Abstract
We report the main characteristics of ‘Olsenella congonensis’ strain Marseille-P3359T (CSUR P3359), which was isolated from the stool sample of a healthy 47-year-old pygmy female.
Keywords: Culturomics, Emerging bacteria, Gut microbiota, Human microbiota, ‘Olsenella congonensis’
The approval of the ethics committee of the Institut Fédératif de Recherche was obtained under the number 09-022 before sample collection from Congo and project start up. Samples collection and analysis were performed with the aim of describing the human gut microbiota using the culturomics approach [1].
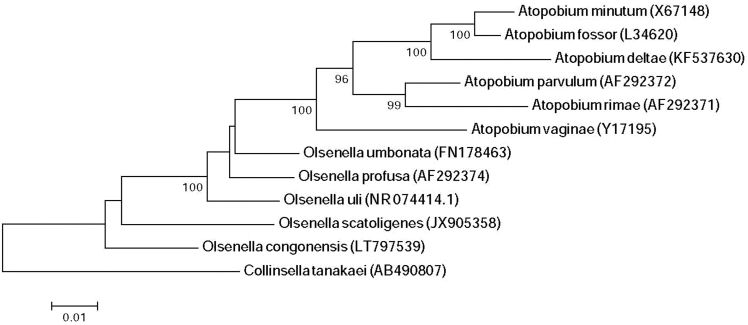
As a first step, stool sample (1 g) was diluted with 1 mL of phosphate buffered saline then incubated in an anaerobic culture bottle, supplemented with 5% filtered rumen and 5% sheep blood, at 37°C for a 30-day follow up. At day 15 a ‘Olsenella congonensis’ colony was purely isolated on 5% blood-enriched Columbia agar (bioMérieux, Marcy l'Etoile, France). The first method used for the identification of Strain Marseille-P3359 was matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) using a Microflex spectrometer (Bruker Daltonics, Leipzig, Germany) [2]. However, identification failed and 16S rRNA gene sequencing was performed, as previously reported, for further analysis using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France) with fD1-rP2 primers (Eurogentec, Seraing, Belgium) [3]. Strain Marseille-P3359 exhibited a 95% sequence identity with Olsenella scatoligenes, the phylogenetically closest species with standing nomenclature (Fig. 1). Hence, it can be classified as a new species within the genus Olsenella [4]. When comparing the 16S rRNA gene sequences of the different closely related species of Olsenella, we found a range of similarity between 93% and 97%. Hence, we propose that strain Marseille-P3359 is a new species of the genus Olsenella.
Fig. 1.
Phylogenetic tree showing the branching of ‘Olsenella congonensis’ strain Marseille-P3359 among the other phylogenetically close neighbours. ClustalW and Mega software were used for alignment and phylogenetic tree generation, respectively. The neighbour joining method was adapted with 500 bootstraps and only scores of at least 90% were kept on the nodes. The scale bar indicates a 1% nucleotide sequence divergence.
Description of the new species ‘Olsenella congonensis’
Colonies of the strain Marseille-P3359T were smooth with a mean diameter of 0.8–1 mm. Bacterial cells were Gram-positive bacilli, catalase and oxidase negative with a mean length of 1.08 μm. We propose the discovery of the new species ‘Olsenella congonensis’ (co.ng.on.en'sis. N.L. masc. adj. congonensis to refer to Congo, from where the stool's donor came). Strain Marseille-P3359T is the type strain of the new species ‘Olsenella congonensis’.
MALDI-TOF MS spectrum accession number
The MALDI-TOF MS spectrum of ‘O. congonensis’ is available online (http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database).
Nucleotide sequence accession number
The 16S rRNA gene sequence was deposited in GenBank under Accession number LT797539.
Deposit in a culture collection
Strain Marseille-P3359 was deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under number P3359.
Conflicts of interest
None to declare.
Funding sources
This work was funded by Mediterrannée Infection Foundation.
References
- 1.Lagier J.-C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.-C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakamoto M., Benno Y. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int J Syst Evol Microbiol. 2006;56:1599–1605. doi: 10.1099/ijs.0.64192-0. [DOI] [PubMed] [Google Scholar]