Abstract
The lack of a cell culture system to support hepatitis C virus (HCV) replication has hampered studies of this frequent cause of chronic liver disease. However, pseudotyped retroviral particles (pp) bearing the HCV envelope glycoproteins have provided a different approach to HCV studies. We used genotype 1a pp to detect neutralizing antibodies (NtAb) in eight chimpanzees and four humans infected with 1a strains, and developed pp of genotypes 2a, 3a, 4a, 5a, and 6a to study crossreactivity. NtAb was detected in one of four chimpanzees and none of three humans with acute resolving infection, suggesting that NtAb is not required for HCV clearance. NtAb were detected at high titer in two of four chimpanzees and, in Patient H, all with persistent infection; responses paralleled humoral responses to envelope 1 and 2 proteins and, in some cases, correlate also with antibodies to the hypervariable region 1, previously thought to be the primary site of neutralization. NtAb raised during 1a infections could neutralize HCVpp of genotypes 4a, 5a, and 6a but had only limited reactivity against 2a and 3a. The detection of high-titer NtAb with cross-genotype reactivity has important implications for the development of active and passive immune-prophylaxis strategies against HCV. Finally, we found that HCVpp infectivity was enhanced by human or chimpanzee sera; apolipoprotein C1 alone or as a component of high-density lipoproteins caused this enhancement. Future studies of the in vivo role of apolipoprotein C1 might provide additional insights into the infection process of HCV.
Keywords: neutralizing antibodies, high-density lipoproteins
Hepatitis C virus (HCV) is a major cause of chronic liver disease worldwide. The HCV genome encodes two envelope (E) glycoproteins, E1 and E2, that associate in the endoplasmic reticulum of the host cell to form heterodimers (1). Numerous studies suggest that these oligomers might comprise the native virion-associated envelope that interacts with the cell receptor during virus infection (1).
The mechanisms that determine the outcome of HCV infection are not well understood, although there is growing evidence that cellular immune responses play an important role (reviewed in ref. 2). The role of the humoral immune response is less well defined. Nevertheless, it has been shown that an antibody response against the envelope proteins, in particular the hypervariable region 1 (HVR1) of E2, correlated with viral clearance (3, 4). In vitro and in vivo studies demonstrated that certain antibodies neutralized HCV and/or blocked attachment of HCV to target cells and prevented infection (reviewed in ref. 5). Recently, the construction of infectious HCV pseudotyped retroviral particles (pp), bearing intact E1 and E2 proteins, was reported (6, 7). Using the HCVpp assay developed by Bartosch et al. (6), we previously identified HCV-specific neutralizing antibodies (NtAb) in sera from persistently infected humans and chimpanzees and showed that they could neutralize HCVpp representing genotypes 1a and 1b, respectively (5). Similar findings subsequently were obtained by Logvinoff et al. (8) by using the assay developed by Hsu et al. (7). In this study, we analyzed sera for antibodies against E1, E2, and HVR1 in consecutive samples collected from HCV-infected chimpanzees and a human and compared the kinetics of their appearance to that of NtAb. In addition, we extended our study of crossreactivity of NtAb to include pp representing genotypes 2a, 3a, 4a, 5a, and 6a.
Materials and Methods
Serum/Plasma Samples. Samples were obtained from four patients with posttransfusion hepatitis C, genotype 1a. Informed consent was obtained from the patients. Three patients had acute resolving infection. The fourth, Patient H, became persistently infected with HCV; strain H77 was obtained from this patient during the acute phase of the infection (9).
Five chimpanzees (Ch1530, Ch1494, Ch1558, Ch1590, and Ch96A008) were experimentally infected with HCV strain H77. Three additional chimpanzees (Ch1422, Ch1581, and Ch1579) were infected with HCV strain HC-TN of genotype 1a that originated from a patient with fulminant HCV infection (10). Chimpanzees 1530, 1581, 1558, and 1590 developed a chronic infection (2, 5). It should be noted that Ch1581 was transiently depleted of CD4+ T cells at week 53 (unpublished data). Chimpanzees 1422, 1494, 1579, and 96A008 resolved their infections (refs. 2 and 5 and unpublished data). The housing, maintenance, and care of the chimpanzees met or exceeded all relevant guidelines and requirements.
HCV RNA Detection and Quantification. Total RNA extracted from 100 μl of serum was tested for HCV RNA in RT-PCR by amplification of the 5′ untranslated region with nested primer pairs (11). The genome titer was determined by using the AMPLICOR HCV Monitor version 2.0 test (Roche Diagnostics).
HCV Antibody Testing. Chimpanzee sera were analyzed for antibodies against core and nonstructural proteins with the Abbott HCV EIA-2, a second-generation ELISA.
Sera collected from chimpanzees were analyzed for anti-E1 by an in-house ELISA by using a secreted truncated form of the E1 protein of strain H77 (12) (amino acids 192–329) in which the signal sequence had been replaced by the signal sequence of murine Ig κ-chain V-J2-C and a polyhistidine tail had been added at the C terminus. Huh-7 cells were infected with a recombinant vaccinia virus (Copenhagen strain) expressing the E1 protein. The E1 protein was purified from the medium by using the TALON Metal Affinity Resin (Clontech) according to the manufacturer's instructions and characterized by immunoprecipitation with an anti-E1 monoclonal antibody. Serum (1:50 dilution), tested by ELISA by using standard procedures, was considered positive when the signal to cut-off was ≥1. The cutoff of each test was calculated as four times the mean OD obtained with individual sera collected from 13 naïve chimpanzees.
Anti-E2 in a 1:100 dilution of serum was detected by ELISA based on a truncated recombinant E2 protein of strain H77 (amino acids 388–664; differs at six amino acid sites from consensus H77 sequence) (13). Every test plate included three negative controls, consisting of a serum pool from seven naïve chimpanzees, and a serial dilution of an anti-E2 positive chimpanzee serum. The cutoff point for each test was set at the negative control mean OD plus the y intercept of the straight line derived from dilutions of the positive control. This cutoff was about six times the negative mean.
Anti-HVR1 was detected by ELISA based on a synthetic 21-mer biotinylated peptide representing a truncated form (amino acids 390–410) of HVR1 of strain H77 (12) or a synthetic 27-mer biotinylated peptide representing the complete sequence of HVR1 (amino acids 384–410) of strain HC-TN. A 1:100 dilution of serum was tested by using standard procedures with streptavidin-coated microplates (Pierce). A cutoff was generated for each test plate based on a standard curve and the three negative controls described above. Serial 2-fold dilutions of positive-control chimpanzee serum yielded a straight line, the y intercept of which, plus the mean negative control value, represented the positive cutoff point.
Neutralization Assay By Using Retroviral HCVpp. The pp were produced as described in refs. 5 and 6. All procedures were performed in the presence of 5–10% FCS. Test sera from humans and chimpanzees were incubated for 1 h at room temperature with HCVpp, added to Huh-7 cells, and incubated at 37°C. Supernatants were removed after ≈8 h, and cells were incubated in DMEM/10% FCS for 72 h at 37°C. GFP-positive cells were quantified by FACS analysis. The percentage of neutralization for each chimpanzee sample (dilution: 1/50) was calculated by comparison with a preinoculation serum from the same animal. For human sera, the percentage of neutralization was calculated by comparison with the mean of three normal human sera. Neutralization titers were determined by serial 2-fold dilutions of sera in DMEM, followed by incubation with the HCVpp. Each dilution was compared with the equivalent dilution of normal serum. Neutralization was defined as ≥50% reduction of the number of GFP-positive cells.
To produce the pp of genotypes 2–6, we replaced the HCV sequence of phCMV-7a (6) with that of the C-terminal core and the entire E1 and E2 genes from HCV isolates representing the consensus sequence of these genotypes (Fig. 9, which is published as supporting information on the PNAS web site). For the 2a construct [pCMV-J6CF(2a)], the HCV sequence of the infectious clone pJ6CF was used (14). For the 3a [pCMV-S52(3a-11)], 4a [pCMV-ED43(4a-1)], 5a [pCMV-SA13(5a-12)], and 6a [pCMV-HK(6a-2.1)] constructs, the consensus sequence obtained from the acute phase chimpanzee plasma pools containing HCV strains S52, ED43, SA13, and HK-6a, respectively (refs. 11, 15, and 16 and unpublished data) were used.
Methods and Reagents Used in the Analysis of Enhancement of HCVpp Infectivity. To determine the percentage of enhancement of pp infectivity for Huh-7 cells resulting from incubation with serum samples from different animal species, pp were incubated 45 min at room temperature with the test serum (concentration 1/50) before infection of Huh-7 cells. Percentage of enhancement was calculated by comparison with infection with pp similarly incubated but without the addition of test serum. To identify the factors involved with enhancement, we first subtracted albumin, transferrin, α1 anti-trypsin, haptoglobin, and IgG from human sera by using an immunosubtraction column (Supporting Materials and Methods, which is published as supporting information on the PNAS web site). Fractionations of the subtracted sample were next performed by HPLC and subsequent protein analysis was performed by nanoflow liquid chromatography-tandem mass-spectrometry (Supporting Materials and Methods).
Lipoproteins and apolipoproteins were obtained from Athens Research and Technology (Athens, GA). Monoclonal and polyclonal antibodies against ApoC1 were obtained from BioDesign (Kennebunk, ME) and Abcam (Cambridge, U.K.), respectively. Briefly, pp were incubated 45 min at room temperature with lipoproteins or apolipoproteins at the concentration found in normal plasma (or as indicated) before infection of Huh-7 cells. Antibodies against ApoC1 (concentration 1/50) were preincubated with lipoproteins or apolipoproteins for 45 min at room temperature before incubating the mixture with pp for 45 min at room temperature and infection of Huh-7 cells.
Results
Sera collected throughout acute or chronic infections with genotype 1a (strains H77C and HC-TN) in chimpanzees (Figs. 1 and 2) and Patient H (Fig. 3) were examined for anti-E1 and anti-E2 with H77-derived antigens. Because the E2 protein contained a mutated HVR1 sequence, the anti-HVR1 ELISA test was based on a consensus H77 peptide and/or an HC-TN peptide. Sera were tested for NtAb by the neutralization assay developed by Bartosch et al. (6) with ppH77(1a); the assay had excellent intertest reproducibility when performed at the National Institutes of Health (Fig. 4). We confirmed that HCVpp were neutralized by antibodies, rather than inactivated nonspecifically, because IgG purified from a serum found to neutralize pp had neutralizing activity equivalent to that of the unfractionated serum (data not shown).
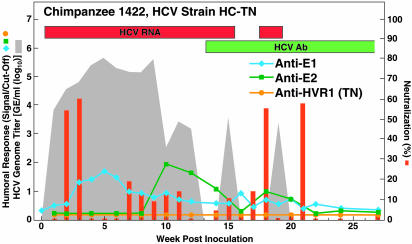
Fig. 1.
Humoral immune response in one chimpanzee (Ch1422, strain HC-TN) with acute resolving HCV infection that developed NtAb. Qualitative detection of HCV RNA in RT-nested PCR is indicated (on top) by red bars. Detection of antibody to core and nonstructural proteins by using the anti-HCV 2.0 assay is indicated (on top) by green bars. The shaded area indicates HCV genome titers as determined by Roche Monitor 2.0; values below the detection limit of ≈500 genome equivalents per ml are shown as 0. The blue, green, and orange lines and the red bars correspond to levels of anti-E1, anti-E2, anti-HVR1, and NtAb, respectively. Percentage of neutralization was calculated by comparison with a preinoculation sample from the same animal. Neutralization values ≤1 are indicated as equal to 1.
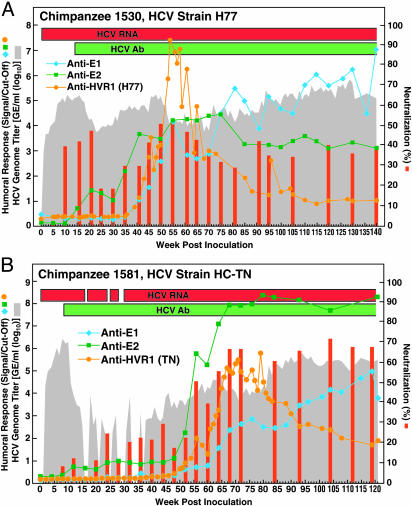
Fig. 2.
Humoral immune response in two chronically HCV-infected chimpanzees that developed NtAb. (A) Ch1530, strain H77. (B) Ch1581, strain HC-TN. See also Fig. 1 legend.
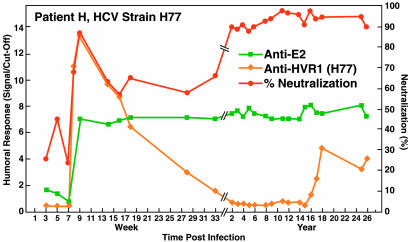
Fig. 3.
Humoral immune response in Patient H, who developed a chronic HCV infection with genotype 1a (strain H77). Because the patient had been vaccinated against smallpox, we could not test for anti-E1 (see Materials and Methods). Because of suboptimal storage of samples, most genome titers could not be determined. However, recent samples contained ≈106 genome copies per ml, which is ≈1.5 logs lower than the peak titer during the acute infection. Samples were not available between week 33 and year 2. See also Fig. 1 legend.
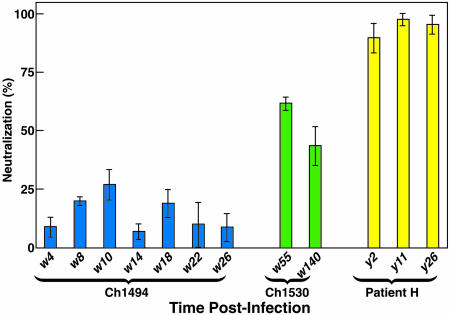
Fig. 4.
Interassay reproducibility of neutralization assay. Result of four independent neutralization tests with sera obtained from Ch1494, Ch1530, and Patient H at the indicated weeks/years after infection. Results are expressed as the average of percentages of neutralization ± SD relative to incubation with the corresponding preinoculation serum for chimpanzees and with three normal human sera for Patient H.
Lack of Correlation Between Outcome and Presence of NtAb. All samples in a series were tested for NtAb against ppH77(1a) in the same test. Data for animals negative for NtAb are in Figs. 10 and 11. which are published as supporting information on the PNAS web site. NtAb were detected in one of four chimpanzees with resolving infection (Fig. 1), and in two of four chimpanzees with chronic infection (Fig. 2). Among five chimpanzees without detectable NtAb, the three with resolving infection did not develop anti-E1 or anti-E2 antibodies (Fig. 10), whereas the two with persistent infection developed anti-E2 late in the infection (Fig. 11). In the one chimpanzee, Ch1422, which cleared HCV and produced NtAb, low-titer NtAb was detected at weeks 2 and 3 but not at week 1, indicating that this NtAb was not passively acquired (Fig. 1). Transient anti-E1 and anti-E2 responses were also detected in this chimpanzee. In the two persistently infected chimpanzees with neutralizing activity, significant titers of NtAb did not develop until after more than one year of followup (Fig. 2 and Table 1), and the virus titer remained relatively high in both animals. In Ch1530 (Fig. 2A and Table 1), the NtAb response paralleled most closely that for anti-E2. A second peak of NtAb correlated also with an H77-specific anti-HVR1 response. Anti-E1 was not detected until week 41, and its titer increased over time. The first and second peaks of NtAb coincided with decreases of ≈1.5 log10 and ≈1 log10 in the HCV genome titer, respectively.
Table 1. Reciprocal titers of NtAb, anti-E1, anti-E2, and anti-HVR1 in persistent HCV genotype 1a infections.
| Patient H
|
Ch1581
|
Ch1530
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Week 9 | Month 8 | Year 2 | Year 26 | Week 20 | Year 1 | Year 2 | Week 21 | Year 1 | Year 3 | |
| NtAb | 200 | 100 | 400 | 3,200 | <50 | 50 | 800 | 50 | 400 | ≈50 |
| Anti-E1 | ND | ND | ND | ND | <50 | <50 | 1,600 | <50 | 200 | 1,600 |
| Anti-E2 | 2,000 | 2,000 | 128,000 | 512,000 | <100 | 100 | 3,200 | 100 | 12,800 | 6,400 |
| Anti-HVR1 | 1,000 | 100 | <100 | 100 | <100 | <100 | <100 | <100 | 3,200 | 400 |
Patient H and Ch1530 were infected with strain H77, and Ch1581 was infected with strain HC-TN. NtAb titers were determined by serial 2-fold dilutions of chimpanzee or human sera and subsequent incubation with ppH77(1a). Value for neutralization of ppH77(1a) with sample collected from Ch1530 at year 3 is indicated as ≈50 because it was repeatedly detected either slightly more or less than 50. Anti-E1, anti-E2, and anti-HVR1 titers were determined by testing serial 2-fold and/or 10-fold dilutions of chimpanzee or human sera by ELISA with H77-specific antigens. ND, not determined.
In Ch1581, NtAb developed only during the chronic phase, at week 55 and thereafter, and the humoral response was somewhat different from that observed in Ch1530 (Fig. 2B and Table 1); we detected anti-E1, anti-E2 and anti-HVR1 (HC-TN specific) responses later in this animal. This animal did not develop H77-specific anti-HVR1, so the HC-TN specific anti-HVR1 antibodies would not have contributed to the neutralization of ppH77(1a) used in our test.
In an attempt to evaluate the relevance of the NtAb responses in chimpanzees, we examined the responses of selected humans with posttransfusion HCV genotype 1a infection. NtAb against ppH77(1a) was not detected in the three patients with acute resolving infection (data not shown). However, in Patient H, who developed a persistent infection, we detected NtAb against ppH77(1a) during weeks 8 and thereafter (peak at week 9), followed by an initial decrease (Fig. 3). However, at years 2–26, NtAb increased to high levels (almost 100% neutralization). NtAb titers obtained by testing dilutions of selected sera indicated that the reciprocal titers increased over time and reached 3,200 at year 26 (Table 1). Despite the high titer of NtAb, the genome titer in Patient H has remained at ≈106 genome equivalents per ml (data not shown). The pattern of NtAb generally paralleled the anti-E2 response, but the NtAb response detected in the acute phase sera correlated with an H77-specific anti-HVR1 response (Fig. 3 and Table 1).
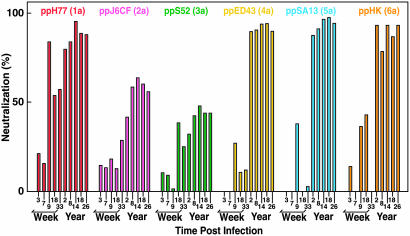
Evidence of Cross-Genotype Neutralization. Acute-phase sera from Patient H, which neutralized ppH77(1a), did not demonstrate significant neutralization of pp-bearing HCV glycoproteins of other major genotypes (Fig. 5). In contrast, chronic phase sera strongly neutralized the pp of genotypes 1a, 4a, 5a, and 6a but demonstrated more limited neutralization of the pp of genotypes 2a and 3a (Fig. 5 and Table 2). Sera taken at year 2 from Ch1581, infected with the HC-TN strain of genotype 1a, neutralized pp of genotype 1a (strain H77) and genotypes 4a, 5a, and 6a (at lower titers) but not genotypes 2a or 3a (Table 2). Sera taken at years 1 and 3 from Ch1530 neutralized the pp of genotypes 1a and 4a but not 2a, 3a, 5a, or 6a (Table 2). Cross-neutralization was not detected with acute-phase serum from Ch1530. However, low-titer cross-neutralization of pp of genotype 3a was repeatedly detected with serum from Ch1422 taken at week 18 (Table 2).
Fig. 5.
Cross-neutralization of different HCVpp genotypes with sera obtained from Patient H at the indicated time points. pp were from genotype 1a (H77), 2a (J6CF), 3a (S52), 4a (ED43), 5a (SA13), and 6a (HK). The percentage of neutralization of the genotype 2a pp with serum from patient H taken at year 2 ranged from 41 to 52 in repeat assays.
Table 2. Reciprocal NtAb titers against homotypic and heterotypic pp in HCV genotype 1a infections.
| Patient H
|
Ch 1530
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Week 9 | Year 2 | Year 26 | Ch1581, year 2 | Week 21 | Year 1 | Year 3 | Ch1422, year 18 | |
| ppH77(1a) | 200 | 400 | 3,200 | 800 | 50 | 400 | ≈50 | 50 |
| ppJ6CF(2a) | <50 | ≈50 | 100 | <50 | <50 | <50 | <50 | <50 |
| ppS52(3a) | <50 | <50 | <50 | <50 | <50 | <50 | <50 | 50 |
| ppED43(4a) | <50 | 400 | 6,400 | 50 | <50 | 100 | 50 | <50 |
| ppSA13(5a) | <50 | 800 | 6,400 | 200 | <50 | <50 | <50 | <50 |
| ppHK(6a) | <50 | 800 | 6,400 | 200 | <50 | <50 | <50 | <50 |
Titers were determined by serial 2-fold dilutions of chimpanzee or human sera from genotype 1a infection and subsequent incubation with the indicated HCVpp. Values for neutralization of ppH77(1a) with sample collected from Ch 1530 at year 3 and cross-neutralization of ppJ6CF(2a) with sample collected from patient H at year 2 are indicated as ≈50 because it was repeatedly detected either slightly more or less than 50.
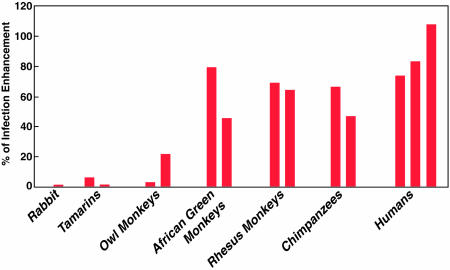
Human and Chimpanzee Sera Enhance Infectivity of HCVpp. When ppH77(1a) were preincubated with preinoculation sera from the eight chimpanzees tested in this study, the number of Huh-7 cells infected was enhanced by 40–70% compared with the number observed without preincubation of pp with chimpanzee serum (Fig. 6 and data not shown). An equivalent or greater enhancement was detected with normal human sera (Fig. 6). ppH77(1a) infectivity was also enhanced by sera from Old World primates but not by sera from New World primates or by rabbit or mouse sera (Fig. 6 and data not shown). Similar enhancement of infectivity by one of the human sera (range 70–130%) was observed for pp of genotypes 1a, 2a, 3a, 4a, and 5a (pp of genotype 6a not tested). In contrast, the sera tested did not enhance infectivity for Huh-7 cells when pp were enveloped with the feline virus RD114 glycoproteins instead of HCV envelope glycoproteins (data not shown).
Fig. 6.
Infectivity-enhancing activity of sera from different animal species. Shown are the results of a single neutralization assay of ppH77(1a). Percentage of enhancement was calculated by comparison with pp infection without preincubation with the indicated serum.
Enhancement of viral infectivity by different mechanisms has been described for several other viruses: antibody-dependent complement-independent enhancement for Dengue virus, complement-dependent antibody-independent enhancement for simian immunodeficiency virus or antibody-dependent complement-dependent enhancement for several members of the Flaviviridae family of viruses (reviewed in refs. 17–19). None of these mechanisms seemed to be involved in the enhancement of HCVpp infectivity, because the infectivity of pp was enhanced with chimpanzee serum but not with derivative IgG and, furthermore, heat-treated human serum (30 min at 65°C) still enhanced, suggesting that complement also was not involved.
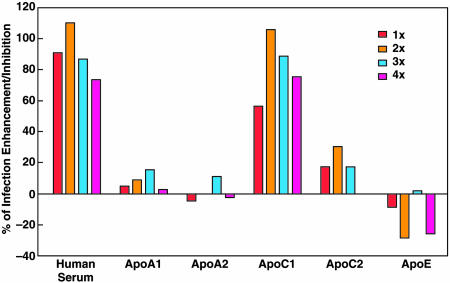
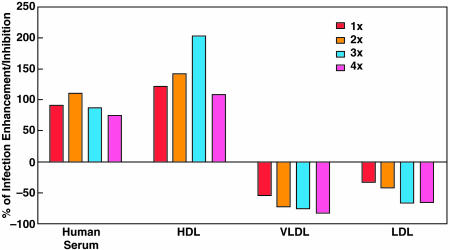
HCVpp Infectivity Is Enhanced by ApoC1. We found that a human serum sample depleted of five major proteins (IgG, α-1 antitrypsin, haptoglobin, albumin, and transferrin) enhanced the infectivity of ppH77(1a) by ≈50%. The subtracted serum was subjected to consecutive rounds of fractionation by HPLC, and the fraction showing the strongest enhancing activity was analyzed by mass spectrometry. Most of the obtained peptide sequences were related to apolipoproteins (data not shown). Therefore, we examined the ability of different apolipoproteins to enhance the infectivity of ppH77(1a) for Huh-7 cells (Fig. 7). We did not detect significant enhancing activity after preincubation of pp with ApoA1, ApoA2, ApoC2, or ApoE. However, infectivity was enhanced with ApoC1, at a level equivalent to that detected with human sera. ApoC1 is associated with lipoproteins. Thus, we next examined the ability of high density lipoproteins (HDL), low-density lipoproteins, and very low-density lipoproteins to enhance the infectivity of ppH77(1a) for Huh-7 cells (Fig. 8). When pp were preincubated with HDL, the percentage of enhancement was equal to or greater than when pp were preincubated with human sera. In contrast, low-density lipoproteins and very low-density lipoproteins inhibited pp infectivity. Preincubation of human sera, HDL, or ApoC1 with monoclonal or polyclonal anti-ApoC1 antibodies, before incubation with pp, abrogated the enhancement of HCVpp infectivity (data not shown). Thus, ApoC1, alone or through HDL, appears to play a critical role in the enhancement of HCVpp infectivity for Huh-7 cells.
Fig. 7.
Influence of apolipoprotein on ppH77(1a) infection of Huh-7 cells. A normal human serum sample was tested in the same assay. 1× represents the concentration of serum usually used in pp assay (1:50) or concentration of apolipoprotein in normal sera at 1:50 dilution. Pseudotyped virus was preincubated with serum or apolipoprotein for 45 min at room temperature before infection of Huh-7 cells.
Fig. 8.
Influence of lipoproteins on ppH77(1a) infection of Huh-7 cells. See also Fig. 7 legend.
Discussion
The pp assay, using infectious retroviral pseudoparticles displaying intact E1 and E2 proteins of HCV (6, 7), provides a practical way to identify NtAb to HCV. Antibodies that had neutralized infectivity of clinical isolates of HCV for chimpanzees or for lymphocytes similarly neutralized HCVpp (5, 20), thus validating this assay as a surrogate assay for the very expensive chimpanzee model or for the insensitive lymphocyte assays. Herein, we have used the pp assay to show that viral clearance in acute HCV infections was not correlated with the development of NtAb. However, high titers of NtAb against the homotypic 1a pp, and also in some instances against pp of other genotypes, in particular genotypes 4a, 5a, and 6a, were found in some humans and chimpanzees with persistent infections. In addition, we found that ApoC1 enhanced pp infectivity for Huh-7 cells, a discovery that may shed light on the infection process of HCV.
Neutralization of pp in our hands was mediated by IgG, as also shown by others (8, 20). However, additional factors might influence the assay. It is possible that IgM, not IgG, caused the low level of neutralization observed during acute-phase infection in two chimpanzees and in Patient H. It is also possible that some NtAb are not detected in the neutralization assay because they are already bound to circulating virions in the patient (21).
Resolution of an HCV infection is related to host intrahepatic CD4+ and CD8+ T-cell responses (2), and antibodies against the HCV envelope glycoproteins are not always detected in chimpanzees or humans who clear the virus (22–25). However, it has also been shown that a humoral immune response against HCV envelope glycoproteins, in particular against the HVR1, correlated with the clearance of this virus (3, 4). It is often not possible to obtain sufficiently early and frequent samples from humans to study the temporal course of viremia, cell-mediated immune responses, and antibody responses and, therefore, experimentally infected chimpanzees offer a significant advantage for studying HCV. Logvinoff et al. (8) found that none of three chimpanzees with acute resolving infection developed NtAb. Similarly, we found that only one of four chimpanzees with acute resolving genotype 1a infection developed NtAb against a genotype 1a pp. We did detect a transient low-titer NtAb response in one chimpanzee (Ch1422) that resolved the infection, but it was not clear whether the NtAb affected the outcome of this infection. In contrast, we previously demonstrated that these same four animals had significant cell-mediated immune responses during the acute infection (ref. 2 and unpublished data). We also did not detect NtAb against the genotype 1a pp in three posttransfusion hepatitis patients with acute resolving genotype 1a infection. Overall, therefore, the cellular immune response, and not NtAb, appears to play the more important role in the resolution of an acute HCV infection.
NtAb developed primarily during chronic infection, and in some cases, continued to rise in titer many years after the onset of infection. This delayed enhancement of B cell-mediated NtAb responses might represent a compensatory response to a deficit of T cell responses during chronic HCV infection, as suggested by Klenerman et al. (26). Clearly, these antibodies were incapable of inducing full viral clearance. Nonetheless, they might play a role in containing the level of viremia within a narrow range as is characteristic of some chronic HCV infections, and antibody-bound virus may be less capable of infecting others. Logvinoff et al. (8) detected NtAb in six of seven chimpanzees that were persistently infected with HCV strain H77. Among the three animals that became persistently infected with strain H77 in our study, only one developed NtAb against ppH77(1a). The two animals that did not develop NtAb also did not appear to have significant intrahepatic cellular immune responses during the acute infection (2). Thus, we cannot explain why the viral titers remained at relatively low levels in these animals. In Ch1581, infected with another genotype 1a strain (HC-TN), NtAb against ppH77(1a) were slow to develop, but NtAb titers increased over time. This chimpanzee had significant intrahepatic cellular immune responses during the first year of followup (ref. 2 and unpublished data) and, coinciding with these responses, there was effective viral control initially. However, during the period of rising NtAb titers in this animal, virus titers actually increased. Similarly, relatively high genome titers persisted in Patient H despite the presence of very high titers of NtAb. Thus, we could not establish any clear correlation between NtAb status and the overall virus titer in persistent HCV.
We previously found that high titer NtAb from Ch1581 and Patient H, infected with genotype 1a, could cross-neutralize pp-bearing glycoproteins of a genotype 1b strain (5). In this study, we examined the ability of NtAb from 1a infections to cross-neutralize pp of other major genotypes (Table 2). The patterns were not straightforward. The titer of cross-neutralizing antibodies against pp 4a, 5a, and 6a in sera collected from Patient H during the chronic phase was at least as high as the homologous neutralization titer, whereas the titer against 4a, 5a, and 6a pp in sera collected from Ch1581 was lower than the homologous titer. Surprisingly, although NtAb from Ch1530, collected even during the chronic phase, did not cross-neutralize pp of another subtype (1b) within the same genotype (5), it did cross-neutralize pp of genotype 4a. In contrast, pp of genotypes 2a and 3a were only minimally neutralized by NtAb induced by 1a infections. The NtAb that developed in Patient H during the acute phase of infection did not cross-neutralize the pp of other genotypes. Thus, it appears that the crossreactivity of NtAb may broaden over time in HCV infections. This cross-genotype reactivity was unlikely to be the result of new infections in Patient H, who was infected at the time of transfusion and had no other known exposures, or in the chimpanzees that were maintained under experimental conditions. NtAb that developed in genotype 1a infections in different hosts had differing degrees of crossreactivity, suggesting that different epitopes in the envelope proteins were targeted. Finally, our data indicated that genotypes 1, 4, 5, and 6 are serologically more closely related, with a more distant serologic relationship to genotypes 2 and 3.
The epitopes responsible for cross-neutralization remain to be identified. The HVR1 sequence (amino acids 384–410) of the 2a, 3a, 4a, 5a, and 6a pp differed from the 1a sequence at 12–16 amino acid positions (Fig. 9). In the putative principal neutralization epitope in HVR1 (amino acids 396–407) (7) the 2a, 3a, 4a, 5a, and 6a pp differed from the 1a sequence at six to eight amino acid positions (Fig. 9). Thus, it is unlikely that the observed cross-neutralization was against HVR1. In another putative neutralization epitope identified in E2 (amino acids 432–447) (7) the number of amino acid differences between 2a, 3a, 4a, 5a, and 6a sequences and the 1a sequence was 6, 5, 4, 5, and 9, respectively (Fig. 9). Thus, it also appears unlikely that the observed crossreactivity was against this E2 epitope. Therefore, as we concluded previously (5), one or more additional neutralization epitopes appear to exist within the HCV glycoproteins. However, an inspection of the pp sequences did not readily identify amino acids conserved among genotypes 1a, 4a, 5a, and 6a sequences but not 2a and 3a sequences (Fig. 9) that could explain the crossreactivity of genotype 1a NtAb against pp of genotypes 4a, 5a, and 6a.
Infectivity of pp was enhanced by preincubation with serum of Old World nonhuman primates or humans. Consequently, the effect of a given serum on pp infectivity was the result of counter-forces, one enhancing and one neutralizing. To correct for enhancing activity, we normalized the neutralization of postinfection samples to that of a presample, or alternatively, to samples from uninfected individuals. We showed that HDL and ApoC1 could replace human or chimpanzee serum for enhancing the number of Huh-7 cells infected by HCVpp. Moreover, antibodies against ApoC1 dramatically inhibited the enhancing activity of human serum, HDL, and ApoC1. Thus, we have demonstrated that ApoC1 plays a central role in the enhancing activity of human serum, alone or associated with HDL. In plasma, this protein is found not only associated with HDL but also with low-density lipoproteins and very low-density lipoproteins. The difference we observed between these lipoproteins for the enhancement of HCVpp infection might be explained by their different roles in the lipid transport system, their respective ratio of apolipoproteins, and their receptor specificities (for review, see refs. 27 and 28). The actual mechanism responsible for the enhancing activity of ApoC1 remains to be determined. However, it should be noted that we, and others, have recently demonstrated that HDL can facilitate HCVpp entry through the scavenger receptor class B type 1, a putative HCV receptor (ref. 29 and unpublished data).
In conclusion, our study demonstrates the complexity of mechanisms influencing HCV entry into cells as measured in a highly reproducible retroviral pseudoparticle system. Although NtAb does not appear to play a significant role in the outcome of acute HCV infection, the detection of cross-reactive NtAb during persistent infections has important implications for the development of active and passive immune-prophylaxis strategies against this important human pathogen. In the final analysis, it will require further studies to determine whether the observed cross-genotype reactivity of NtAb in the pp assay translates into cross-genotype neutralization in vivo. The role of NtAb in influencing the course of chronic hepatitis C remains unknown. It is possible, but not yet demonstrated, that a reduction in the level of NtAb during immunosuppression contributes to the accelerated course of liver disease in the transplant setting.
Supplementary Material
Acknowledgments
We thank Dr. Larry Lantz (National Institute of Allergy and Infectious Diseases Custom Antibodies Services Facility, National Institutes of Health) for performing IgG purifications, Dr. Patrizia Farci (University of Cagliari, Cagliari, Italy) for providing human serum samples, Dr. Jean Dubuisson (Institut de Biologie de Lille, Lille, France) for providing the E1 expression vector pTMI, Dr. Bernard Moss (National Institute of Allergy and Infectious Diseases, National Institutes of Health) for providing vaccinia viruses, Dr. Isa Mushahwar (Abbott) for providing recombinant E2 protein, and Dr. Harry Greenberg (Stanford University, Stanford, CA) for providing the A4 monoclonal antibody.
Author contributions: J.-C.M., S.U.E., R.H.P., and J.B. designed research; J.-C.M., R.E.E., K.F., M.Z., and J.B. performed research; B.B., H.A., and F.-L.C. contributed new reagents/analytic tools; J.-C.M., R.E.E., S.U.E., R.H.P., and J.B. analyzed data; and J.-C.M., H.A., S.U.E., R.H.P., and J.B. wrote the paper.
Abbreviations: E, envelope; HCV, hepatitis C virus; HDL, high-density lipoproteins; HVR1, hypervariable region 1; NtAb, neutralizing antibodies; pp, pseudotyped retroviral particles.
References
- 1.Op De Beeck, A., Cocquerel, L. & Dubuisson, J. (2001) J. Gen. Virol. 82, 2589–2595. [DOI] [PubMed] [Google Scholar]
- 2.Thimme, R., Bukh, J., Spangenberg, H. C., Wieland, S., Pemberton, J., Steiger, C., Govindarajan, S., Purcell, R. H. & Chisari, F. V. (2002) Proc. Natl. Acad. Sci. USA 99, 15661–15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allander, T., Beyene, A., Jacobson, S. H., Grillner, L. & Persson, M. A. (1997) J. Infect. Dis. 175, 26–31. [DOI] [PubMed] [Google Scholar]
- 4.Zibert, A., Schreier, E. & Roggendorf, M. (1995) Virology 208, 653–661. [DOI] [PubMed] [Google Scholar]
- 5.Bartosch, B., Bukh, J., Meunier, J. C., Granier, C., Engle, R. E., Blackwelder, W. C., Emerson, S. U., Cosset, F. L. & Purcell, R. H. (2003) Proc. Natl. Acad. Sci. USA 100, 14199–14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartosch, B., Dubuisson, J. & Cosset, F. L. (2003) J. Exp. Med. 197, 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu, M., Zhang, J., Flint, M., Logvinoff, C., Cheng-Mayer, C., Rice, C. M. & McKeating, J. A. (2003) Proc. Natl. Acad. Sci. USA 100, 7271–7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Logvinoff, C., Major, M. E., Oldach, D., Heyward, S., Talal, A., Balfe, P., Feinstone, S. M., Alter, H., Rice, C. M. & McKeating, J. A. (2004) Proc. Natl. Acad. Sci. USA 101, 10149–10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feinstone, S. M., Alter, H. J., Dienes, H. P., Shimizu, Y., Popper, H., Blackmore, D., Sly, D., London, W. T. & Purcell, R. H. (1981) J. Infect. Dis. 144, 588–598. [DOI] [PubMed] [Google Scholar]
- 10.Farci, P., Munoz, S. J., Shimoda, A., Govindarajan, S., Wong, D. C., Coiana, A., Peddis, G., Rubin, R. & Purcell, R. H. (1999) J. Infect. Dis. 179, 1007–1011. [DOI] [PubMed] [Google Scholar]
- 11.Bukh, J., Apgar, C. L., Engle, R., Govindarajan, S., Hegerich, P. A., Tellier, R., Wong, D. C., Elkins, R. & Kew, M. C. (1998) J. Infect. Dis. 178, 1193–1197. [DOI] [PubMed] [Google Scholar]
- 12.Yanagi, M., Purcell, R. H., Emerson, S. U. & Bukh, J. (1997) Proc. Natl. Acad. Sci. USA 94, 8738–8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lesniewski, R., Okasinski, G., Carrick, R., Van Sant, C., Desai, S., Johnson, R., Scheffel, J., Moore, B. & Mushahwar, I. (1995) J. Med. Virol. 45, 415–422. [DOI] [PubMed] [Google Scholar]
- 14.Yanagi, M., Purcell, R. H., Emerson, S. U. & Bukh, J. (1999) Virology 262, 250–263. [DOI] [PubMed] [Google Scholar]
- 15.Bukh, J., Purcell, R. H. & Miller, R. H. (1993) Proc. Natl. Acad. Sci. USA 90, 8234–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chamberlain, R. W., Adams, N., Saeed, A. A., Simmonds, P. & Elliott, R. M. (1997) J. Gen. Virol. 78, 1341–1347. [DOI] [PubMed] [Google Scholar]
- 17.Takada, A. & Kawaoka, Y. (2003) Rev. Med. Virol. 13, 387–398. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan, N. J. (2001) Curr. Top Microbiol. Immunol. 260, 145–169. [DOI] [PubMed] [Google Scholar]
- 19.Freissmuth, D., Dierich, M. P. & Stoiber, H. (2003) Front. Biosci. 8, S733–S739. [DOI] [PubMed] [Google Scholar]
- 20.Yu, M.-Y. W., Bartosch, B., Zhang, P., Guo, Z. P., Renzi, P. M., Shen, L.-M., Granier, C., Feinstone, S. M., Cosset, F.-L. & Purcell, R. H. (2004) Proc. Natl. Acad. Sci. USA 101, 7705–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hijikata, M., Shimizu, Y. K., Kato, H., Iwamoto, A., Shih, J. W., Alter, H. J., Purcell, R. H. & Yoshikura, H. (1993) J. Virol. 67, 1953–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takaki, A., Wiese, M., Maertens, G., Depla, E., Seifert, U., Liebetrau, A., Miller, J. L., Manns, M. P. & Rehermann, B. (2000) Nat. Med. 6, 578–582. [DOI] [PubMed] [Google Scholar]
- 23.Prince, A. M., Brotman, B., Lee, D. H., Ren, L., Moore, B. S. & Scheffel, J. W. (1999) J. Infect. Dis. 180, 987–991. [DOI] [PubMed] [Google Scholar]
- 24.Bassett, S. E., Thomas, D. L., Brasky, K. M. & Lanford, R. E. (1999) J. Virol. 73, 1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hadlock, K. G., Gish, R., Rowe, J., Rajyaguru, S. S., Newsom, M., Warford, A. & Foung, S. K. (2001) J. Med. Virol. 65, 23–29. [PubMed] [Google Scholar]
- 26.Klenerman, P., Lechner, F., Kantzanou, M., Ciurea, A., Hengartner, H. & Zinkernagel, R. (2000) Science 289, 2003. [DOI] [PubMed] [Google Scholar]
- 27.Fidge, N. H. (1999) J. Lipid Res. 40, 187–201. [PubMed] [Google Scholar]
- 28.Wang, M. & Briggs, M. R. (2004) Chem. Rev. 104, 119–137. [DOI] [PubMed] [Google Scholar]
- 29.Voisset, C., Callens, N., Blanchard, E., Op De Beeck, A., Dubuisson, J. & Vu-Dac, N. (2005) J. Biol. Chem. 280, 7793–7799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.