oligomers correlates with the development of PMD. Based on these findings, we propose that the premature oligomerization of PLP in the ER of oligodendrocytes contributes to the pathology of PMD.
Keywords: oligomer, Pelizaeus–Merzbacher disease, endoplasmic reticulum, quality control, membrane protein
Myelin sheaths in the CNS are formed by the oligodendrocyte plasma membrane, which wraps around the axon. CNS myelin contains a number of specific lipids and membrane-associated proteins, of which two predominate. Myelin basic protein is a soluble protein that binds to the cytoplasmic surface of the myelin membrane and brings the cytoplasmic faces close together, allowing a tight spiral to form around the axon (1). Proteolipid protein (PLP) and its splice variant DM-20 are integral membrane proteins that together constitute ≈50% of the protein in CNS myelin (2). PLP is synthesized at the endoplasmic reticulum (ER) and then transported to the cell surface where it is incorporated into the myelin membrane. Although some evidence suggests that PLP may function as a channel-forming protein (3, 4), the primary role of PLP in myelin formation is currently thought to be the adhesion and stabilization of the extracellular surfaces of the myelin membrane (5, 6). Consistent with this finding, biochemical and biophysical studies suggest that native PLP forms homooligomers (7–9). However, it is not known how or where PLP oligomers are assembled within the cell, or what role the different domains play in oligomerization.
Mutations in the PLP gene are associated with debilitating diseases of the CNS, including Pelizaeus–Merzbacher disease (PMD) and X-linked spastic paraplegia (10). The amino acid sequence of PLP is highly conserved, and a large number of naturally occurring mutations are associated with disease. Most of these mutations prevent PLP from reaching the cell surface (11, 12), and evidence suggests that the mutant PLP is misfolded (13, 14). Mutant forms of PLP are retained in the ER, and the resulting accumulation of mutant PLP in the ER is thought to be a direct cause of the oligodendrocyte cell death that is the primary clinical feature of PMD (12, 15).
Despite its obvious importance to the pathology of PMD, the molecular mechanisms that mediate the ER retention of misfolded PLP are unknown. We have recently shown that the ER chaperone calnexin plays a key role in this process (16), but it is not clear what underlies the recognition of mutant PLP and whether this interaction is the causal basis for retention of PLP mutants. Given that the assembly of oligomeric membrane protein complexes often occurs in the ER, and that in many such instances oligomer formation is a prerequisite for ER exit, one attractive possibility is that the impaired ability of mutant forms of PLP to assemble appropriately into oligomers causes these proteins to be recognized and retained by the ER quality control machinery (17).
However, in contrast to such predictions, we now provide evidence that a central characteristic that distinguishes disease-linked mutant forms of PLP from the WT protein is in fact the accelerated rate at which they form stable, SDS-resistant homooligomers. The WT protein forms such oligomers only after an extended maturation period, and well after it has been transported to the cell surface. These findings have implications for our understanding of the pathology of human PMD and provide a basis for the gain-of-function phenotype of PLP-associated diseases.
Methods
Reagents and Antibodies. Brefeldin A was from Alexis (San Diego). Cross-linking reagents were from Pierce. All other chemicals were from Sigma or BDH/Merck. Rabbit α-ha and mouse α-myc (9E10) were from Sigma. Serum from rabbits immunized with peptide AFPSKTSASIGSLC from PLP was used for detection of untagged PLP. Horseradish peroxidase-conjugated secondary antibodies were from DAKO.
Molecular Cloning and DNA Manipulations. The cDNA encoding mouse PLP was inserted into pcDNA3.1(-)/Myc-His as described (16). QuikChange mutagenesis (Stratagene) was used to generate the six point mutations, add an N-terminal ha tag and generate TM1-3ha by inserting an ha tag and stop codon after residue 200 of PLP mutant 5 (Table 1). Constructs were confirmed by DNA sequencing.
Table 1. Missense mutations used in this study.
| Mutation | Substitution | Phenotype |
|---|---|---|
| 1 | H36P | Severe |
| 2 | T181P | Severe |
| 3 | A242V (msd) | Severe |
| 4 | T155I | Mild |
| 5 | I186T | Mild |
| 6 | P215S | Mild |
Cell Culture and Transfection. COS-7 and HeLa cells were grown in DMEM containing 10% FCS. Clontech Tet-On HeLa cells were grown in DMEM containing 100 μg/ml geneticin plus 100 μg/ml hygromycin. For biochemical studies, cells were transiently transfected by using Lipofectamine 2000 (Invitrogen) and analyzed after 18–24 h. For immunofluorescence, cells were transfected by using FuGENE 6 (Roche Diagnostics) and fixed after 18–24 h. HeLa cell lines stably expressing WT or msd myc his-tagged PLP (PLPmh) under the control of a tetracyclin inducible promoter were generated as described (16). Expression was induced by 1 μg/ml deoxycyclin, and cells were analyzed after 18–24 h.
SDS/PAGE and Western Blotting. Crude myelin membranes were prepared from porcine brains as described (18). Cells were grown and transfected or induced in 12-well dishes and harvested after 18–24 h unless stated otherwise. Cells were rinsed twice with PBS, incubated with 10 mM iodoacetamide in PBS for 15 min at 4°C, and then rinsed twice with PBS and lysed in SDS/PAGE sample buffer [25 mM Tris, pH 6.8/1% SDS/0.05% bromophenol blue/5% (wt/vol) glycerol/100 mM DTT]. Samples were incubated at 95°C for 5 min or 70°C for 10 min and then loaded onto 12% polyacrylamide gels containing 0.1% SDS. Gels were transferred to Hybond C membrane (Amersham Pharmacia) and immunoblotted by using an enhanced chemiluminescence system.
Blue Native Gel Electrophoresis. Cells expressing PLPmh were rinsed twice with PBS and solubilized with 0.5–2% N-dodecylmaltoside. Samples were incubated on ice for 1 h with vortexing and then centrifuged at 16,000 × g for 10 min. One microliter of sample buffer {100 mM Bistris ([bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane), pH 7.0/500 mM 6-aminocaproic acid/5% (wt/vol) Coomassie brilliant blue G250} was added to 10 μl of the cell lysate, and samples were loaded onto a 6–16% polyacrylamide gradient gel (19).
Coimmunoprecipitation. Cells stably expressing PLPmh were transfected with hemagglutinin-tagged PLP (PLPha) in 10-cm dishes and harvested 18–24 h later. Cells were rinsed twice with PBS, incubated with 10 mM iodoacetamide in PBS for 15 min at 4°C, or cross-linked as described below, and then rinsed twice with PBS and solubilized with standard immunoprecipitation (IP) buffer (16). Samples were incubated on ice for 1 h with vortexing and then centrifuged at 16,000 × g for 10 min. The supernatant was split in half, incubated with α-epitope tag antibody or species-matched control overnight at 4°C, and then 20 μl of protein A Sepharose was added and samples were rotated at 4°C for 3 h. Beads were washed three times, and samples were analyzed by SDS/PAGE and Western blotting.
Gel Filtration. Cell lysates were prepared as above, and 50 μl were separated on a Superdex 200 PC 3.2/30 column (Amersham Pharmacia) in IP buffer. Twenty-seven fractions (80 μl) were collected and analyzed by SDS or blue native gel electrophoresis, or immunoprecipitated with α-myc or control antibody as described above.
Cross-Linking. Cells were rinsed twice with PBS and then incubated for 10 min at room temperature with 0.25 mM 1,4-di-(3′-[2′pyridyldithio]-propionamido) butane (DPDPB) in PBS. DPDPB cross-links adjacent proteins by means of the –SH groups of free cysteines and is cleavable by reducing agents. Cells were rinsed twice with PBS and then incubated for 10 min on ice with 5 mM cysteine to quench the cross-linking reactions, before IP as described above.
Indirect Immunofluorescence Microscopy. Cells were fixed by using cold methanol, and immunofluorescence microscopy was performed as described (16).
Results
Endogenous PLP Forms Stable SDS-Resistant Oligomers in Myelin. We first examined the oligomeric status of WT endogenous PLP in myelin. When analyzed by reducing SDS/PAGE followed by Western blotting with PLP antisera, we found that a large proportion of PLP in a crude brain membrane extract was present in higher molecular weight complexes (Fig. 1A, lane 2). The existence of high molecular weight forms of PLP has been noted by several investigators, and purified PLP monomers can assemble into SDS-resistant homooligomers (7). Assuming that these higher molecular weight species represent PLP homooligomers, their apparent molecular masses of ≈40 and 60 kDa would correspond to dimers and trimers respectively.
Fig. 1.
Structure and subcellular distribution of WT PLP. (A) Crude myelin extracts were analyzed by SDS/PAGE and Western blotting with a PLP-specific serum or the corresponding preimmune sera. (B) Diagram depicting the topology of PLP. Palmitoylation sites are shown by squiggles, disulfide bonds are shown by lines, and point mutations used in this study are shown by numbers 1–6. Mutations associated with mild PMD are in filled circles, and those associated with severe disease are in open circles. (C) COS-7 cells transiently transfected with WT PLPmh were stained with α-myc followed by a FITC-conjugated secondary antibody. (Scale bar: 20 μm.) (D) Lysates of cells transfected with WT PLPmh were analyzed by Western blotting with α-myc.
Formation of SDS-Resistant Oligomers Occurs Slowly. To examine the effect of disease-associated mutations on the oligomerization of PLP, we used an established cellular expression system (16). Myc/His-6 (mh) or ha epitope tags were added to the C terminus of PLP (Fig. 1B) to facilitate analysis. We have previously shown that the topology and folding of epitope-tagged PLP is similar to that of the untagged protein (16). When expressed in COS-7 cells, WT PLPmh was transported to the cell surface as expected (Fig. 1C and ref. 16). However, when lysates of cells expressing WT PLPmh were analyzed by SDS/PAGE and Western blotting, the expressed protein migrated almost exclusively as a monomer (Fig. 1D, lane 1). Similar results were obtained when WT PLPmh was expressed in HeLa cells (Fig. 1D, lane 2), and with WT PLP possessing a C-terminal ha tag (not shown).
To determine whether WT PLP forms less stable oligomers that are labile during SDS/PAGE when expressed in cultured cells, blue native gel electrophoresis was performed (Fig. 2A). This analysis revealed that the majority of WT PLPmh expressed in COS-7 cells was indeed present in higher molecular weight complexes (Fig. 2 A, lanes 1–3) as seen in myelin by SDS/PAGE (Fig. 1 A). These complexes most likely also correspond to dimers and trimers, although the different conditions of electrophoresis and a significant shift in molecular weight due to the epitope tag do not allow direct comparison. To substantiate that these complexes were indeed PLP homooligomers, we performed coimmunoprecipitation experiments under nondenaturing conditions. When WT PLPmh and WT PLPha were coexpressed and the PLPha was immunoisolated by means of its ha tag, PLPmh was specifically coimmunoprecipitated with anti-ha and not with a control antibody (Fig. 2B, compare lanes 1 and 2). The efficiency of coimmunoprecipitation increased significantly if PLP oligomers were stabilized by covalent cross-linking before solubilization by using the thiol-cleavable cross-linker DPDPB (Fig. 2B, lane 4), indicating that PLP oligomers are normally present in intact membranes but lack the distinctive stability of the endogenous PLP oligomers present in myelin.
Fig. 2.
Oligomerization of WT PLPmh. (A) COS-7 cells transfected with WT PLPmh were solubilized with the indicated concentration of N-dodecylmaltoside (DDM) and analyzed by blue native gel electrophoresis and Western blotting with α-myc. (B) Cells stably expressing WT PLPmh were transfected with WT PLPha, solubilized, and immunoprecipitated with α-ha or control (ctl) antibody. Fifty percent of the lysate used for IP is present in lane 3. Lanes 4 and 5 show cells that were cross-linked with DPDPB before IP. Immunoprecipitated material was analyzed by Western blotting with α-myc. Fragments of the IP antibody heavy chain are visible at the top of the panel. (C) A stable cell line was induced to express WT PLPmh for the time indicated, and lysates were analyzed by SDS/PAGE and Western blotting with α-myc. Several different exposures were quantified by densitometry, and the dimer:monomer ratio was calculated.
This reduction in stability could reflect a requirement for oligodendrocyte-specific factor(s) necessary for the stabilization of PLP oligomers. Alternatively, the formation of stable oligomers may be a slow process requiring a prolonged maturation period. To address this issue, we generated stable cell lines expressing WT PLPmh under the control of an inducible promoter. Cells were induced to express WT PLPmh for increasing periods of time, and the formation of SDS-resistant oligomers was assessed by SDS/PAGE and Western blotting. As was seen in transiently transfected cells, 24 h after induction, WT PLPmh migrated almost exclusively as a monomer (Fig. 2C, lane 2). However, after 48 h, SDS-resistant oligomers of WT PLPmh could be clearly seen (Fig. 2C, lane 3), and, by 96 h, this proportion had increased further (Fig. 2C, lane 4). The majority of these oligomers were putative dimers, although some trimers were also observed (Fig. 2C, lane 4). Immunofluorescence microscopy confirmed that essentially all of the WT PLPmh was localized to the cell surface at these times (not shown). Given the very rapid rate at which PLP is transported (20), it is likely that oligomeriztion occurred well after arrival of the protein at the cell surface. In addition, biotinylation experiments revealed that the stable oligomers of WT PLP were present at the cell surface, suggesting that they were formed by correctly folded protein (Fig. 5, which is published as supporting information on the PNAS web site). These results demonstrate that, in this system at least, the assembly of SDS-resistant oligomers of WT PLP occurs relatively slowly. Because the endogenous PLP found in myelin is also present in SDS-resistant oligomers (although a greater proportion is in trimers), these complexes are most likely en route to the final native conformation of PLP.
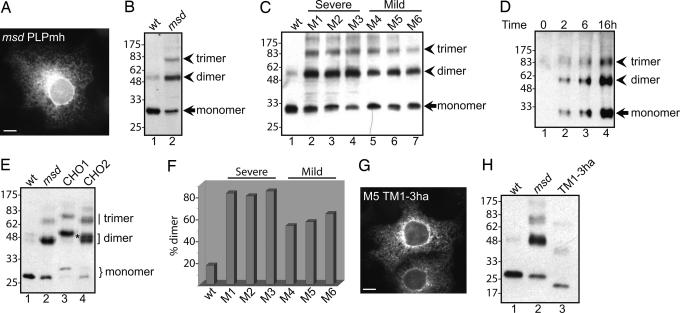
Disease-Associated Mutant Forms of PLP Rapidly Form SDS-Resistant Oligomers. A large number of naturally occurring missense mutations in PLP have been identified, and these are associated with disease of varying severity (10). A particularly well studied mutation, the msd allele, which causes severe disease in transgenic mice, is the substitution of a valine for an alanine at position 242 in the fourth transmembrane (TM) domain (Fig. 1B and Table 1, mutation 3). In contrast to WT PLPmh, msd PLPmh was retained in the ER of transfected COS-7 cells (Fig. 3A and ref. 16). We next examined the ability of this mutant form of PLPmh to form SDS-resistant oligomers when expressed in COS-7 cells. Remarkably, in contrast to WT PLPmh (Fig. 3B, lane 1), the majority of the expressed msd PLPmh was present in SDS-resistant oligomers within 24 h of transfection (Fig. 3B, lane 2). These oligomers were most likely dimers and trimers, similar to those formed by endogenous PLP in myelin (compare Fig. 1A; note that the myc/His tag causes a significant shift in molecular weight).
Fig. 3.
Oligomerization of PLP mutants. (A) COS-7 cells transiently transfected with msd PLPmh were stained with α-myc followed by a FITC-conjugated secondary antibody. (Scale bar: 20 μm.) (B) Lysates of COS-7 cells transfected with msd PLPmh were analyzed by SDS/PAGE and Western blotting with α-myc. (C) COS-7 cells were transfected with WT or mutant forms of PLPmh and then analyzed as in B. (D) A stable cell line was induced to express msd PLPmh for the time indicated and then analyzed as in B. Note that approximately three times more cell lysate was used here than in Fig. 2C. (E) Lysates of COS-7 cells transfected with WT or msd PLPha, or with msd PLPha containing a glycosylation site in the first (msd CHO1) or second (msd CHO2) luminal loop, were analyzed by SDS/PAGE and Western blotting with α-ha. The position of glycosylated and nonglycosylated monomeric PLPha is shown by a curly bracket, and dimers are indicated by a square bracket. A dimer composed of glycosylated and nonglycosylated PLP is shown by an asterisk. (F) COS-7 cells were transfected as in C and then analyzed by SDS/PAGE and Western blotting with α-myc followed by [125I]protein A. PLPmh was quantified by autoradiography, and the amount of SDS-resistant dimer is expressed as a percentage of the total amount of dimer plus monomer. The chart shows the means of two to three independent experiments. The significance of the difference between mutants associated with mild and severe disease was determined by using the independent-samples t test. (G) COS-7 cells were transfected with TM1-3ha and stained with α-ha followed by a FITC-conjugated secondary antibody. (Scale bar: 20 μm.) (H) COS-7 cells were transfected with WT or msd PLPha, or with mutant 5 TM1-3ha and analyzed as in E.
These data suggested that, in contrast to our original hypothesis, mutant PLP forms stable oligomers much more rapidly than the WT protein. To determine whether this propensity to oligomerize into stable structures is an inherent property of all mutant forms of PLP, or a specific feature of the msd mutant, we generated five additional disease-associated mutations (Fig. 1B and Table 1). These mutations are located at different positions throughout the primary sequence of PLP (Fig. 1B) and ensured that we examined the effect of mutating different regions of the polypeptide. Immunofluorescence microscopy confirmed that all mutants were located in the ER of transfected COS-7 cells (not shown). Strikingly, each of the different mutant proteins migrated primarily as higher molecular weight complexes by SDS/PAGE (Fig. 3C, lanes 2–7), in contrast to WT PLPmh (Fig. 3C, lane 1). Therefore, the tendency to form SDS stable oligomers is characteristic of a range of different disease-associated PLP mutants.
To examine the kinetics with which mutant PLP forms stable oligomers, a stable cell line was induced to express msd PLPmh for short periods of time, and the extent of oligomer formation was examined by SDS/PAGE and Western blotting. SDS-resistant oligomers were apparent after just 2 h and appeared with a similar time course to the monomer (Fig. 3D, lanes 1–4). Moreover, the ratio of oligomer:monomer did not alter substantially over a 16-h period. These results clearly show that msd PLPmh assembles into stable oligomers much more rapidly than the WT protein. It is important to note that the cell lines expressing WT and msd PLPmh synthesize similar levels of these proteins (16), ruling out any nonspecific effect of differing protein concentration.
To demonstrate that the SDS-resistant complexes formed by mutant PLP are indeed homooligomers, we generated a higher molecular weight version of the protein by introducing artificial N-glycosylation sites into the first (CHO1, Asn 47) or second (CHO2, Asn 222) luminal loop of msd PLPha. As shown in Fig. 3E, both PLP CHO1 and CHO2 formed SDS-resistant complexes very efficiently. PLP CHO1 was fully glycosylated, resulting in a shift in the molecular weight of the monomeric protein and a corresponding increase in the size of the putative dimer (Fig. 3E, lane 3). PLP CHO2 was less efficiently glycoslyated and produced a mixture of glycosylated and nonglycosylated protein (Fig. 3E, lane 4). Hence, some of the dimers formed by PLP CHO2 had an intermediate molecular weight (Fig. 3E, lane 4, asterisk), being smaller than dimers of glycosylated PLP CHO1 (Fig. 3E, lane 3), but larger than dimers of nonglycosylated PLP (Fig. 3E, lane 2). Thus, we conclude that these SDS-resistant complexes are homodimers of PLP composed of one glycosylated and one nonglycosylated polypeptide.
Formation of Stable Oligomers Is Correlated with Disease. Allofthe mutant proteins we examined are associated with PMD, and all form SDS-resistant oligomers in the ER. Three of these mutations are associated with severe PMD, and three with milder forms of the disease (Table 1). Enhanced chemiluminescent Western blotting suggested that the mutant proteins might differ in their ability to form stable oligomers (Fig. 3C). To provide a more quantitative assessment, we performed Western blotting with [125I]protein A. This analysis revealed that >80% of each severe PMD-associated mutant protein was present in SDS-resistant dimers (mean 82.2 ± 1.3%, Fig. 3F). Intriguingly, mutants associated with milder versions of the disease formed these stable oligomers significantly less efficiently [Sig (2-tailed) 0.009], with ≈60% of each mutant protein migrating as a dimer on SDS/PAGE (mean 57.7 ± 3.2%, Fig. 3F). These results strongly suggest that there may be a link between the formation of stable oligomers of mutant PLP and disease phenotype. To explore this issue further, we constructed a truncated version of PLP mutant 5 (Table 1) lacking the fourth TM domain and most of the large extracellular loop (TM1-3ha). Transgenic mice expressing a slightly shorter polypeptide (lacking TM4, all of the large extracellular loop, and half of TM3), PLPneo, have a very mild phenotype and do not exhibit symptoms of PMD (21, 22). Like full-length PLP mutants, TM1-3ha was localized to the ER when expressed in COS-7 cells (Fig. 3G). However, in marked contrast to the full-length mutant PLP, this truncated mutant did not form SDS-resistant oligomers to any significant degree (Fig. 3H, compare lanes 2 and 3). This is an important finding that provides further evidence that the formation of stable PLP oligomers in the ER, rather than ER retention, per se, is related to the development of PMD.
SDS-Resistant Oligomers of Mutant PLP Resemble Properly Folded Oligomers. These results suggest that disease-linked mutations of PLP are associated with the accelerated formation of SDS-resistant oligomers that resemble mature, native PLP. However, an alternative explanation is that these mutants are misfolded and/or aggregated, and coincidentally exhibit similar properties to the WT protein when analyzed under the denaturing conditions of SDS/PAGE. To exclude this possibility, we characterized in detail the nature of WT and mutant PLP under native conditions.
Detergent solubilized extracts of cells expressing WT PLPmh (for 96 h) or msd PLPmh (for 24 h) were separated on a Superdex 200 gel filtration column, and fractions were analyzed by SDS/PAGE and Western blotting. The SDS-resistant dimers formed by properly folded WT PLPmh eluted predominantly in fractions 9 and 10, whereas the protein that migrated as a monomer on SDS/PAGE had a slightly lower apparent molecular mass and eluted in fractions 10 and 11 (Fig. 4A). Although mutant PLP had a more complex elution profile (Fig. 4B), the bulk of the protein was present in the form of SDS-resistant dimers that were again concentrated in fractions 10 and 11 (Fig. 4B; see also Fig. 6A, which is published as supporting information on the PNAS web site). This important finding shows that mutant PLP does not aggregate, and demonstrates that the large majority of mutant PLP oligomers are similar in size to those formed by the properly folded WT protein, even when analyzed under native conditions. Consistent with this finding, virtually all msd PLPmh was solubilized in a range of nondenaturing detergents (Fig. 6B). Interestingly, a significant peak of larger SDS-resistant dimers and monomers of msd PLPmh eluted in fraction 5 (Figs. 4B and 6A). This peak represented a small fraction (≈10%) of the total PLP and may correspond to protein with a distinct conformation. Blue native gel electrophoresis (Fig. 4C) further confirmed that msd PLPmh present in fractions 9 and 10 was in similar complexes to the WT protein (Fig. 2A), and very little material was in higher molecular weight aggregates.
Fig. 4.
Stable oligomers are not due to prolonged ER retention or aberrant disulfide bonds. (A and B) Stable cell lines were induced to express WT or msd PLPmh for 96 or 24 h, respectively. Lysates were separated on a Superdex 200 column, and fractions were analyzed by SDS/PAGE and Western blotting with α-myc. The amount of monomer and dimer was quantified by densitometry and is expressed as a percentage of the total amount of monomer/dimer. The chart shows the average of three to four independent experiments. The position of three soluble molecular weight markers is indicated. (C) Lysates of cells expressing msd PLPmh were separated as in B, and fractions 9 and 10 were analyzed by blue native gel electrophoresis and Western blotting with α-myc. (D) Lysates of cells expressing msd PLPmh were separated as in B, and fractions 4–7 and 8–11 were pooled and immunoprecipitated with α-myc or control (ctl) antibody. Immunoprecipitated material was analyzed by Western blotting with α-calnexin. Lanes 5 and 6 show one-thirtieth of the total lysate used for IP. (E) Stable cell lines were induced to express WT or msd PLPmh overnight in the presence of 5 μg/ml brefeldin A, and lysates were analyzed by SDS/PAGE and Western blotting with α-myc. (F) COS-7 cells were transfected with WT or msd PLPmh, or with WT or msd PLPmh in which the ER luminal cysteine residues were replaced with glycine (ΔCys). Lysates were analyzed as in A.
One characteristic of misfolded proteins is that they associate strongly with components of the ER quality control machinery (17). We have previously shown that some of these components, including calnexin, bind stably to mutant PLP but not the WT protein (16). An explanation of these findings that would be consistent with our gel filtration data is that calnexin binds predominantly to the higher molecular weight pool of msd PLP, which corresponds to nonnative protein, but binds less well to the more substantial pool of msd PLP that is similar in size to the WT protein. To test this idea, lysates of cells expressing msd PLPmh were separated by gel filtration. Fractions 4–7 and 8–11 were pooled and immunoprecipitated with anti-myc or control antibodies, and the immunoisolated material was analyzed by Western blotting with anti-calnexin antibodies. As anticipated, very little calnexin was coimmunoprecipitated with msd PLPmh in fractions 8–11 (Fig. 4D, lane 4), even though these fractions contained the vast majority of PLP (Figs. 4B and 6A). In marked contrast, a significant amount of calnexin was coimmunoprecipitated with the minor population of PLP in fractions 4–7 (Fig. 4D, lane 2). This result demonstrates that the majority of mutant PLP oligomers do not bind stably to calnexin, supporting our hypothesis that they adopt a conformation that resembles the properly folded protein. In contrast, the stable oligomers that elute in fraction 5 have a distinct “nonnative” conformation that is recognized by calnexin. The small amount of calnexin associated with the lower molecular weight fractions probably represents a failure to completely resolve the two pools of PLP by gel filtration chromatography, although we cannot rule out a weak interaction between calnexin and the lower molecular weight pool of msd PLP.
Formation of SDS-Resistant Oligomers Is Not Due to Prolonged ER Retention or Aberrant Intermolecular Disulfide Bonds. Because PLP mutants are retained in the ER, it was important to establish whether the formation of stable oligomers was simply a consequence of prolonged exposure to the lumen of the ER, which is both an oxidizing environment and contains a variety of molecular chaperones that could promote oligomerization (17). To do this, we artificially retained WT PLPmh in the ER by adding brefeldin A to cells during the induction of protein expression. Although this treatment caused all of the WT PLPmh to be retained in the ER, as judged by immunofluorescence microscopy (data not shown), it did not result in the formation of SDS-resistant oligomers (Fig. 4E, lane 1). Under the same conditions, a large proportion of msd PLPmh had formed SDS-resistant oligomers (Fig. 4E, lane 2).
The large extracellular/ER luminal loop of PLP contains four cysteine residues thought to form intramolecular disulfide bonds in the mature protein (Fig. 1B). Misfolded proteins have a tendency to form disulfide bonded aggregates (ref. 23 and references therein). This is not the case for mutant PLP, which has the same mobility under reducing and nonreducing conditions (Fig. 6C). However, some misfolded proteins form aberrant intermolecular disulfide bonds that are stable during reducing SDS/PAGE (23). Therefore, it was possible that the stable oligomers we detected resulted from such reduction-resistant disulfide bonds. To address this issue, we replaced all four luminally oriented cysteines with glycine (PLPmh ΔCys). As shown in Fig. 4F, msd PLPmh ΔCys forms SDS-resistant oligomers with a similar efficiency to msd PLPmh (compare lanes 2 and 4), demonstrating that these stable oligomers are not an aberrant disulfide-linked species. Interestingly, we also found that WT PLPmh ΔCys was retained in the ER (data not shown) and efficiently formed SDS-resistant oligomers (Fig. 4F, lane 3), confirming that these residues are essential for the polypeptide to follow an appropriate folding pathway.
Discussion
Several lines of evidence suggest that PMD is due to a toxic “gain of function” associated with mutations in the PLP gene rather than a loss of functional protein. For example, PLP knockout mice have a very mild phenotype (5), whereas increased expression and many missense mutations are associated with severe forms of PMD (24, 25). The underlying pathogenic mechanism of the missense mutations is thought to be the accumulation of mutant PLP, which is harmful to oligodendrocytes and causes programmed cell death (15). Eukaryotic cells have evolved an efficient quality control system that functions to eliminate unwanted proteins and maintain ER homeostasis (17). In the face of this constitutive activity, it is unclear why the synthesis of mutant PLP is so damaging to oligodendrocytes. One explanation might be that the rate of mutant protein synthesis is sufficiently rapid to overwhelm the ER degradation machinery, allowing large amounts of mutant PLP to accumulate and impair cell function. The inefficient degradation of msd PLP compared with other misfolded proteins (16), would exacerbate such a buildup of mutant protein. However, an intriguing alternative possibility is that a specific property or function of mutant PLP is responsible for its toxicity. Two recent observations indirectly support this notion. First, the expression of a mutant form of PLP, but not a control membrane protein, has been shown to cause a severe reduction in the survival of immortalized oligodendrocytes (26). Second, as noted by Simons et al. (27), overexpression of PLP in oligodendrocyte precursors has a more negative effect on cell viability than overexpression of myelin oligodendrocyte glycoprotein, another myelin protein synthesized at the ER.
Our results demonstrate that mutant forms of PLP rapidly assemble into stable oligomers in the ER of transfected cells. These oligomers adopt a “native” conformation that resembles that of WT PLP as judged by several criteria: size under both denaturing and nondenaturing conditions; stability in SDS; and, most significantly, a lack of interaction with the ER quality control machinery. The extent to which this conformation precisely matches that of mature PLP must await high-resolution structural studies, and, of course, ER-associated PLP may not be able to participate in all of the heterooligomeric interactions that normally occur at the cell surface. Nevertheless, our data strongly indicate that at least the majority of mutant PLP is not retained in the ER simply because it is aggregated and misfolded. It is not yet clear what mediates the ER retention of mutant PLP in the absence of an interaction with the quality control machinery. One attractive possibility is that the rapid formation of stable oligomers masks a signal required for ER exit.
In contrast to mutant PLP, WT PLP is transported to the cell surface and forms stable oligomers much more slowly. We speculate that this slow maturation may provide a mechanism to ensure that PLP achieves its native conformation only after it has reached the cell surface where its function is required. In light of its proposed roles as an adhesion molecule (5, 6), ion channel (3, 4), and/or signaling molecule (28), it is easy to envisage that the inappropriate formation of mature PLP oligomers in the ER of oligodendrocytes could have a deleterious effect on cell viability. We have observed that vesicular stomatitis virus glycoprotein (VSVG) is still transported to the surface of COS-7 cells expressing mutant PLP (unpublished data), indicating that the synthesis of mutant PLP does not cause a complete block in traffic from the ER. However, myelinating cells are thought to be particularly sensitive to perturbations in the secretory pathway (29), and therefore, in the context of the myelinating oligodendrocyte, more subtle changes in ER structure or function may well prove catastrophic.
Our results provide evidence that there is a link between the formation of stable PLP oligomers in the ER and the development of PMD. We find that a truncated version of PLP, similar to the PLPneo polypeptide, which does not cause PMD, does not form stable oligomers, despite being retained in the ER. In contrast, six different PLP mutants that are associated with disease rapidly form stable oligomers. Furthermore, we show that PLP mutants associated with severe PMD have a greater tendency to form stable oligomers than mutants associated with mild forms of the disease. Together, these data provide strong evidence that the toxicity of mutant PLP is closely related to the inappropriate formation of stable oligomers in the oligodendrocyte ER and is not simply a result of retention of misfolded protein.
Supplementary Material
Acknowledgments
We thank Maylis Vergnolle for assistance with blue native gels, Sandra Bunning for technical assistance, and Martin Pool for helpful discussions. This work was supported by the Biotechnology and Biological Sciences Research Council and by cooperative group funding from the Medical Research Council (Grant G9722026). S.H. is a Biotechnology and Biological Sciences Research Council Professorial Fellow.
Author contributions: E.S., S.H., and P.W. designed research; E.S. and A.H. performed research; E.S., S.H., and P.W. analyzed data; and E.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, endoplasmic reticulum; PMD, Pelizaeus–Merzbacher disease; DPDPB, 1,4-di-(3′-[2′pyridyldithio]-propionamido) butane; TM, transmembrane; PLP, proteolipid protein; PLPmh, myc his-tagged PLP; PLPha, hemagglutinin-tagged PLP.
References
- 1.Readhead, C., Popko, B., Takahashi, N., Shine, H. D., Saavedra, R. A., Sidman, R. L. & Hood, L. (1987) Cell 48, 703–712. [DOI] [PubMed] [Google Scholar]
- 2.Lees, M. & Brostoff, S. L. (1984) Proteins of Myelin (Plenum, New York).
- 3.Kitagawa, K., Sinoway, M. P., Yang, C., Gould, R. M. & Colman, D. R. (1993) Neuron 11, 433–448. [DOI] [PubMed] [Google Scholar]
- 4.Diaz, R. S., Monreal, J. & Lucas, M. (1990) J. Neurochem. 55, 1304–1309. [DOI] [PubMed] [Google Scholar]
- 5.Klugmann, M., Schwab, M. H., Puhlhofer, A., Schneider, A., Zimmermann, F., Griffiths, I. R. & Nave, K. A. (1997) Neuron 18, 59–70. [DOI] [PubMed] [Google Scholar]
- 6.Bizzozero, O. A., Bixler, H. A., Davis, J. D., Espinosa, A. & Messier, A. M. (2001) J. Neurochem. 76, 1129–1141. [DOI] [PubMed] [Google Scholar]
- 7.Smith, R., Cook, J. & Dickens, P. A. (1984) J. Neurochem. 42, 306–313. [DOI] [PubMed] [Google Scholar]
- 8.Brophy, P. J., Horvath, L. I. & Marsh, D. (1984) Biochemistry 23, 860–865. [DOI] [PubMed] [Google Scholar]
- 9.Sinoway, M. P., Kitagawa, K., Timsit, S., Hashim, G. A. & Colman, D. R. (1994) J. Neurosci. Res. 37, 551–562. [DOI] [PubMed] [Google Scholar]
- 10.Yool, D. A., Edgar, J. M., Montague, P. & Malcolm, S. (2000) Hum. Mol. Genet. 9, 987–992. [DOI] [PubMed] [Google Scholar]
- 11.Gow, A., Friedrich, V. L., Jr., & Lazzarini, R. A. (1994) J. Neurosci. Res. 37, 574–583. [DOI] [PubMed] [Google Scholar]
- 12.Gow, A. & Lazzarini, R. A. (1996) Nat. Genet. 13, 422–428. [DOI] [PubMed] [Google Scholar]
- 13.Gow, A., Gragerov, A., Gard, A., Colman, D. R. & Lazzarini, R. A. (1997) J. Neurosci. 17, 181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung, M., Sommer, I., Schachner, M. & Nave, K. A. (1996) J. Neurosci. 16, 7920–7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gow, A., Southwood, C. M. & Lazzarini, R. A. (1998) J. Cell Biol. 140, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swanton, E., High, S. & Woodman, P. (2003) EMBO J. 22, 2948–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellgaard, L. & Helenius, A. (2003) Nat. Rev. Mol. Cell Biol. 4, 181–191. [DOI] [PubMed] [Google Scholar]
- 18.Swanton, E., Sheehan, J., Bishop, N., High, S. & Woodman, P. (1998) Mol. Biol. Cell 9, 1633–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schagger, H. & von Jagow, G. (1991) Anal. Biochem. 199, 223–231. [DOI] [PubMed] [Google Scholar]
- 20.Colman, D., Kreibich, G., Frey, A. & Sabatini, D. (1982) J. Cell Biol. 95, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boison, D., Bussow, H., D`Urso, D., Muller, H. W. & Stoffel, W. (1995) J. Neurosci. 15, 5502–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boison, D. & Stoffel, W. (1994) Proc. Natl. Acad. Sci. USA 91, 11709–11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cannon, K. S., Hebert, D. N. & Helenius, A. (1996) J. Biol. Chem. 271, 14280–14284. [DOI] [PubMed] [Google Scholar]
- 24.Readhead, C., Schneider, A., Griffiths, I. & Nave, K. A. (1994) Neuron 12, 583–595. [DOI] [PubMed] [Google Scholar]
- 25.Hodes, M. E., Pratt, V. M. & Dlouhy, S. R. (1993) Dev. Neurosci. 15, 383–394. [DOI] [PubMed] [Google Scholar]
- 26.Bongarzone, E. R., Jacobs, E., Schonmann, V., Kampf, K., Campagnoni, C. W. & Campagnoni, A. T. (2001) J. Neurosci. Res. 65, 485–492. [DOI] [PubMed] [Google Scholar]
- 27.Simons, M., Kramer, E. M., Macchi, P., Rathke-Hartlieb, S., Trotter, J., Nave, K. A. & Schulz, J. B. (2002) J. Cell Biol. 157, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gudz, T. I., Schneider, T. E., Haas, T. A. & Macklin, W. B. (2002) J. Neurosci. 22, 7398–7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baerwald, K. D., Corbin, J. G. & Popko, B. (2000) J. Neurosci. Res. 59, 160–169. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.