Abstract
Faithful chromosome segregation is essential for the maintenance of genetic stability during cell division and it is at least partly monitored by the spindle checkpoint, a surveillance mechanism preventing the cell from prematurely entering anaphase. The adenomatous polyposis coli (Apc) gene also plays an important role in regulating genomic stability, as mutations of Apc cause aneuploidy. Here we show that whereas ApcMin/+ mice developed many adenomatous polyps, mostly in the small intestine, by 3 mo of age; BubR1+/–ApcMin/+ compound mutant mice developed 10 times more colonic tumors than ApcMin/+ mice. The colonic tumors in BubR1+/–ApcMin/+ mice were in higher grades than those observed in ApcMin/+ mice. Consistently, BubR1+/–ApcMin/+ murine embryonic fibroblasts (MEFs) contained more β-catenin and proliferated at a faster rate than WT or BubR1+/– MEFs. Moreover, BubR1+/–ApcMin/+ MEFs slipped through mitosis in the presence of nocodazole and exhibited a higher rate of genomic instability than that of WT or BubR1+/– or ApcMin/+ MEFs, accompanied by premature separation of sister chromatids. Together, our studies suggest that BubR1 and Apc functionally interact in regulating metaphase–anaphase transition, deregulation of which may play a key role in genomic instability and development and progression of colorectal cancer.
Keywords: polyposis, genetic instability, colon cancer, spindle checkpoint, mitosis
Genetic instability is an integral component of human neoplasia. High-fidelity DNA replication and faithful chromosome segregation are fundamental processes that allow cells to transmit their genetic information to progeny. Failures in maintaining genetic stability inevitably cause either cell death or abnormal phenotypes such as malignancy. Missegregation of chromosomes may result from various causes, including defects of spindle checkpoint components and abnormal centrosomal duplication. In fact, a significant fraction of cancer cells that exhibit aneuploidy either harbor defects in cell cycle checkpoint pathways (1, 2) or contain an abnormal centrosome number (3).
Aneuploidy occurs frequently in colorectal cancer. Colorectal tumors exhibit a defect in chromosome segregation, leading to frequent gains or losses of chromosomes (>102 per chromosome per generation) (4). Chromosome instability has been detected in the smallest adenoma, suggesting that chromosome instability may occur at very early stages of colorectal carcinogenesis (5). Extensive research during the past has led to the identification of genes that play a major role in the development of colorectal cancer. For example, mutations or deletions of the adenomatous polyposis coli (Apc) gene, encoding a 310-kDa cytoplasmic protein (6, 7), are commonly found in inherited familial adenomatous polyposis patients and in sporadic colorectal cancers (8, 9). Such mutations appear to be an early event during colorectal tumorigenesis (10). Mouse models have been developed to study the role of Apc mutations (truncated Apc) in affecting cell proliferation or loss of Apc as a contributing factor in formation of polyps (11, 12). The importance of the Apc gene in intestinal cancer development by regulating the stability of β-catenin has been well established by efforts of many laboratories (13, 14). However, Apc mutations are detected in prenoeplastic lesions, the earliest stages of polyp formation, 7–15 years ahead of the malignant tumor formation (15). It is therefore likely that loss of Apc at the early stage promotes tumor formation at least in part by mechanism(s) independent of the proliferation pathway mediated by β-catenin. Recent studies suggest that a loss of WT Apc correlates with chromosome instability (16, 17), a hallmark of colon polyposis. C-terminal Apc truncations similar to those found in colon tumors correlate with chromosome instability in mouse embryonic cells (17). Although the molecular mechanism by which Apc regulates chromosomal segregation remains to be established, the existing evidence that Apc associates with spindle checkpoint components and localizes to kinetochores and centrosomes (16, 18) strongly suggests a role of this protein in controlling metaphase–anaphase transition.
The spindle checkpoint plays an essential role in the maintenance of chromosomal segregation (19). Haploinsufficiency of many spindle checkpoint components results in enhanced genomic instability and tumor formation (20–22). A couple of recent studies show that a compromised BubR1 activity due to specific germ-line mutations causes aneuploidy, infertility, and/or early onset of malignancies (23) and strongly suggest that spindle checkpoint failure is an underlying cause for the development of certain diseases. In this paper, we demonstrate that BubR1+/–ApcMin/+ (BubR1 is also termed Bub1B) compound mutant mice developed colonic tumors at an accelerated rate and that this enhanced tumorigenecity was associated with premature separation of sister chromatids and greatly elevated chromosomal instability.
Materials and Methods
Generation of ApcMin/+BubR1+/– Mice. We used our previously developed BubR1+/– mice (21) and ApcMin/+ mice purchased from The Jackson Laboratory to generate ApcMin/+BubR1+/– genotype mice. All of the mice were housed in a pathogen-free barrier environment for the duration of the study. We intercrossed female BubR1+/– with male ApcMin/+ to obtain mouse progenies having genotypes of BubR1+/+Apc+/+, BubR1+/+ ApcMin/+, BubR1+/–Apc+/+, and BubR1+/–ApcMin/+. BubR1 genotyping of each animal was carried by PCR according to previously published procedures (25). Apc genotyping was carried out by using primers obtained from The Jackson Laboratory.
Mouse Intestinal Tumor Analysis. Age-matched mice with genotypes of BubR1+/+Apc+/+, BubR1+/+ApcMin/+, BubR1+/–Apc+/+, or BubR1+/–ApcMin/+ were selected to assess the small and large intestinal tumorigenesis patterns. In brief, at 5 wk of age, WT mice or mice deficient in either Apc or BubR1 or both were fed semipurified AIN-76A diet. At 12 wk of age, all of the mice were killed by CO2, and their intestines were removed and flushed with Krebs–Ringer solution. Intestines were opened and analyzed for number, location, and size of tumors with the help of a dissecting microscope. Small intestinal and colonic tumors were further histologically analyzed by the hematoxylin/eosin staining procedure.
Maintenance and Treatment of Murine Embryonic Fibroblasts (MEFs). MEFs were derived from embryonic day 14.5 fetuses produced from intercrosses of BubR1+/– and ApcMin/+ mice as described (25). Four different genotypes of MEFs (namely, WT, BubR1+/–, ApcMin/+, and BubR1+/–ApcMin/+) were obtained. MEFs were maintained at 37°C with 5% CO2 in DMEM supplemented with 10% FBS, 100 μg/ml penicillin, 50 μg/ml streptomycin sulfate, and 0.2 mM 2-mercaptoethanol. MEFs were also treated with nocodazole (0.4 μg/ml) for various times.
Western Blot Analysis. Total proteins were prepared from MEFs treated with vehicle or with various agents as above. Equal amounts of total proteins were analyzed by SDS/PAGE. Proteins fractionated on the denaturing gels were transferred to nitrocellulose membranes, which were blotted with antibodies to Apc (Abcam, Cambridge, MA), BubR1, β-catenin (Cell Signaling Technology, Beverly, MA), and β-actin (Sigma). Immunoreactive bands were detected with an appropriate second antibody and visualized with a chemiluminescence kit (Pierce).
Fluorescence Microscopy and Immunohistochemistry. Immunostaining of MEFs was carried out as described (21). In brief, cells fixed in 4% paraformaldehyde were treated with 0.1% Triton X-100 on ice and then washed three times with ice-cold PBS. After blocking with 2.0% BSA in PBS for 15 min on ice, cells were stained with DAPI (1 μg/ml, Fluka). Fluorescence microscopy was performed on a Nikon microscope, and images were captured by using a digital camera (Optronics International, Chelmsford, MA). For immunohistochemistry, sections from formalin-fixed, paraffin-embedded mouse colon tissues were prepared and incubated with 0.3% hydrogen peroxide for 10 min and then stained with the primary antibody to proliferating cell nuclear antigen, cyclin D1, Apc, or BubR1 for 1 h. After incubation with an appropriate second antibody, the sections were washed and incubated with a preformed avidin–biotinylated enzyme complex for 30 min by using an ABC kit (Vector Laboratories) followed by color development with diaminobenzidine tetrahydrochloride. The stained colon sections were counterstained with hematoxylin. For TUNEL assays, formalin-fixed, paraffin-embedded sections from the intestines of various genotypes were prepared. In situ end labeling (or TUNEL) was performed by using an apoptosis assay kit (Oncogene Science) according to the manufacturer's instructions. For TUNEL assays, sections from lymph nodes of WT mice were used as positive controls, whereas sections incubated with nonimmune sera were used as negative controls. Cells undergoing apoptosis manifested as in situ end labeling were examined under a microscope. Three slides per group were reviewed.
Cytogenetics. MEFs seeded for 24 h were treated with Colcemid (0.04 μg/ml) for 3 h to arrest cells in metaphase. Cells detached from the culture plates with trypsin/EDTA were incubated in 75 mM KCl for 20 min at 37°C. These cells were then fixed in three changes of methanol/acetic acid (3:1), and the fixed cell pellets were used for slide spread. Slides were air dried for at least 2 d at 37°C before examination. For MEFs of each genotype, at least 50 metaphase spreads were examined.
Cell Viability Assay. Cell viability was assayed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) method. MEFs (2 × 104 cells per well) were seeded on 96-well plate in triplicates. After a 24-h culture at 37°C, the culture medium was aspirated and a fresh medium was added to a final volume of 200 μl. MTT (20 μl, Sigma, 2 mg/ml in PBS) was then added to each well. After the cells were incubated at 37°C for 4 h, the medium was removed. MTT formazan precipitates were dissolved in 100 μl of DMSO with mechanical shaking for 10 min. The absorbance of dissolved samples was measured at 570 nm by using a plate reader.
Results
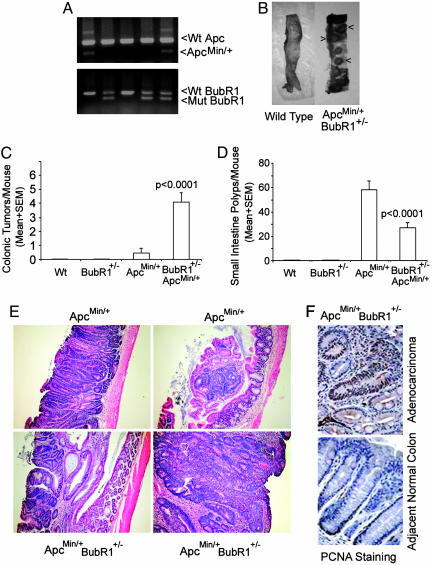
To determine whether BubR1 is directly involved in colon carcinogenesis, we introduced the BubR1 mutation into ApcMin/+ mice through cross-breeding BubR1+/– mice with ApcMin/+ mice. The genotypes of offspring were determined through PCR as described (25), and typical results of genotyping are shown in Fig. 1A. As expected, no mice with a homozygous mutation of either BubR1 or Apc were obtained. The detected frequency of newborn mice heterozygous for both BubR1 and Apc was 13%, that is approximately one-half of the percentage predicted by the Mendelian segregation rule, indicating a certain extent of embryonic lethality. Because ApcMin/+ mice typically develop numerous adenomas in the small intestine by 3 mo of age, we asked whether BubR1+/–ApcMin/+ mice would be more susceptible to tumor development, especially in the large intestine. An examination of intestines obtained from mice of various genetic backgrounds revealed that WT or BubR1+/– mice developed no visible polyps, whereas some ApcMin/+ mice sporadically developed one or two visible polyps (Table 1). On the other hand, age-matched BubR1+/–ApcMin/+ compound mutant mice developed many tumor masses in the large intestine (Fig. 1B). In fact, the average number (4.1 ± 1.7) of colonic tumor masses in BubR1+/–ApcMin/+ compound mutant mice was ≈10 times of that (0.4 ± 0.6) in ApcMin/+ mice (Table 1 and Fig. 1C). Interestingly, there were significantly fewer polyps in small intestines of BubR1+/–ApcMin/+ mice than in ApcMin/+ mice (Fig. 1D). Histological analysis revealed that tumors from ApcMin/+ mice were either well defined tubular adenomas or tubular adenomas within the lamina propria (Fig. 1E Upper). On the other hand, tumor masses from BubR1+/–ApcMin/+ colons were adenocarcinomas that were either poorly or moderately differentiated with cribriform tumor glands, stratified epithelial cells, and necrotic tissues (Fig. 1E Lower), indicating that colonic tumors from BubR1+/–ApcMin/+ compound mutant mice had progressed to much higher grades than those from ApcMin/+ mice. Immunohistochemical analysis revealed that tumor cells from BubR1+/–ApcMin/+ colons uniformly exhibited strong nuclear staining for proliferating cell nuclear antigen, whereas normal colonic epithelia showed proliferating cell nuclear antigen staining primarily in stem cells of basal crypts (Fig. 1F Upper), suggesting that mutations in both BubR1 and Apc genes confer the growth advantage for colonic tumors.
Fig. 1.
Haploinsufficiency of BubR1 and Apc results in an increased formation of colonic tumor polyps. (A) Genomic DNA samples isolated from mouse tails were subjected to genotyping by PCR using primers that detect both WT (Wt) and mutant alleles of Apc or BubR1. (B) Dissection micrographs of representative colons from WT and BubR1+/–ApcMin/+ mice at 3 mo of age. Arrows denote the colonic tumor masses. (C and D) Mice of various genotypes at 3 mo of age were killed. Intestines from each mouse were examined under a dissection microscope for tumor polyps. Average numbers of tumor masses in colon (C) and the small intestine (D) from WT, BubR1+/–, ApcMin/+, and BubR1+/–ApcMin/+ mice are shown. No visible intestinal tumor masses were detected in WT or BubR1+/– mice. (E) Hematoxylin/eosin-stained sections of tumor masses from ApcMin/+ (Upper) and BubR1+/–ApcMin/+ (Lower) mice. (×400.) (F) Sections of colonic tumors from BubR1+/–ApcMin/+ mice were subjected to immunohistochemical studies after staining with IgGs to proliferating cell nuclear antigen. (×400.) Typical malignant tissues (Upper) and normal colon tissues adjacent to the adenocarcinomas (Lower) are presented.
Table 1. Colonic tumor numbers in individual mice of various genotypes.
| Genotype | No. of tumors in individual mice | Mean ± SEM | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BubR1+/- | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ApcMin/+ | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0.4 ± 0.6 |
| BubR1+/- ApcMin/+* | 7 | 4 | 6 | 3 | 3 | 4 | 2 | 2 | 5 | 5 | 4.1 ± 1.7 | ||||||||
There were fewer mice in the BubR1+/- ApcMin/+ compound mutant group because of a limited availability of age-matched mice as a result of embryonic lethality.
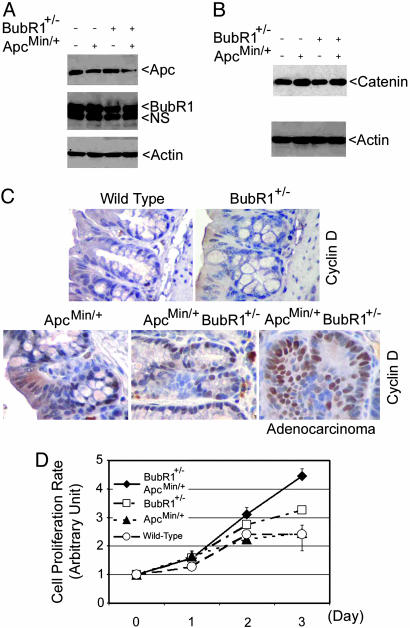
To examine levels of BubR1 and Apc expression in mutant mice, we derived murine fibroblasts from embryos with various genetic backgrounds. As expected, both ApcMin/+ and BubR1+/–ApcMin/+ MEFs expressed ≈50% less Apc protein than WT or BubR1+/– MEFs; similarly, BubR1+/– and BubR1+/–ApcMin/+ cells contained at least 50% less BubR1 protein (Fig. 2A). Because the stability of β-catenin, a key component in the Wnt pathway, is regulated by the Apc complex, we examined β-catenin levels in various MEFs. Immunoblot analysis revealed that MEFs with an Apc mutation contained more β-catenin (Fig. 2B), thus in agreement with the role of Apc in negative regulation of this molecule. Moreover, because cyclin D1 is a target of β-catenin that was elevated in Apc-deficient cells, we analyzed cyclin D1 levels in colonic tissues of various genetic backgrounds by immunocytochemistry. We observed that whereas no significant cyclin D1 staining was seen in normal colonic epithelia from WT or BubR1+/– mice, strong cyclin D1 staining was detected in basal crypts of colonic epithelia from BubR1+/– ApcMin/+ colons as well as ApcMin/+ colons (Fig. 2C). Furthermore, a high level of cyclin D1 was present in colonic adenocarcinomas from BubR1+/–ApcMin/+ compound mutant mice (Fig. 2C). Consistently, cell proliferation assays showed that BubR1+/–ApcMin/+ cells grew at an accelerated rate compared with WT MEFs or MEFs with a single gene mutation (Fig. 2D).
Fig. 2.
BubR1+/–ApcMin/+ MEFs proliferate at an accelerated rate. (A) Paired MEFs were lysed and an equal amount of cell lysates were blotted for Apc, BubR1, or β-actin. Arrow NS denotes a nonspecific cross-reactive band. (B) Equal amount of cell lysates from MEFs of various genotypes were blotted for β-catenin and β-actin. (C) Sections of normal colons from mice of various genotypes were subjected to immunohistochemical studies after staining with IgGs to cyclin D1. A typical adenocarcinoma section from BubR1+/–ApcMin/+ mice that was stained with cyclin D1 is also presented. (D) MEFs of various genotypes were subjected to cell proliferation assays using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) method. The data are summarized from three independent experiments.
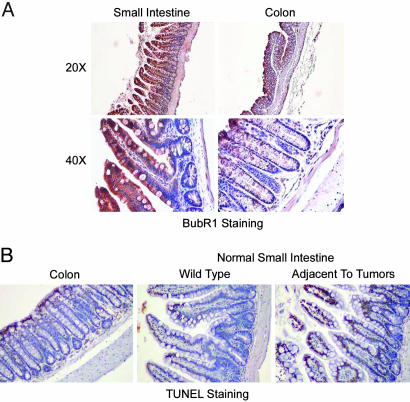
We were rather puzzled about the reduced number of small intestinal tumors in BubR1+/–ApcMin/+ compound mutant mice. One possible reason is a difference of BubR1 expression between small intestine and colon. Immunohistochemistry revealed that although epithelia from both small intestine and colon were stained with BubR1, both the cytoplasm and the nucleus of epithelial cells from the small intestine but not from the colon were strongly stained with BubR1 (Fig. 3A), suggesting that the small intestine may tolerate BubR1 deficiency better than the colon because of a higher level of BubR1 expression. A second possibility is that chromosomal instability that occurred because of BubR1 haploinsufficiency would induce apoptosis at an accelerated rate in the small intestine. Our studies showed that some apoptosis, as revealed by TUNEL staining, occurred in the apical portion of epithelia of both colon and small intestine and that there was no significantly enhanced apoptosis in normal small intestinal tissues compared with those of colon (Fig. 3B). However, we observed that small intestinal tissues adjacent to the tumors exhibited significantly elevated apoptotic signals, suggesting that the enhanced programmed cell death may partly mediate the reduction in the tumor number in mice deficient in BubR1 and Apc.
Fig. 3.
Analysis of BubR1 expression and apoptosis in small intestines by immunohistochemistry. (A) Sections of paraffin-embedded small intestine and colon from WT mice were stained with antibody to BubR1. Representative images at various magnifications are presented. Samples from at least three mice were examined. (B) Sections of paraffin-embedded small intestine and colon samples from BubR1+/–ApcMin/+ mice were examined for apoptosis by using a TUNEL kit. Representative images from three independent samples are presented.
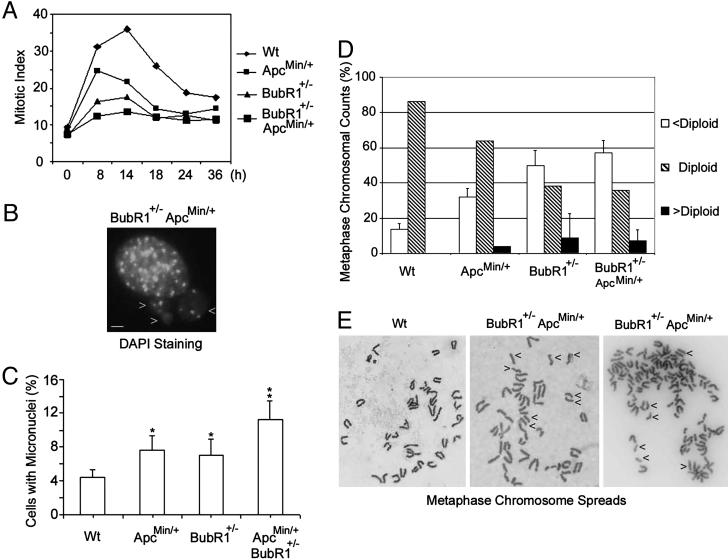
Because the spindle checkpoint failure causes mitotic slippage (21), we next determined the mitotic index in various MEFs treated with nocodazole for various times. We observed that whereas a significant fraction of WT MEFs were arrested at mitosis 14 h after nocodazole treatment, MEFs with either an Apc or a BubR1 mutation significantly compromised the mitotic arrest induced by the spindle poison; consistent with the enhanced proliferation rate as shown in Fig. 2 C and D, MEFs with both Apc and BubR1 mutations exhibited little mitotic arrest in the presence of nocodazole (Fig. 4A). Enhanced mitotic slippage often results in genomic instability (21). Micronuclei analysis revealed that there was an increased rate of micronuclei formation in BubR1+/–ApcMin/+ MEFs (Fig. 4 B and C) compared with WT MEFs. The enhanced genomic instability was also confirmed by chromosomal counts on metaphase spreads (Fig. 4D). BubR1+/–ApcMin/+ MEFs showed a significant increase in the frequency of aneuploid metaphase compared with WT MEFs, which have a very stable karyotype of 40 (diploid); although many BubR1+/– and ApcMin/+ MEFs also exhibited an unstable karyotype, there were fewer BubR1+/– ApcMin/+ MEFs that exhibited a normal karyotype (Fig. 4D). These results are thus consistent with the reported role of BubR1 and Apc in the maintenance of genomic stability (17, 21, 24, 26). Our further analysis revealed that a significant fraction (≈30%) of BubR1+/– ApcMin/+ MEFs contained prematurely separated sister chromatids (Fig. 4E Center and Right). This precocious separation of sister chromatids was less frequently (<10%) observed in BubR1+/– mitotic figures (data not shown) and rarely (<1%) observed in WT (Fig. 4E Left) and ApcMin/+ mitotic figures, suggesting that the spindle checkpoint is severely compromised in BubR1+/–ApcMin/+ compound mutant cells.
Fig. 4.
Enhanced genomic instability in MEFs with compound mutations in BubR1 and Apc. (A) MEFs of various genotypes cultured on chamber slides were treated with nocodazole for the indicated times. At the end of the treatment, cells were fixed, stained with DAPI, and examined for the mitotic index. (B) A typical DAPI-stained cell with micronuclei. Arrows point to the micronuclei. (Bar, 2 μm.) (C) Percentage of micronuclei in MEFs of various genotypes. *, The difference between this group and WT or BubR+/–ApcMin/+ is statistically significant (P < 0.05); **, the difference between this group and the rest of the groups is statistically significant (P < 0.05). (D) The percentage of metaphase chromosomal counts from MEFs of various genotypes. Individual chromosomal counts were divided into three groups, namely, cells with a normal karyotype (diploid), a low chromosomal count (<diploid), and a high chromosomal count (>diploid). (E) Metaphase spreads from Wt MEFs (Left) and BubR1+/–ApcMin/+ MEFs (Center and Right). Arrows point to some separated sister chromatids.
Discussion
Our current studies show that both BubR1 and Apc genes play an important role in suppression of colonic tumorigenesis in mice. In mice with Apc deficiencies, polyps are primarily developed in the small intestine and few colonic polyps are formed in these mice. BubR1+/–ApcMin/+ mice develop many tumors in the colon (Fig. 1), a phenotype similar to that observed in humans with familial adenomatous polyposis. Given that haploinsufficiency of both BubR1 and Apc promotes the formation of micronuclei and aneuploidy, which is associated with premature separation of sister chromatids, it is reasonable to speculate that an accelerated chromosomal instability may be the underlying cause for the dramatic increase in the development of colonic cancer in BubR1+/–ApcMin/+ mice.
It is intriguing that BubR1 mutation is infrequently detected in cancer. One explanation is that BubR1, as well as other spindle checkpoint genes, is pivotal to the maintenance of accurate chromosomal segregation during metaphase–anaphase transition; mutational inactivation of BubR1 would greatly compromise chromosomal stability, which often triggers mitotic catastrophe. In other words, normal cells do not tolerate well severe chromosomal missegregation, a consequence of spindle checkpoint failure. However, certain mutations that do not completely inactivate BubR1 function may survive the selection pressure during early growth. For example, a recent study (23) demonstrated that germ-line mutations of BubR1 exist in a rare recessive condition called mosaic variegated aneuploidy. Patients with this disease are often growth retarded and prone to the development of childhood cancer. This study thus provides a strong link between BubR1 deficiency and cancer predisposition in humans (27), consistent with our observations described in this report.
Although the Apc target(s) in the spindle checkpoint pathway remains obscure, BubR1 is known to be a potent inhibitor of APC/CCdc20. Recent studies show that Apc exhibits a kinetochore localization as well (17). Apc forms complexes with spindle checkpoint proteins Bub1 and Bub3, and its mutated form loses the ability to bind to Bub1 (26). Therefore, Apc may be directly involved in monitoring the metaphase–anaphase transition by regulating the integrity of bipolar spindle and activating the spindle checkpoint. This notion is supported by the observation that Apc physically interacts with microtubules and that this interaction enhanced microtubule stability in vivo and in vitro (28).
It is generally agreed that colorectal cancers develop as a consequence of accumulation of mutations in key genes such as K-Ras, Apc, and p53 that are critical for regulating cell proliferation or cell cycle checkpoint control. In humans, the development from early adenomas to metastatic carcinomas takes somewhere from 20 to 40 years; it is believed that genetic instability plays a key role in accelerating the rate of mutation in cancerous cells (4). Our studies show that haploinsufficiency of BubR1 not only significantly increases the frequency of formation of colonic polyps but also enhances their progression toward more malignant phenotypes in ApcMin/+ mice (Fig. 1). Given that BubR1 primarily functions in the spindle checkpoint that monitors chromosomal segregation, it is likely that chromosomal instability due to BubR1 deficiency is the driving force for both expansion of proliferative cell population and accelerated progression of colonic tumors in Apc-deficient mice.
It is unexpected that enhanced colonic tumor development in BubR1+/–ApcMin/+ mice is correlated with a significant decrease in the number of adenomas in the small intestine (Fig. 1D). Although the exact mechanism for the suppression of tumor formation by BubR1 deficiency in the small intestine of ApcMin/+ mice remains unclear it is most likely due to enhanced cell death as a consequence of accelerated chromosomal instability. Theoretically, cells with chromosomal instability have two fates. (i) Most cells with chromosomal instability will undergo apoptosis because of severe imbalance of genetic content within the cell. Many in vivo and in vitro studies support this notion. For example, enhanced apoptosis occurs in normal small intestinal tissues adjacent to tumors from BubR1+/–ApcMin/+ mice (Fig. 3B). Ectopic expression of Bub1 dominant-negative mutant induces apoptosis in vitro (29). In addition, homozygous deletions of spindle checkpoint genes such as BubR1 and Mad2 result in embryonic lethality, which is accompanied by enhanced apoptosis (25, 30). (ii) A small fraction of cells with missegregated chromosomes will survive and proliferate at an accelerated rate because of a right combination of genetic contents; these cells eventually give rise to tumors in vivo. Somehow, the microenvironment in ApcMin/+ colon either tolerates chromosomal instability better or confers more resistance to apoptosis when the spindle checkpoint is compromised (i.e., BubR1 deficiency), resulting in dramatic increase in colonic tumor formation.
In conclusion, our studies demonstrate that there is a functional interaction between BubR1 and Apc genes in vivo and that BubR1 deficiency confers the susceptibility of ApcMin/+ mice to develop colonic tumors, supporting the notion that enhanced chromosomal instability due to spindle checkpoint failure may play a key role in the development and progression of colorectal cancer.
Acknowledgments
We thank Drs. Rulong Sheng and Gita Ramaswamy for advice on our histological studies and Dr. Christoph Lengauer for critical reading of the manuscript. This work was supported in part by grants from the National Institutes of Health to W.D. (CA90658) and to C.V.R. (CA80003). Y.-M.Y. was supported in part by an R03 grant (CA110057) from the National Institutes Health.
Author contributions: C.V.R., Y.-M.Y., and W.D. designed research; Y.-M.Y., M.V.S., T.L., Y.F., and R.M. performed research; C.V.R., Y.-M.Y., Y.F., and W.D. analyzed data; W.D. wrote the paper; and M.J.-U. helped discussion.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Apc, adenomatous polyposis coli; MEF, murine embryonic fibroblast.
References
- 1.Storchova, Z. & Pellman, D. (2004) Nat. Rev. Mol. Cell Biol. 5, 45–54. [DOI] [PubMed] [Google Scholar]
- 2.Motoyama, N. & Naka, K. (2004) Curr. Opin. Genet. Dev. 14, 11–16. [DOI] [PubMed] [Google Scholar]
- 3.Doxsey, S. (2002) Mol. Cell 10, 439–440. [DOI] [PubMed] [Google Scholar]
- 4.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1997) Nature 386, 623–627. [DOI] [PubMed] [Google Scholar]
- 5.Rajagopalan, H., Nowak, M. A., Vogelstein, B. & Lengauer, C. (2003) Nat. Rev. Cancer 3, 695–701. [DOI] [PubMed] [Google Scholar]
- 6.Groden, J., Thliveris, A., Samowitz, W., Carlson, M., Gelbert, L., Albertsen, H., Joslyn, G., Stevens, J., Spirio, L., Robertson, M., et al. (1991) Cell 66, 589–600. [DOI] [PubMed] [Google Scholar]
- 7.Kinzler, K. W., Nilbert, M. C., Su, L. K., Vogelstein, B., Bryan, T. M., Levy, D. B., Smith, K. J., Preisinger, A. C., Hedge, P., McKechnie, D., et al. (1991) Science 253, 661–665. [DOI] [PubMed] [Google Scholar]
- 8.Bodmer, W., Bishop, T. & Karran, P. (1994) Nat. Genet. 6, 217–219. [DOI] [PubMed] [Google Scholar]
- 9.Bodmer, W. F., Bailey, C. J., Bodmer, J., Bussey, H. J., Ellis, A., Gorman, P., Lucibello, F. C., Murday, V. A., Rider, S. H., Scambler, P., et al. (1987) Nature 328, 614–616. [DOI] [PubMed] [Google Scholar]
- 10.Powell, S. M., Zilz, N., Beazer-Barclay, Y., Bryan, T. M., Hamilton, S. R., Thibodeau, S. N., Vogelstein, B. & Kinzler, K. W. (1992) Nature 359, 235–237. [DOI] [PubMed] [Google Scholar]
- 11.Bienz, M. & Clevers, H. (2000) Cell 103, 311–320. [DOI] [PubMed] [Google Scholar]
- 12.Oshima, M., Dinchuk, J. E., Kargman, S. L., Oshima, H., Hancock, B., Kwong, E., Trzaskos, J. M., Evans, J. F. & Taketo, M. M. (1996) Cell 87, 803–809. [DOI] [PubMed] [Google Scholar]
- 13.Polakis, P. (2001) Cell 105, 563–566. [DOI] [PubMed] [Google Scholar]
- 14.Fearnhead, N. S., Britton, M. P. & Bodmer, W. F. (2001) Hum. Mol. Genet. 10, 721–733. [DOI] [PubMed] [Google Scholar]
- 15.Shih, I. M., Zhou, W., Goodman, S. N., Lengauer, C., Kinzler, K. W. & Vogelstein, B. (2001) Cancer Res. 61, 818–822. [PubMed] [Google Scholar]
- 16.Green, R. A. & Kaplan, K. B. (2003) J. Cell Biol. 163, 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fodde, R., Kuipers, J., Rosenberg, C., Smits, R., Kielman, M., Gaspar, C., van Es, J. H., Breukel, C., Wiegant, J., Giles, R. H., et al. (2001) Nat. Cell Biol. 3, 433–438. [DOI] [PubMed] [Google Scholar]
- 18.Dikovskaya, D., Newton, I. P. & Nathke, I. S. (2004) Mol. Biol. Cell 15, 2978–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1998) Nature 396, 643–649. [DOI] [PubMed] [Google Scholar]
- 20.Michel, L. S., Liberal, V., Chatterjee, A., Kirchwegger, R., Pasche, B., Gerald, W., Dobles, M., Sorger, P. K., Murty, V. V. & Benezra, R. (2001) Nature 409, 355–359. [DOI] [PubMed] [Google Scholar]
- 21.Dai, W., Wang Q, Liu, T.-Y., Swamy, M., Fang, Y.-Q., Xie, S.-Q., Mahmood, R., Yang, Y. M., Xu, M. & Rao, C. V. (2004) Cancer Res. 15, 440–445. [DOI] [PubMed] [Google Scholar]
- 22.Babu, J. R., Jeganathan, K. B., Baker, D. J., Wu, X., Kang-Decker, N. & van Deursen, J. M. (2003) J. Cell Biol. 160, 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanks, S., Coleman, K., Reid, S., Plaja, A., Firth, H., Fitzpatrick, D., Kidd, A., Mehes, K., Nash, R., Robin, N., et al. (2004) Nat. Genet. 36, 1159–1161. [DOI] [PubMed] [Google Scholar]
- 24.Baker, D. J., Jeganathan, K. B., Cameron, J. D., Thompson, M., Juneja, S., Kopecka, A., Kumar, R., Jenkins, R. B., de Groen, P. C., Roche, P., et al. (2004) Nat. Genet. 36, 744–749. [DOI] [PubMed] [Google Scholar]
- 25.Wang, Q., Liu, T.-Y., Fang, Y.-Q., Xie, S.-Q., Huang X, Ramaswamy, G., Sakamoto, K., Darzynkiewicz, Z., Xu, M. & Dai, W. (2004) Blood 103, 1278–1285. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan, K. B., Burds, A. A., Swedlow, J. R., Bekir, S. S., Sorger, P. K. & Nathke, I. S. (2001) Nat. Cell Biol. 3, 429–432. [DOI] [PubMed] [Google Scholar]
- 27.Lengauer, C. & Wang, Z. (2004) Nat. Genet. 36, 1144–1145. [DOI] [PubMed] [Google Scholar]
- 28.Zumbrunn, J., Kinoshita, K., Hyman, A. A. & Nathke, I. S. (2001) Curr. Biol. 11, 44–49. [DOI] [PubMed] [Google Scholar]
- 29.Taylor, S. S. & McKeon, F. (1997) Cell 89, 727–735. [DOI] [PubMed] [Google Scholar]
- 30.Dobles, M., Liberal, V., Scott, M. L., Benezra, R. & Sorger, P. K. (2000) Cell 101, 635–645. [DOI] [PubMed] [Google Scholar]