Abstract
Viral epitranscriptomics is a newly emerging field that has identified unique roles for RNA modifications in modulating life cycles of RNA viruses. Despite the observation of a handful of modified viral RNAs five decades ago, very little was known about how these modifications regulate viral life cycles, until recently. Here we review the pro- and anti-viral effects of methyl-6-adenosine in distinct viral life cycles, the role of 2′ O-methyl modifications in RNA stability and innate immune sensing, and functions of adenosine to inosine modifications in retroviral life cycles. With roles for over 100 modifications in RNA still unknown, this is a rapidly emerging field that is destined to suggest novel antiviral therapies.
Keywords: Epitranscriptomics, methyltransferase writers, demethylase erasers, YTHDF readers, RNA viruses, flaviviruses, retroviruses, methyl-6-adenonsine, 2′-O-methyl, A-to-I editing
Viral epitranscriptomics has identified unique roles for RNA modifications. Gonzales-van Horn and Sarnow review how adenosine modifications play a role in the life cycle of certain RNA viruses. The host machineries that modulate these modifications may offer putative targets for antiviral interventions.
Main Text
Introduction
While the epigenetics field identified important roles for DNA and histone modifications, epitranscriptomics has only begun to characterize and legitimize the importance of RNA modifications in all three kingdoms of life. With respect to eukaryotes, RNA modifications play a role in regulating transfer RNA (tRNA) and ribosomal RNA (rRNA) structure and function, messenger RNA (mRNA) translation efficiency and stability, microRNA maturation, cell differentiation, and innate sensing pathways of pathogens, to name a few (Li and Mason, 2014). The most abundantly modified RNAs in eukaryotes are those present on cellular rRNAs and tRNAs, which regulate RNA structure and function (Choi et al., 2016, Motorin and Helm, 2010, Sloan et al., 2016). Several modifications have been identified in mRNAs and non-coding RNAs (ncRNAs) such as N1-methyladenosine (m1A), N6-methyladenosine (m6A), 5-methylcytidine (m5C), inosine (I), pseudouridine (Ψ), and the 5′ cap modifications N 6,2-O-dimethyladenosine (m6Am) and 2′-O-methylation (2′-O-me). Methods to detect such modifications and their interactions with RNA binding proteins are summarized in a recent comprehensive review (Helm and Motorin, 2017). Recent studies in the viral epitranscriptomics field have established an equally important role for these chemical modifications during viral infection. For example, m6A-modifications can positively affect RNA replication or negatively affect RNA assembly, dependent on the lifestyle of the virus. Thus, m6A-modifications can have pro- or antiviral functions and, thus, offer new potential targets for antiviral intervention.
These advances in epitranscriptomics have been powered by improvements of several techniques to study RNA modifications and RNA-binding proteins. First, the refinement of high-performance liquid chromatography coupled with tandem mass spectrometry (HPLC-MS/MS) has led to greater confidence in the detection of RNA modifications and their abundances. Further, transcriptome sequencing has been paired with three classic techniques to identify specific modifications within unique transcripts: (a) surveying altered base-pairing properties at the site of modification; (b) identification of truncated products caused by reverse transcription at the modified site; and (c) antibody-based isolation and enrichment of RNAs with specific modifications. Using a combination of these techniques has been instrumental in validating chemical modifications present in host and viral RNAs. For further information about the usefulness and limitations of each of these techniques, please refer to the excellent reviews by Schwartz and Motorin (2016) and Li et al. (2016).
In this review, we focus on the recent work describing how adenosine modifications, m6A, m6Am, 2′-O-me, and A-to-I editing (Figure 1 ) play a role in the life cycle of certain RNA viruses. We discuss the role of these modifications with respect to viral cap structures, viral replication, innate sensing pathways, and modulation of the innate immune response. The content presented here will give a current understanding of how a handful of different viral RNAs are modified, how the changes in RNA alter the virus life cycle, and what remains to be clarified in the field.
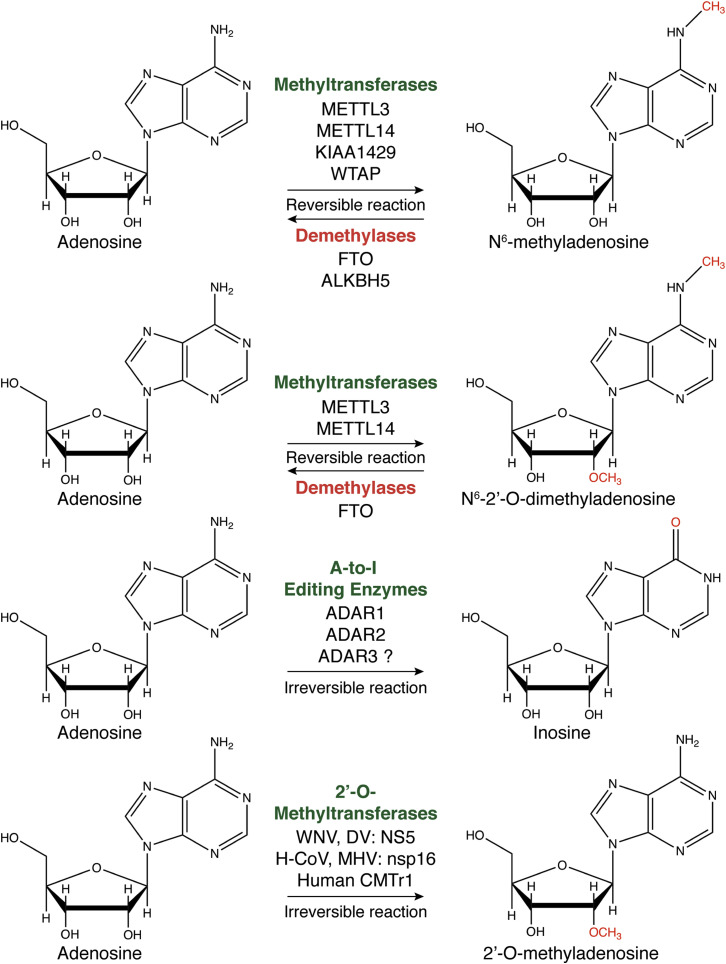
Figure 1.
Adenosine Modifications and the Respective Enzymes that Facilitate These Chemical Modifications
N6-methyladenosine and N6,2-O-dimethyladenosine are reversible RNA modifications, while inosine and 2′-O-methyladenosine seem to be irreversible. Of the modifications listed, only 2′-O-methyltransferases are encoded by viral genes (WNV, West Nile virus; DV, dengue virus; H-CoV, human coronavirus), further supporting the important role for this modification in modulating viral sensing by innate immune pathways.
Roles for N6-Methyladenosine Modification in the Life Cycle of Retroviridae and Flaviviridae
It has been known for a long time that RNAs transcribed both in DNA virus- and RNA virus-infected cells can be modified on adenosine residues. For example, Lavi and Shatkin reported that RNAs from simian virus 40, a small DNA virus, are modified on adenosines in infected cells (Lavi and Shatkin, 1975). Krug et al. provided an example that the viral RNA genomes of influenza virus are modified as well (Krug et al., 1976). However, it was not until 1985 when Kane and Beemon reported that specific adenosines were methylated in viral RNAs in cells infected with Rous sarcoma virus (Kane and Beemon, 1985). More recently, m6A has been the focus of intense study in elucidating the role of this RNA modification in viral epitranscriptomics (Gokhale and Horner, 2017, Kennedy et al., 2017).
The Machinery—Writers, Erasers, Readers—That Performs and Modulates m6A Modifications
The identification of methyltransferases and demethylases demonstrated that m6A modification was reversible and sensitive to external signals. The methyltransferase “writer” complex consists of Methyltransferase Like 3 (METTL3) (Bokar et al., 1994), Methyltransferase Like 14 (METTL14) (Liu et al., 2014), KIAA1429 (Schwartz et al., 2014), and Wilms’ Tumor 1-Associating Protein (WTAP) (Ping et al., 2014) (Figure 1). WTAP aids METTL3/METTL14 to interact with mRNAs in the nucleus to improve m6A modification efficiency (Liu et al., 2014, Ping et al., 2014). The importance of each of these factors has been validated using depletion and gene knockout approaches. Depletion of writers METTL3, METTL14, or WTAP reduces m6A abundances by approximately 2-, 2.5-, and 6.5-fold in eukaryotic cells, respectively (Liu et al., 2014, Schwartz et al., 2014). Although not as dramatic as WTAP depletion, depletion of KIAA1429 in human cells results in a 4-fold decrease of m6A abundance (Schwartz et al., 2014). Recent structural studies have demonstrated that METTL3 is the catalytic subunit of the writer complex, while METTL14 plays a role in complex stability and RNA recruitment (Śledź and Jinek, 2016). Most recently, RNA-binding motif protein 15 (RBM15) and its paralog RBM15B have been identified as members of the m6A writer complex that recruits the METTL3/14 protein complex to long ncRNAs (Patil et al., 2016). Future studies are needed to define the role, if any, of RBM15 and RBM15B in the context of other RNA species, including viral RNAs.
Reversal of m6A is performed by at least two demethylase “erasers” that have been identified in mammalian cells. Fat mass and obesity-associated protein (FTO) is associated with human body weight regulation (Jia et al., 2011), while ALKBH5 is associated with fertility in mice (Zheng et al., 2013) (Figure 1). The reversibility of the m6A modification suggests a dynamic role for the modification in controlling the fate of modified RNAs. Indeed, m6A modifications have been reported to modulate eukaryotic translation efficiency (Lin et al., 2016, Wang et al., 2015), cap-independent translation (Meyer et al., 2015), mRNA stability (Schwartz et al., 2014), RNA splicing (Dominissini et al., 2012), and miRNA biogenesis (Alarcón et al., 2015). More globally, m6A abundance in specific RNAs correlates with their functioning in cellular reprogramming (Chen et al., 2015), heat shock response (Zhou et al., 2015), circadian cycle (Fustin et al., 2013), cell development (Ping et al., 2014), and fertility (Zheng et al., 2013). Most likely, the function of m6A-containing RNAs is determined by “readers,” including the YTH domain-containing family proteins (YTHDF1–YTHDF3), each of which recognize and bind m6A moieties in RNAs with their conserved carboxy-terminal YTH motif (Meyer and Jaffrey, 2014).
In addition, it is known that m6A can alter local RNA structures, for example, by destabilizing RNA helices. Thus, m6A can affect RNA stability or possibly RNA localization. Additionally, heterogeneous ribonucleoproteins (hnRNP) have recently been shown to interact with m6A-modified RNA. The presence of m6A modifies local RNA structure, which promotes the nuclear protein hnRNP C to bind to this region and to modulate pre-mRNA processing (Liu et al., 2015). Further, hnRNP A2B1 interacts with m6A to similarly promote micoRNA processing and alternative splicing (Alarcón et al., 2015). The nuclear localization of these RNA-binding proteins suggests a role in the life cycle of viruses that replicate their genomes in the nucleus, such as influenza virus.
Roles for m6A in the Life Cycle of Human Immunodeficiency Virus
Three recent publications revealed some common and some controversial roles for m6A modifications in the HIV life cycle (Lichinchi et al., 2016a, Kennedy et al., 2016, Tirumuru et al., 2016) (Figure 2 ). A common observation is that writers METTL3/METTL14 promote HIV replication, while erasers ALKBH5 and FTO suppress it (Lichinchi et al., 2016a, Tirumuru et al., 2016) (Figure 2). In addition, the compound 3-deazaadenosine, which blocks m6A methylation, has been shown to inhibit the growth of HIV at concentrations that did not affect cell viability (Kennedy et al., 2016). However, it is not known yet whether this inhibitor has additional unspecific effects. More directly, mutations of identified m6A residues reduced viral RNA abundances and viral titers (Kennedy et al., 2016). Using a CD4+ T cell model, Lichinichi et al. reported that HIV infection enhances m6A modifications of both viral and host RNAs, and depletion of the m6A writers and erasers decreased and increased HIV infection, respectively (Lichinchi et al., 2016a). Interestingly, several host mRNAs are modified at m6A during HIV infection such as TRAF2, PABPC3, and ETS2, all of which are known to have pro-viral functions. It will be interesting to identify and to mutate the modified adenosine residues in these mRNAs to examine whether they exert their pro-viral functions at the level of mRNA stability or mRNA translation. Overall, there is agreement that m6A in HIV RNA modulates viral gene expression.
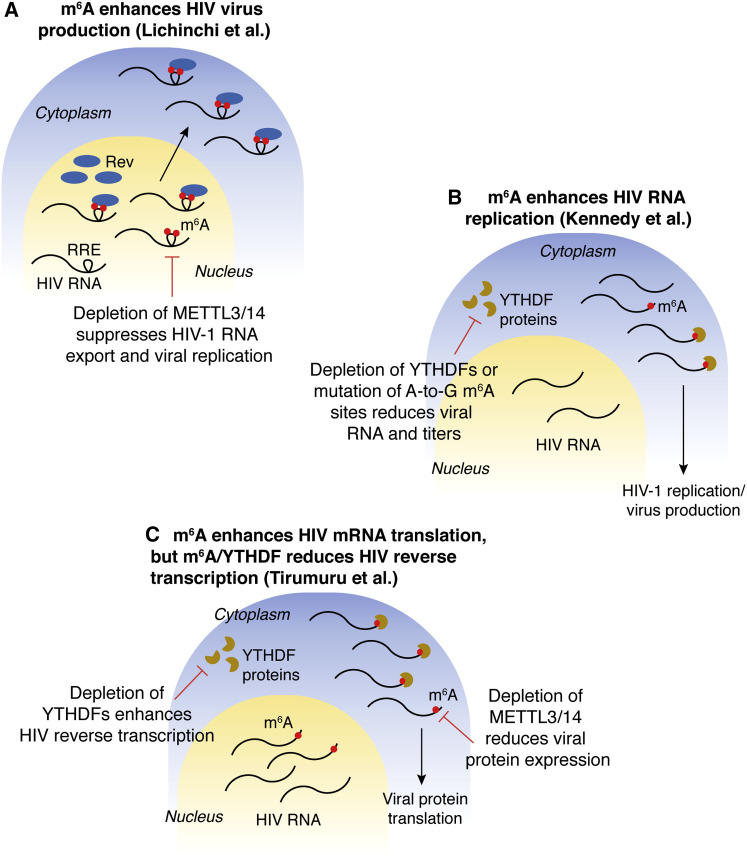
Figure 2.
Three Current Models for m6A-Mediated Regulation of the HIV-1 Life Cycle
(A) m6A enhances HIV virus production (Lichinchi et al., 2016a). Stemloop II within the HIV Rev response element (RRE) is m6A methylated at two sites, which promotes the HIV-encoded RNA binding protein, Rev, to bind to the RRE and enhance RNA nuclear export into the cytoplasm. Depletion of the methyltransferases METTL3/14 suppressed HIV-1 RNA export and viral replication, suggesting that m6A modifications within the RRE are important for the HIV life cycle.
(B) m6A enhances HIV RNA replication (Kennedy et al., 2016). Like the model presented in (A), this second study confirmed that the presence of m6A enhances viral replication. Cellular m6A-modified HIV RNAs are bound by the reader YTHDF proteins, which promote HIV replication. Depletion of YTHDF proteins reduced viral RNA abundance and virus titers, as did mutating certain m6A-modified adenosine residues to guanines.
(C) m6A enhances HIV mRNA translation, but m6A/YTHDF readers reduce HIV reverse transcription (Tirumuru et al., 2016). In contrast to the models presented in (A) and (B), m6A was found to enhance translation, but to suppress reverse transcription, suggesting a modular role in the HIV life cycle depending on the replication step. Depletion of METTL3/14 resulted in a reduction of viral protein expression, while the depletion of YTHDF proteins enhanced HIV reverse transcription.
The three reports disagree by what mechanisms m6A modulates HIV gene expression. First, the number and locations of m6A sites in the HIV RNA genome is controversial. Kennedy et al. (2016) identified m6A sites exclusively within the 3′ region of the RNA genome; in contrast, Lichinchi et al. (2016a) identified 14 m6A modifications throughout the entire HIV RNA genome, including 2 in the Rev responsive element (RRE), a structured region that is bound by the viral Rev protein (Figure 2). These modified residues modulate nuclear export of RRE-containing RNAs and enhance the binding of the viral Rev protein to the RRE (Figure 2). Curiously, 11 of the identified modified sites locate 5′ to the sites identified by Kennedy et al. (2016).
Why did mapping m6A residues yield such different results? Some obvious reasons could be the use of different viral strains, cell types, and reagents. Other differences between the authors’ techniques could account for some of the observed differences. For example, the amount of RNA in the m6A-RNA immunoprecipitation and crosslinking-immunoprecipitation experiments likely play a role in the accuracy of the analysis. Furthermore, changes in the sensitivity of the bioinformatics pipeline could have large effects on output. In addition, different viral replication kinetics could also influence the dynamic behavior of readers, writers, and eraser proteins. One main difference between the m6A mapping strategy is that Lichinchi et al. (2016a) immunoprecipitated fragmented RNA with an antibody directed against m6A. In contrast, Kennedy et al. (2016) labeled viral RNA with 4-thiouridine and then crosslinked the m6A antibody to the full-length RNA. In addition, cell-based crosslinking of tagged YTHDF1–YTHDF3 reader proteins to viral RNA revealed very similar sites of crosslinking in both approaches, except that the YTHDF proteins could be crosslinked in addition to a region in the viral nef gene. Because accessibility of m6A antibodies to m6A sites could be different in fragmented versus full-length RNAs, these different approaches may yield different results should high-order RNA structures play a role in the antibody-assisted recognition of m6A residues. Crosslinking of YTHDF proteins to target RNAs before m6A analysis should thus minimize the potential artifact that distinct YTHDF interaction affinities occur at specific m6A sites in RNAs.
In addition, the effect of each YTHDF on HIV transcription and replication remains controversial. Kennedy et al. observe that YTHDF reader protein overexpression enhances HIV replication and HIV p24, p55, and Nef protein abundance, while Tirumuru et al. observe that overexpression of YTHDF proteins suppress reverse transcription and HIV replication (Kennedy et al., 2016, Tirumuru et al., 2016). Both studies utilized overexpression and depletion approaches using a similar VSV-pseudotyped HIV infection model. It is currently unclear what caused the large discrepancy between the two studies. Non-physiological overexpression or non-quantitative depletion of YTHDF reader proteins could reveal results that are not physiologically relevant. Because m6A modification is reversible, it is possible that distinct m6A residues are targeted by writers, erasers, and readers in a dynamic process that may be influenced by the growth state of the cell, or by cellular events that accompany HIV infection. A recent review has suggested that the presence of a Firefly luciferase reporter sequence in the HIV genome used by Tirumuru et al. (2016) might bias the abundance of YTHDF and effects on HIV replication, because this non-viral RNA sequence can be heavily modified by m6A (Kennedy et al., 2017).
In addition to the role of YTHDF proteins in regulating the HIV life cycle, hnRNP proteins might interact with HIV RNA in an m6A-dependent manner. hnRNP C has been shown to interact with m6A-modified RNAs, promoting pre-splicing events (Liu et al., 2015). Previous studies have shown that other hnRNP proteins affect the HIV life cycle, including numerous hnRNP proteins that interact with the HIV Rev protein (Hadian et al., 2009). Thus, the binding of Rev to the m6A-containing RRE element present in HIV RNA (Lichinchi et al., 2016a) could be modulated by hnRNP C.
Roles for m6A in the Life Cycle of Hepatitis C Virus and Zika Virus
Until recently, there was little evidence that functional m6A methyltransferases operated outside of the nucleus. Recent studies with viruses that have exclusively cytoplasmic life cycles have demonstrated a broader role for the writer enzymes in the cytoplasm as well. Two recent studies describe the role of m6A modifications in regulating different members of Flaviviridae, whose gene expression takes place in the cytosol. To date, m6A analysis has been performed in cells infected with HCV, dengue virus (DV), West Nile virus (WNV), yellow fever virus (YFV), and three strains of Zika virus (ZV) (Gokhale et al., 2016, Lichinchi et al., 2016b).
The regulation of HCV RNA by small microRNAs and by RNA structures has been widely appreciated. Liver-specific microRNA miR-122 is required for HCV replication and has been shown to bind at two sites at the 5′ end of the HCV genome to protect the viral RNA from degradation by host cell exoribonucleases (Jopling et al., 2005). Further, using selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE), a number of structures and ribonucleotide interactions have been shown to play an important role in regulating the virus life cycle, as mutating these sites led to the inhibition or enhancement of viral replication (Mauger et al., 2015, Pirakitikulr et al., 2016). An additional level of regulation at the RNA level has been recently provided by Stacy Horner’s laboratory. Gokhale and colleagues identified 19 sites of m6A enrichment within the HCV RNA genome, including sites spanning the 5′ UTR, the core gene, as well as other genes within the HCV genome (Gokhale et al., 2016). m6A present in 5′ UTRs has been shown to enhance mRNA translation in a cap-independent manner (Meyer et al., 2015). This observation may explain how viruses, such as HCV, utilize cap-independent mechanisms to initiate the production of viral proteins. HCV contains an internal ribosome entry site (IRES) that directly binds the 40S subunit of the ribosome without the use of the cap binding protein complex eIF4F (Kieft et al., 2001, Pestova et al., 1998). Although Gokhale et al. did not detect a defect in HCV translation in the absence of m6A writers (Gokhale et al., 2016), other viral genomes that can be translated when eIF4F abundance is low could be affected by the presence of m6A.
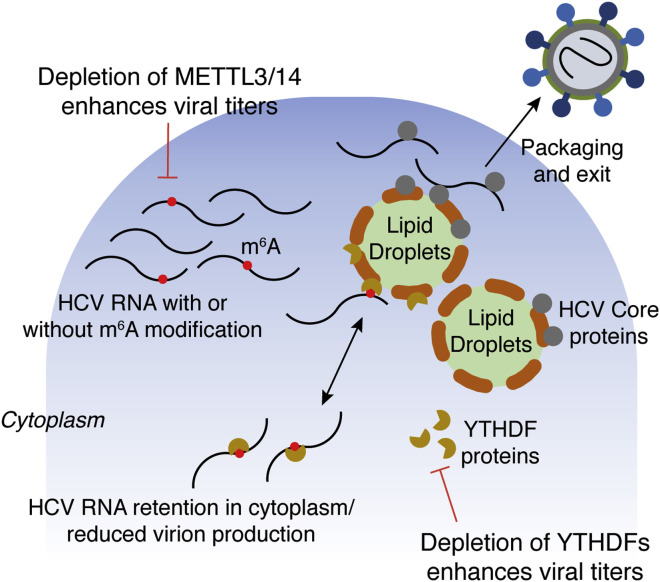
The importance of m6A modifications in the HCV viral life cycle was demonstrated by siRNA-mediated depletion of the methyltransferases METTL3, METTL14, and FTO, which showed that the methyl writers METTL3/14 negatively regulate HCV protein expression, while the methyl eraser FTO enhanced viral protein expression (Gokhale et al., 2016) (Figure 3 ). In addition, the methyl YTHDF readers have been implicated in modulating viral assembly. YTHDF1–YTHDF3 reader proteins colocalize with HCV RNA to lipid droplets, which are the sites for assembly of HCV virions (Figure 3), and negatively regulate virus assembly without affecting viral replication (Gokhale et al., 2016). YTDFH proteins interact with one m6A site in particular that is present within the envelope protein-encoding gene of HCV RNA. This site contains a DRAmCH motif that is a characteristic target for adenosine methylation. Mutational analysis of this site correlated with a 3-fold enhancement of HCV viral titer, without affecting viral replication or protein production (Gokhale et al., 2016). This region of RNA containing the m6A motif has been shown to be bound by HCV Core protein to promote virion packaging and assembly (Shimoike et al., 1999). Mutation of this site reduced YTHDF protein binding and enhanced HCV Core protein binding (Figure 3). Perhaps the role of YTHDF is antiviral, in which enhanced binding results in reduced nascent virion production, while Core binding prevents YTHDF binding, enhancing virion assembly within lipid droplets. It will be important to determine what signaling pathways activate METTL3/14-mediated methylation at distinct sites within the viral RNA genome to identify potential antiviral pathways that can be targeted for antiviral intervention.
Figure 3.
The Role of Methyltransferases and Methyl Reader Proteins in Regulating the HCV Life Cycle
HCV RNA is modified at several adenosines (m6A) throughout the genome, and ablation of these modifications promotes viral replication and release from the cell. This process is modulated by YTHDF methyl readers sequestering m6A-modified HCV RNAs, preventing the localization and interaction with HCV Core protein at the lipid droplets and subsequent virus particle production. See text for further details.
Another member of the Flaviviridae family that has received recent attention by the scientific community is Zika virus. 2′-O-methyl modifications were abundant on adenosine, cytosine, and uracil ribonucleotides during Zika virus infection, while m6A was found to be present on strikingly 3% of all adenosines present within the ZV RNA genome (Lichinchi et al., 2016b). Further, 12 m6A unique enrichment sites exist within the MR766 African strain of ZV, half of which are present in the region encoding the nonstructural protein, NS5, of the viral RNA and the 3′ UTR region (Lichinchi et al., 2016b). Similarly to the role of m6A on regulating the HCV life cycle, writers METTL3 and METTL14 suppress replication, while erasers ALKBH5 and FTO enhance replication of ZV. Further, the methyl reader proteins YTHDF1–YTHDF3 bind m6A in ZV RNA, which correlates with viral RNA decay and reduced viral replication (Lichinchi et al., 2016b). These data are corroborated by experiments demonstrating that the absence of METTL3/14 prevents YTHDF reader proteins from negatively regulating ZV replication (Lichinchi et al., 2016b). Thus, m6A modifications within ZV negatively regulate the viral life cycle, similarly to HCV, albeit through different mechanisms.
To date, several studies have determined the host gene expression profiles in response to ZV infection (Bayer et al., 2016, Tang et al., 2016, Zhang et al., 2016). ZV modulates the interferon response, cell cycle, protein localization, ER stress, and apoptosis in the infected host cell, among other pathways. Antiviral responses require fine-tuned regulation to optimally control viral infections without inducing immune-mediated pathology. RNA modifications in host cellular RNAs can play important roles during viral infection as described above. Indeed, ZV infection caused enrichment of m6A modifications of host mRNAs (Lichinchi et al., 2016b). Because m6A acts at the post-transcriptional level to alter downstream pathways, it was not surprising that there was no significant change in the abundances of RNAs encoding antiviral proteins, including IRF8, INFAR1, IFNAR2, MAP3K3, and IFIH1, despite becoming enriched with m6A in response to ZV infection. Of course, it is likely that some of these genes are modulated at the post-transcriptional level. For example, m6A enhancement in 5′ UTRs of stress-induced host transcripts play a role in enhancing cap-independent translation of these mRNAs (Zhou et al., 2015). Indeed, members of the Flaviviridae virus family are known to modulate stress response pathways and the unfolded protein response (Ambrose and Mackenzie, 2013, Peña and Harris, 2011, Sir et al., 2008). Finally, YTHDF2 modulates the ability of FTO to remove m6A modifications in the 5′ UTR of heat shock-induced mRNAs by binding to these sites in the nucleus of heat shock-treated cells. Thus, it is possible that YTHDF binding to m6A sites within host and viral RNAs prevents host methyltransferases from modulating m6A enrichment. As a result, changes to mRNA stability and translation efficiency could occur in host mRNAs. In the case of ZV RNA, this effect seems to result in controlling ZV replication.
In addition to the importance of m6A modifications repressing the life cycle of the two members of the Flaviviridae, other family members have been observed to contain m6A sites within their RNA genomes. The m6A presence in WNV, DV, and YFV was recently reported, and unique, conserved sites for m6A modifications in NS3, NS5, and the 3′ UTR were discovered (Gokhale et al., 2016). Notably, each of these mosquito-transmitted viruses contains secondary RNA structures that regulate viral translation and replication, many of which are within the 5′ and 3′ UTRs. Recent work determined that the 3′ UTR stem loop structures are important for viral replication and fitness with respect to mosquito-to-human transmission (Villordo et al., 2015). Perhaps the enrichment of m6A in human versus mosquito cells is modulated differently, leading to altered local RNA structures and RNA binding protein recruitment. For example, the methylation at putative sites located near the polypyrimidine-rich motif in the 3′ UTR may modulate RNA folding and the binding of hnRNP C, which is essential for HCV gene amplification (Gontarek et al., 1999). Further studies will be important in identifying the differences between the methylomes of these viruses in different hosts to identify roles in viral pathogenesis.
Roles for N6,2-O-Dimethyladenosine Modification in the Viral Life Cycles
In addition to m6A, N 6,2-O-dimethyladenosine (m6Am) modification is abundant in cap structures, m7GpppN, present at the 5′ end of most viral and all host mRNAs (Figure 1; Keith et al., 1978, Wei et al., 1975). This chemical modification was recently identified to play a role in regulating mRNA stability (Mauer et al., 2017). Mauer et al. (2017) identified this novel cap-proximal modification within eukaryotic mRNAs containing adenosine as their first nucleotide. This modification is the preferential substrate for the eraser demethylase FTO, with a 100-fold greater catalytic efficiency toward m6Am compared to the previously identified substrate, m6A. The preferential interaction between FTO and m6Am was found to be due to the proximity of the adenosine to the RNA m7Gppp cap. Substitution of m7G with a non-methylated guanine reduced the rate of interaction (Mauer et al., 2017). Furthermore, the enzyme DCP2, which removes the m7Gppp cap on mRNAs, was shown to be the enzyme responsible for reading the m6Am modification. Although no formal study has demonstrated a role for the METTL3/14 writer complex for m6Am specifically, it is likely that this methyl-writer complex is responsible for the addition of this cap-specific moiety. This hypothesis is substantiated by the finding by Schwartz et al. that this process is WTAP-independent (Schwartz et al., 2014). Interestingly, the presence of m6Am in RNAs has distinct functions from the presence of m6A. m6A seems to play a role in enhancing mRNA decay, while the presence of m6Am stabilizes RNA (Mauer et al., 2017, Sommer et al., 1978, Wang et al., 2014). m6Am modifications have been observed in influenza virus (Krug et al., 1976) and herpes simplex virus type 1 (Moss et al., 1977); however, the role of this modification in regulating the viral life cycles remains unexplored. It could be that m6Am stabilizes viral RNA similarly to what is observed with host mRNAs, suggesting that viruses might utilize this modification to control the abundance of viral RNAs within the cell.
Roles for 2′-O-Methylation of Ribose Moieties in the Virus Life Cycle
The 2′-O-me modification is a highly abundant chemical modification found on all types of eukaryotic RNAs. 2′-O-me-synthesized RNAs have been shown to act as agonists of the innate immune receptor Toll-like receptor 7 (TRL7), inhibiting the production of inflammatory cytokines, including type I interferon, in both mouse and human cells (Robbins et al., 2007). Thus, many siRNAs that are being used for gene manipulation contain modified bases at the 2′ ribose to prevent aberrant immune responses. Daffis et al. identified that members of the flaviviruses, poxviruses, and coronaviruses, all viruses that replicate in the cytoplasm, utilize a virus-encoded methyltransferase to add a 2′-O-methyl viral cap structure, resulting in the evasion from the antiviral activity exerted by murine Ifit1 (Daffis et al., 2010), a member of the IFIT family that limits viral replication by binding to viral proteins and RNAs and regulating their functions. In human cells, IFIT5, but not IFIT1, IFIT2, or IFIT3, suppressed infection of a WNV mutant that lacks the ability to generate the 2′-O-me group without affecting the N7-methyltransferase activity of NS5 (Ray et al., 2006). These findings suggest that cytoplasmic viruses containing the 2′-O-me RNA modification have enhanced virulence by evading the antiviral effects caused by distinct interferon-induced IFIT proteins. Interestingly, a number of IFIT family members attenuate translation (Hui et al., 2003, Hui et al., 2005, Terenzi et al., 2006), suggesting that utilization of the 2′-O-me modulates viral pathogenicity.
In addition to immune evasion, 2′-O-me on viral RNAs prevents type I interferon production mediated by the dsRNA sensor MDA-5 during infection of cells with coronavirus family members (Züst et al., 2011). Mutant human coronavirus lacking 2′-O-me enzymatic activity within non-structural protein Nsp16 enhanced expression of IFN-β in primary human macrophages compared to wild-type virus, suggesting that 2′-O-me has a biologically significant role in controlling the interferon response (Züst et al., 2011). This process is dependent on MDA-5 because mda5 knockout murine macrophages failed to produce detectable levels of IFN-β upon infection with a murine coronavirus, mouse hepatitis virus (MHV), which lacked enzymatic 2′-O-me function. In vivo infection models demonstrate that interferon signaling is a critical component of controlling MHV infection in the absence of 2′-O-me activity because interferon alpha/beta receptor (Ifnar) knockout mice rescued mutant virus spread and replication similar to wild-type MHV virus infection. Further, the growth of mutant and wild-type virus in Mda5/Tlr7 double-knockout mice mirrored the levels observed in Ifnar knockout mice (Züst et al., 2011), confirming that these sensors are critical in the detection of MHV, and that 2′-O-me serves as a virulence factor for this cytoplasmic virus. In contrast to the mechanism of IFIT-mediated restriction seen against WNV lacking 2′-O-me function, Ifit1 knockout macrophages completely rescued MHV replication to wild-type virus levels, suggesting that this MDA-5-dependent IFN-β production is a distinct mechanism, because IFIT1 and IFIT2 overexpression failed to rescue mutant WNV replication (Daffis et al., 2010).
Roles for Adenosine to Inosine Modification in the Retroviral Life Cycle
The significance of adenosine-to-inosine modifications (A-to-I editing) is appreciated for its abundance in both host and viral RNAs (Samuel, 2011). A-to-I editing is mediated by RNA adenosine deaminases (ADARs), which catalyze the deamination of carbon 6 to produce the inosine modification (Figure 1) within double-stranded RNA (dsRNA). A-to-I editing results in the destabilization of dsRNA structure through the inosine-uracil mismatch, which is less stable than the original adenosine-uracil pairing. Three ADARs exist, ADAR1, ADAR2, and ADAR3, all of which have been found to be sensitive to different cellular cues and whose expressions are cell specific. To date, the presence of inosines has been observed to modulate host-viral interactions in a number of viruses, including respiratory syncytial virus (Liao et al., 2011) and HIV (Doria et al., 2009, Phuphuakrat et al., 2008).
Mutations in the ADAR genes have been linked to aberrant innate immune responses through the activation of the type I interferon signaling pathway by cytosolic sensors of endogenous host RNAs (Liddicoat et al., 2015, Mannion et al., 2014). Using knockout animal models, the absence or mutation of ADAR1 was found to markedly enhance the expression of interferon-induced genes in the fetal liver, leading to heightened apoptosis and inflammation. Interestingly, A-to-I hyper-editing was identified in the 3′ UTR of a handful of specific genes. Based on in silico structural analysis, the substitution of adenosines with inosines inhibited perfect dsRNA stem loop structures, preventing the formation of long matched dsRNA (Liddicoat et al., 2015). These finds suggest that endogenous RNA lacking A-to-I editing can be recognized by cytosolic sensors, activating the innate immune response to double-stranded RNAs.
The role of A-to-I editing has been previously studied in HIV-infected human cell lines. Using siRNA knockdown and overexpression approaches, ADAR1 was found to correlate with reduced and enhanced expression of p24 Gag protein, respectively (Doria et al., 2009, Phuphuakrat et al., 2008). Importantly, the siRNA knockdown-mediated reduction in Gag protein expression could be rescued by an ADAR1 expression vector, suggesting that ADAR1 plays a positive role in regulating the life cycle of HIV in human cells. The importance of these observations is highlighted by the expression patterns of ADAR1 in primary human CD4+ T cells. Only in stimulated T cells did ADAR1 expression increase, while ADAR2 expression remained unchanged. Furthermore, the production of HIV is minimal in resting CD4+ T cells, suggesting a link between ADAR1 expression and HIV viral gene expression in activated CD4+ T cells. Alternative mechanisms, distinct from the RNA editing activity of ADAR1, have been suggested to contribute to HIV and vesicular stomatitis virus (VSV) life cycles (Doria et al., 2009, Nie et al., 2007). Mutating the enzymatic region of ADAR1 enhanced HIV replication (Doria et al., 2009). This observation was correlated with a reduction in phosphorylated PKR and eIF2α in cells transfected with the enzymatically silenced ADAR1 mutant, suggesting that a similar PKR-targeted mechanism might contribute to enhanced HIV replication, similar to what has been observed with VSV infection. The RNA-editing dependent and independent mechanisms may not be mutually exclusive, however. Finally, synthetic HIV constructs containing A-to-G mutations seem to be expressed more efficiently compared to wild-type HIV. However, there is no indication of whether such HIV isolates exist in nature. These A-to-I studies provide further examples of the importance of RNA modifications in regulating viral life cycles and immune response during infection.
Concluding Remarks
In this review, we have highlighted the recent advances in identifying the role for unique adenosine modifications in regulating the viral life cycle and the immune response to viral infection. There are some modifications we did not cover, such as m5C and pseudouridine, because there is insufficient information on their role in regulating viral RNAs or cellular antiviral responses. Curiously, some modifications, such as m6A, have both pro-viral and antiviral functions in distinct viral life cycles. Such opposite effects could be attributed to the intracellular localizations of viral RNAs, and thus availability of host proteins, that respond to m6A-modified RNAs. It is apparent that the field of viral epitranscriptomics is uncovering the importance of these RNA modifications at a rapid pace. Understanding how the dynamics of these modifications in viral and cellular RNAs affect distinct steps in the viral life cycles and viral pathogenesis will provide insight for the development of novel antiviral therapies.
Author Contributions
S.G.-v.H. and P.S. wrote and edited the manuscript.
Acknowledgments
This study was supported by grants from the National Institutes of Health (AI47365, AI069000). S.G.-v.H. was supported by a Dean’s Fellowship from the Stanford School of Medicine and by T32 AI007328.
References
- Alarcón C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose R.L., Mackenzie J.M. ATF6 signaling is required for efficient West Nile virus replication by promoting cell survival and inhibition of innate immune responses. J. Virol. 2013;87:2206–2214. doi: 10.1128/JVI.02097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer A., Lennemann N.J., Ouyang Y., Bramley J.C., Morosky S., Marques E.T.D.A., Jr., Cherry S., Sadovsky Y., Coyne C.B. Type III Interferons Produced by Human Placental Trophoblasts Confer Protection against Zika Virus Infection. Cell Host Microbe. 2016;19:705–712. doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar J.A., Rath-Shambaugh M.E., Ludwiczak R., Narayan P., Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994;269:17697–17704. [PubMed] [Google Scholar]
- Chen T., Hao Y.-J., Zhang Y., Li M.-M., Wang M., Han W., Wu Y., Lv Y., Hao J., Wang L. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- Choi J., Ieong K.-W., Demirci H., Chen J., Petrov A., Prabhakar A., O’Leary S.E., Dominissini D., Rechavi G., Soltis S.M. N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol. 2016;23:110–115. doi: 10.1038/nsmb.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.-Y., Schneller S., Zust R., Dong H. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Doria M., Neri F., Gallo A., Farace M.G., Michienzi A. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res. 2009;37:5848–5858. doi: 10.1093/nar/gkp604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin J.-M., Doi M., Yamaguchi Y., Hida H., Nishimura S., Yoshida M., Isagawa T., Morioka M.S., Kakeya H., Manabe I., Okamura H. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Gokhale N.S., Horner S.M. RNA modifications go viral. PLoS Pathog. 2017;13:e1006188. doi: 10.1371/journal.ppat.1006188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale N.S., McIntyre A.B.R., McFadden M.J., Roder A.E., Kennedy E.M., Gandara J.A., Hopcraft S.E., Quicke K.M., Vazquez C., Willer J. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe. 2016;20:654–665. doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontarek R.R., Gutshall L.L., Herold K.M., Tsai J., Sathe G.M., Mao J., Prescott C., Del Vecchio A.M. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3'NTR of the HCV RNA genome. Nucleic Acids Res. 1999;27:1457–1463. doi: 10.1093/nar/27.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadian K., Vincendeau M., Mäusbacher N., Nagel D., Hauck S.M., Ueffing M., Loyter A., Werner T., Wolff H., Brack-Werner R. Identification of a heterogeneous nuclear ribonucleoprotein-recognition region in the HIV Rev protein. J. Biol. Chem. 2009;27:33384–33391. doi: 10.1074/jbc.M109.021659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M., Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 2017;18:275–291. doi: 10.1038/nrg.2016.169. advance online publication. [DOI] [PubMed] [Google Scholar]
- Hui D.J., Bhasker C.R., Merrick W.C., Sen G.C. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. J. Biol. Chem. 2003;278:39477–39482. doi: 10.1074/jbc.M305038200. [DOI] [PubMed] [Google Scholar]
- Hui D.J., Terenzi F., Merrick W.C., Sen G.C. Mouse p56 blocks a distinct function of eukaryotic initiation factor 3 in translation initiation. J. Biol. Chem. 2005;280:3433–3440. doi: 10.1074/jbc.M406700200. [DOI] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.-G., He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kane S.E., Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol. Cell. Biol. 1985;5:2298–2306. doi: 10.1128/mcb.5.9.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith J.M., Ensinger M.J., Mose B. HeLa cell RNA (2′-O-methyladenosine-N6-)-methyltransferase specific for the capped 5′-end of messenger RNA. J. Biol. Chem. 1978;253:5033–5039. [PubMed] [Google Scholar]
- Kennedy E.M., Bogerd H.P., Kornepati A.V.R., Kang D., Ghoshal D., Marshall J.B., Poling B.C., Tsai K., Gokhale N.S., Horner S.M., Cullen B.R. Posttranscriptional m(6)A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe. 2016;19:675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E.M., Courtney D.G., Tsai K., Cullen B.R. Viral Epitranscriptomics. J. Virol. 2017;91 doi: 10.1128/JVI.02263-16. e02263-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft J.S., Grech A., Adams P., Doudna J.A. Mechanisms of internal ribosome entry in translation initiation. Cold Spring Harb. Symp. Quant. Biol. 2001;66:277–283. doi: 10.1101/sqb.2001.66.277. [DOI] [PubMed] [Google Scholar]
- Krug R.M., Morgan M.A., Shatkin A.J. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J. Virol. 1976;20:45–53. doi: 10.1128/jvi.20.1.45-53.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi S., Shatkin A.J. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc. Natl. Acad. Sci. USA. 1975;72:2012–2016. doi: 10.1073/pnas.72.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Mason C.E. The pivotal regulatory landscape of RNA modifications. Annu. Rev. Genomics Hum. Genet. 2014;15:127–150. doi: 10.1146/annurev-genom-090413-025405. [DOI] [PubMed] [Google Scholar]
- Li X., Xiong X., Yi C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods. 2016;14:23–31. doi: 10.1038/nmeth.4110. [DOI] [PubMed] [Google Scholar]
- Liao J.-Y., Thakur S.A., Zalinger Z.B., Gerrish K.E., Imani F. Inosine-containing RNA is a novel innate immune recognition element and reduces RSV infection. PLoS One. 2011;6:e26463. doi: 10.1371/journal.pone.0026463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G., Gao S., Saletore Y., Gonzalez G.M., Bansal V., Wang Y., Mason C.E., Rana T.M. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 2016;1:16011. doi: 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G., Zhao B.S., Wu Y., Lu Z., Qin Y., He C., Rana T.M. Dynamics of Human and Viral RNA Methylation during Zika Virus Infection. Cell Host Microbe. 2016;20:666–673. doi: 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddicoat B.J., Piskol R., Chalk A.M., Ramaswami G., Higuchi M., Hartner J.C., Li J.B., Seeburg P.H., Walkley C.R. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Choe J., Du P., Triboulet R., Gregory R.I. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion N.M., Greenwood S.M., Young R., Cox S., Brindle J., Read D., Nellåker C., Vesely C., Ponting C.P., McLaughlin P.J. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.-J., Chen Q. Reversible methylation of m(6)Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger D.M., Golden M., Yamane D., Williford S., Lemon S.M., Martin D.P., Weeks K.M. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc. Natl. Acad. Sci. USA. 2015;112:3692–3697. doi: 10.1073/pnas.1416266112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Jaffrey S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.-B., Jaffrey S.R. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B., Gershowitz A., Stringer J.R., Holland L.E., Wagner E.K. 5′-Terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J. Virol. 1977;23:234–239. doi: 10.1128/jvi.23.2.234-239.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y., Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- Nie Y., Hammond G.L., Yang J.-H. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J. Virol. 2007;81:917–923. doi: 10.1128/JVI.01527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil D.P., Chen C.-K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña J., Harris E. Dengue virus modulates the unfolded protein response in a time-dependent manner. J. Biol. Chem. 2011;286:14226–14236. doi: 10.1074/jbc.M111.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Shatsky I.N., Fletcher S.P., Jackson R.J., Hellen C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuphuakrat A., Kraiwong R., Boonarkart C., Lauhakirti D., Lee T.-H., Auewarakul P. Double-stranded RNA adenosine deaminases enhance expression of human immunodeficiency virus type 1 proteins. J. Virol. 2008;82:10864–10872. doi: 10.1128/JVI.00238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping X.-L., Sun B.-F., Wang L., Xiao W., Yang X., Wang W.-J., Adhikari S., Shi Y., Lv Y., Chen Y.-S. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirakitikulr N., Kohlway A., Lindenbach B.D., Pyle A.M. The Coding Region of the HCV Genome Contains a Network of Regulatory RNA Structures. Mol. Cell. 2016;62:111–120. doi: 10.1016/j.molcel.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D., Shah A., Tilgner M., Guo Y., Zhao Y., Dong H., Deas T.S., Zhou Y., Li H., Shi P.-Y. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 2006;80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins M., Judge A., Liang L., McClintock K., Yaworski E., MacLachlan I. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 2007;15:1663–1669. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- Samuel C.E. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology. 2011;411:180–193. doi: 10.1016/j.virol.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Motorin Y. Next-generation sequencing technologies for detection of modified nucleotides in RNAs. RNA Biol. 2016:1–14. doi: 10.1080/15476286.2016.1251543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., Mertins P., Ter-Ovanesyan D., Habib N., Cacchiarelli D. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoike T., Mimori S., Tani H., Matsuura Y., Miyamura T. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 1999;73:9718–9725. doi: 10.1128/jvi.73.12.9718-9725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir D., Chen W.L., Choi J., Wakita T., Yen T.S.B., Ou J.H.J. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śledź P., Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife. 2016;5:e18434. doi: 10.7554/eLife.18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan K.E., Warda A.S., Sharma S., Entian K.-D., Lafontaine D.L.J., Bohnsack M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2016:1–16. doi: 10.1080/15476286.2016.1259781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S., Lavi U., Darnell J.E., Jr. The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J. Mol. Biol. 1978;124:487–499. doi: 10.1016/0022-2836(78)90183-3. [DOI] [PubMed] [Google Scholar]
- Tang H., Hammack C., Ogden S.C., Wen Z., Qian X., Li Y., Yao B., Shin J., Zhang F., Lee E.M. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell. 2016;18:587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenzi F., Hui D.J., Merrick W.C., Sen G.C. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J. Biol. Chem. 2006;281:34064–34071. doi: 10.1074/jbc.M605771200. [DOI] [PubMed] [Google Scholar]
- Tirumuru N., Zhao B.S., Lu W., Lu Z., He C., Wu L. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife. 2016;5:e15528. doi: 10.7554/eLife.15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villordo S.M., Filomatori C.V., Sánchez-Vargas I., Blair C.D., Gamarnik A.V. Dengue virus RNA structure specialization facilitates host adaptation. PLoS Pathog. 2015;11:e1004604. doi: 10.1371/journal.ppat.1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Gershowitz A., Moss B. N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature. 1975;257:251–253. doi: 10.1038/257251a0. [DOI] [PubMed] [Google Scholar]
- Zhang F., Hammack C., Ogden S.C., Cheng Y., Lee E.M., Wen Z., Qian X., Nguyen H.N., Li Y., Yao B. Molecular signatures associated with ZIKV exposure in human cortical neural progenitors. Nucleic Acids Res. 2016;44:8610–8620. doi: 10.1093/nar/gkw765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.-M., Li C.J., Vågbø C.B., Shi Y., Wang W.-L., Song S.-H. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.-B. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J., Szretter K.J., Baker S.C., Barchet W., Diamond M.S. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]