Abstract
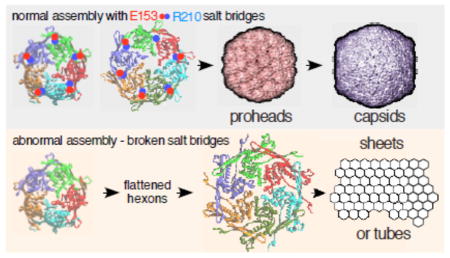
Viruses build icosahedral capsids of specific size and shape by regulating the spatial arrangement of the hexameric and pentameric protein capsomers in the growing shell during assembly. In the T=7 capsids of E. coli bacteriophage HK97 and other phages, sixty capsomers are hexons, while the rest are pentons that are correctly positioned during assembly. Assembly of the HK97 capsid to the correct size and shape has been shown to depend on specific ionic contacts between capsomers. We now describe additional ionic interactions within capsomers that also regulate assembly. Each is between the long hairpin, the “E-loop,” that extends from one subunit to the adjacent subunit within the same capsomer. Glutamate E153 on the E-loop and arginine R210 on the adjacent subunit’s backbone alpha-helix form salt bridges in hexamers and pentamers. Mutations that disrupt these salt bridges were lethal for virus production, because the mutant proteins assembled into tubes or sheets instead of capsids. X-ray structures show that the E153-R210 links are flexible and maintained during maturation despite radical changes in capsomer shape. The E153-R210 links appear to form early in assembly to enable capsomers to make programmed changes in their shape during assembly. The links also prevent flattening of capsomers and premature maturation. Mutant phenotypes and modeling support an assembly model in which flexible E153-R210 links mediate capsomer shape changes that control where pentons are placed to create normal size capsids. The E-loop may be conserved in other systems in order to play similar roles in regulating assembly.
Keywords: bacteriophage assembly, virus capsids, salt bridge, capsid assembly, protein structure
Subject classification: Viruses and bacteriophages
Graphical abstract
Introduction
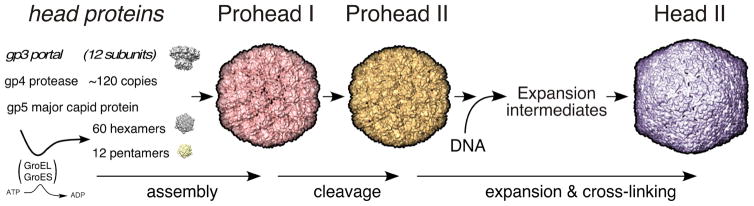
Viral capsids are commonly built using a symmetric icosahedral plan which allows one or only a few proteins to be used over and over to create the protein shell used to protect viral genomes as they travel from one host to the next [1]. These capsids vary widely in size [2] to accommodate viral chromosomes with a broad range of genomic length. However, while each virus usually makes only one appropriate size of capsid, the mechanism by which that capsid size is regulated is poorly understood in most cases. An important aspect of capsid assembly for the tailed phages (Caudoviridae) and herpesviruses (Herpesviridae) is that initial steps produce proheads or procapsids that are obligatory intermediates in an assembly pathway that includes additional maturation steps. Mature capsids are angular, while proheads are more rounded, usually smaller, and contain hexons with distinctly asymmetric shapes. Figure 1 provides an example of this change of capsid shape from prohead to head for Escherichia coli bacteriophage HK97. Capsid protein conformations change radically during maturation causing the shell to become thinner but more stable as the hexons adopt a new, flattened and nearly hexagonal shape [3]. This difference is important because the final geometry and size of a capsid [1] is established at the procapsid stage, when the positions of hexons and pentons in the growing structure are determined. This means that it is the earliest prohead structures that need to be studied in order to understand how capsid size and shape are regulated, although clues may remain in mature capsids. In support of this, we have recently shown that transient inter-capsomer contacts between pairs of residues on the outside of the bacteriophage HK97 prohead (D231 and K178) are essential for correct assembly, even though these same residues move ~20 Å apart in the mature capsid [4]. This was surprising, even though it has long been known from early work on phage T4 [5, 6] that capsid maturation involved large rearrangements of structural elements.
Figure 1. The HK97 capsid assembly pathway.
The HK97 major capsid protein (gp5, also abbreviated “mcp”) folds with the aid of GroEL and GroES and assembles into pentamers and hexamers. These co-assemble with the protease (gp4) and the portal protein (gp3, if present) to form Prohead I. After assembly, the protease removes the N-terminal 102-residue delta domain of gp5 creating the procapsid, Prohead II. The delta domains act like the scaffolding proteins of other phages and Herpesviruses. DNA packaging normally induces maturation to Head II. Maturation includes an expansion in size and conformational changes in all subunits. Stabilization of Head II is aided by the formation of covalent crosslinks between gp5* subunits during expansion.
The HK97 capsid assembly pathway (Figure 1) is well characterized and a near-atomic resolution movie of many parts of that process has been compiled [7, 8]. Especially important are the X-ray structures of early HK97 capsid assembly intermediates, Prohead I at 5.2 Å [9] and Prohead II at 3.7 Å [10], which provide the near-atomic details necessary to formulate hypotheses about how the structures may assemble (Figure 2). Careful analysis of the repertoire of HK97 structures has allowed the identification of specific residues and interactions that play important roles in structural integrity, assembly, maturation and the crosslinking that occurs during maturation[7–16]. Crosslinking only occurs when a triad of adjacent capsomers all adopt an expanded and flattened conformation, bringing together the three residues required for crosslinking [11, 12, 17]. Thus, the formation of crosslinks acts as a local reporter for the expanded conformation for HK97 and allows crosslinking to be used to monitor the course of maturation [7, 8, 13, 16].
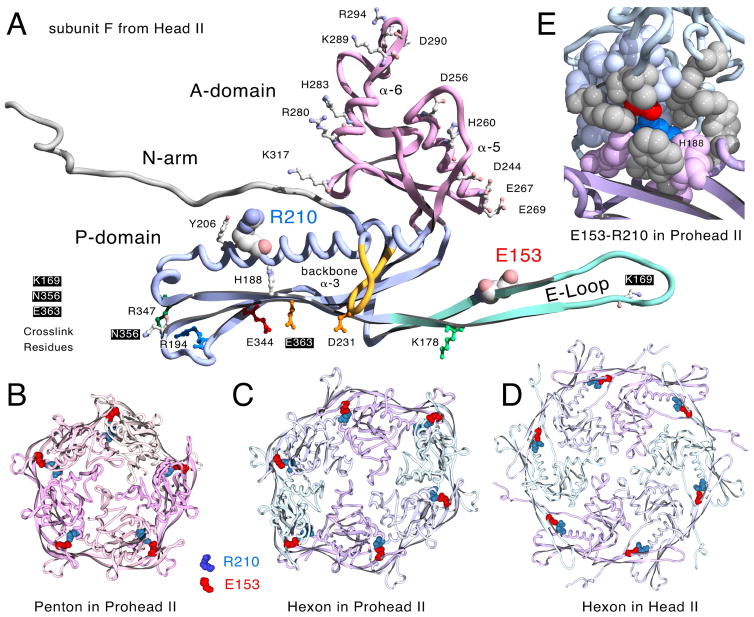
Figure 2. Molecular models displaying the Glu153-Arg210 interaction.
A. One monomer of the HK97 major capsid protein (chain F from HK97 Head II [14], PDB ID: 1OHG) with E153 on the E-loop and R210 on the backbone α-helix in space-filled mode[14]. Residues defining the positively and negatively charged borders of the A-domain, those involved in crosslinking, and other residues mentioned in the text and also shown. B. Prohead II penton (5 G chains from PDB ID: 3E8K, residues 128 to the end). C. Prohead II hexon (chains A – F from PDB ID: 3E8K, residues 128 to the end). D. Head II hexon showing the E153-R210 interaction (PDB ID: 1OHG, residues 128 to the end, but with truncated E-loops matching those from Prohead II (PDB ID: 3E8K)). The E153-R210 interaction is also maintained in expanded pentons (not shown). E. Close-up view revealing the partially buried character of the E153-R210 salt bridge (chains D and E from PDB ID: 3E8K as ribbons) All residues within ~7 Å of E153 and R210 are shown in space-filling mode, including 11 hydrophobic residues in grey (Trp 189, Val 190, Tyr 206, Leu 211, Tyr 239 in chain D and Ala 139, Phe 176, Ile 174, Phe 270, Met 331, Tyr 371 in chain E).
Structure-informed mutational studies have revealed several residues participating in ionic interactions that are important in regulating assembly [18, 19] [12, 20] [4]. We have focused on identifying additional ionic interactions because they are easily identified and because HK97 Prohead I can be disassembled by treatment with high salt [19], suggesting that ionic interactions are important at the beginning of assembly. One intriguing set of ionic interactions identified in X-ray structures of HK97 particles are the salt bridges between glutamic acid 153 (E153) on the E-loop of each major capsid protein monomer and arginine 210 (R210) on the backbone α-helix of the next monomer (counter-clockwise, when viewed from the outside) within both hexons and pentons (Figure 2) [10]. The E153-R210 interactions are found in both proheads [10] and mature heads [16]. Figure 2 shows the arrangement of these salt bridges in Prohead II pentons (panel B) and hexons (panel C), and Head II hexons (panel D). It was suggested earlier that these ionic interactions are important for guiding the E-loops into position to allow cross-linking during the final steps of maturation [10]. However, we suspected that these interactions might also regulate assembly, since they are also present in proheads. Furthermore, the E153-R210 ionic interaction appears relatively well conserved (http://gigapan.com/gigapans/b806a9da80b4a66bcb1dd1b273d5b7a4/), suggesting that these interactions could have general importance. Given the structural and evolutionary conservation of these ionic interactions, we engineered substitution mutations at both E153 and R210 in the HK97 major capsid protein gene to see if disrupting the interactions would reveal more about their roles in HK97 capsid assembly.
Results
The E153-R210 interaction is preserved throughout maturation
We analyzed the available HK97 X-ray models for E153-R210 interactions and found that they were universally present. Clear evidence was found in each of the following: a) the empty mature Head II (PDB ID: 1OHG) [14, 16], b) an incompletely expanded Head in which pentons are slightly recessed (PDB ID: 2FT1) [7], c) Head I, made from a mutant which cannot crosslink and in which pentons are slightly more recessed, PDB ID: 2FS3) [7], d) pepsin-treated Expansion Intermediate IV (PDB ID: 2FSY) [7], similar to Head I, but in which the penton E-loops have been proteolyzed, e) Prohead II (Figure 2 and PDB ID: 3E8K) [10] the normal HK97 procapsid in which the delta domains have been removed, and f) inferred in the 5.2Å Prohead I model (PDB ID: 3QPR) [9], that has no explicit side chains, but which is very similar in structure to Prohead II (~2 Å RMSD). We have illustrated this persistence in a movie (Supplemental Movie S1) which shows the close contacts between E153 and R210 residues in both hexamers and pentamers in the five X-ray structures listed above. The results of this analysis show that the E153-R210 interaction is robust and apparently persists throughout the drastic conformation changes that occur during maturation and thus appears to have an important role in that process. We also found that the local environment of the E153-R210 interaction site seems to be almost completely shielded from exterior solvent (Figure 2E), which could potentially strengthen this ionic interaction. Many of the nearest residues are hydrophobic, including Y371, V190, Y206, F270 and F176, and the positions of these five residues in alignments of related major capsid proteins are mostly occupied by similar hydrophobic residues (http://gigapan.com/gigapans/b806a9da80b4a66bcb1dd1b273d5b7a4/).
Mutants that break the E153 - R210 ionic interaction assemble tubes and sheets of major capsid protein
Amino acid substitutions were made in an expression plasmid containing the HK97 protease and major capsid protein genes (genes 4 and 5 respectively). With no mutations, induction of the capsid protein genes in an expression host makes high yields of HK97 proheads and allows for rapid assays of the effects of the mutations [18, 21]. To disrupt the proposed E153-R210 ionic interaction, we replaced glutamate 153 with a variety of residues: alanine, valine, glutamine and arginine (mutants E153A, E153V, E153Q, and E153R). When tested, all four of the mutated proteins were defective in gene 5 complementation (Table 1), showing that the E153-R210 interaction is apparently essential for producing viable phage.
Table 1.
Complementation of HK97 amber mutant phage by HK97 gene 5 E153 and R210 mutants expressed from plasmids.
| Mutation(s) | Relative EOP for gene 5 amber | Relative EOP for wild-type* |
|---|---|---|
| WT | 1 | 1 |
| E153A | 3 × 10−4 | 0.3 |
| E153V | 3 × 10−4 | 0.3 |
| E153Q | 10−3 | 0.3 |
| E153R | 3 × 10−4 | 0.3 |
| R210E | 10−4 | 0.3 |
| E153R/R210E | 3 × 10−4 | 0.1 |
| E153Q/R210E | < 10−5 | 0.3 |
| E153D | 1 | 1 |
This wild type control was included to test for dominant negative affects. Only very weak effects were observed in these mutants. A strong dominant negative effect would indicate that the mutant protein can poison a wild-type phage infection by interacting assembling with the wild type protein.
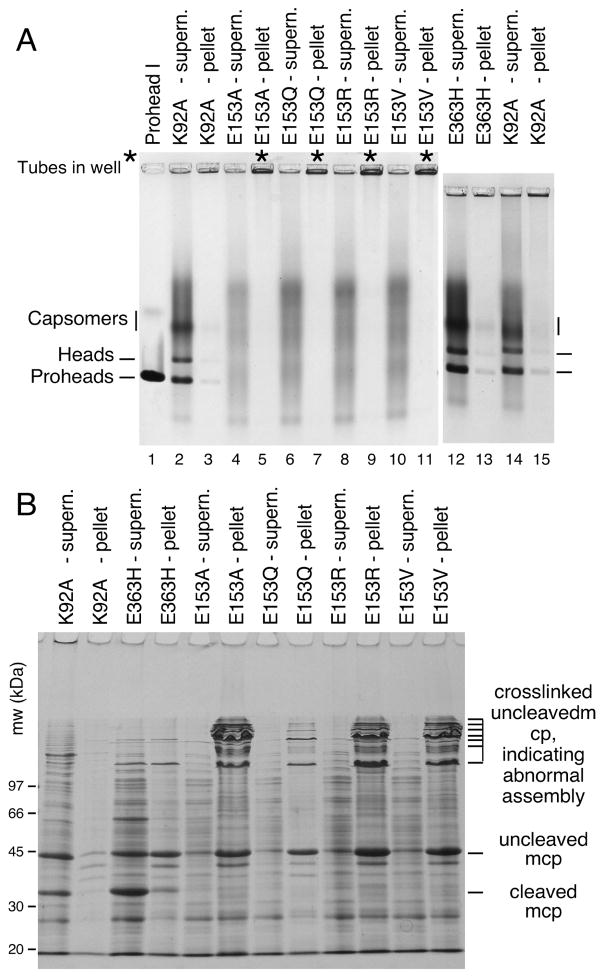
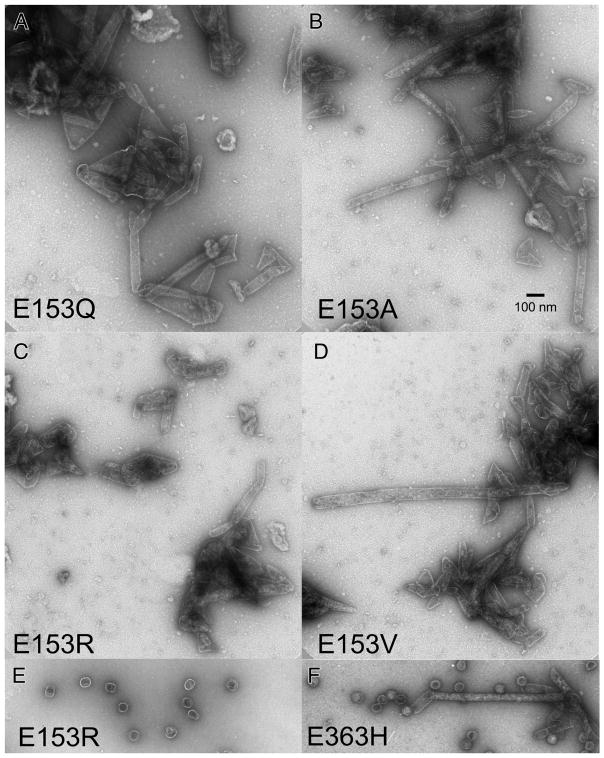
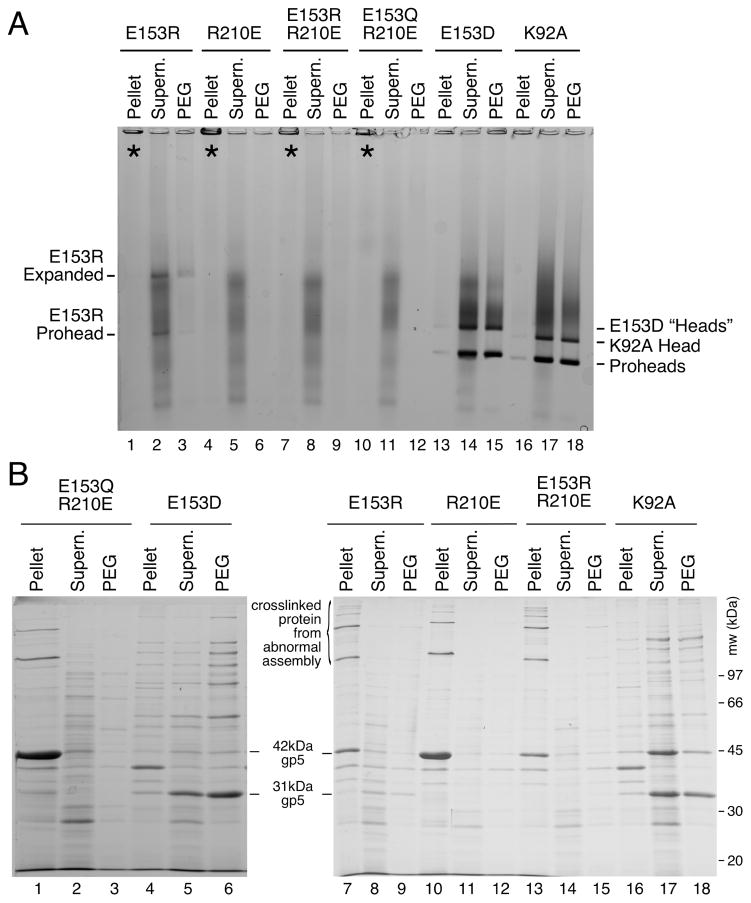
We induced high-level expression of the mutant proteins from plasmids in small cultures and analyzed the proteins and structures produced (Figures 3 and 4). Under these conditions, expression of HK97 capsid components is so efficient, that they can be readily analyzed by electrophoresis of unpurified extracts [4, 22]. To aid in the interpretation of these experiments, we included mutant K92A as a control that yields a wide spectrum of HK97 assembly intermediates to serve as markers for proheads, heads and capsomers. Agarose gel analysis of the four E153 mutants suggested that they were incapable of normal assembly, because no prohead, head or capsomer bands were observed in either the pellet or supernatant fractions (Figure 3A). Intriguingly, a large amount of stained material was retained in the wells of the agarose gel for the pellet fraction of each of the four mutants (marked with asterisks in the Fig. 3A), suggesting that some protein was produced but was perhaps insoluble. SDS-polyacrylamide gels (Figure 3B) confirmed that the bulk of the major capsid protein from the four mutants was in the pellet fractions, where it was present as 42 kDa monomers and as ladders of crosslinked 42 kDa monomers. In normal HK97 assembly, crosslinking only occurs as a late step as Prohead II expands to become Head II [21], well after cleavage of the major capsid protein to 31kDa (Figure 1A), so the presence of crosslinked 42 kDa protein ladders, as observed here, usually indicates that abnormal assembly has occurred [4]. Accordingly, we examined the pellet fractions of the E153 mutants by electron microscopy. The negatively-stained EM images revealed that the mutant proteins had assembled into tubes, or mixtures of tubes and sheets of major capsid protein (Figure 4), explaining why they had become trapped in the cell pellets and appeared to be insoluble. Thus, the four E153 mutants were not completely defective in their ability to assemble, as we had initially inferred. Indeed, assembly in the four E153 mutants occurred very efficiently, but was uncontrolled and produced hexagonal arrays of capsomers that frequently curve into tubes. This clearly demonstrates that the interaction we sought to disrupt is an important component of the assembly process for HK97 capsids and plays a role in regulating how that assembly proceeds. The fact that all of these samples had crosslinked 42 kDa protein ladders (instead of ladders of cleaved, 31 kDa gp5*) also indicates that these structures are in an “expanded” conformation that is more like Head II than Prohead II, since crosslinking can only occur in the expanded state. Proteolytic removal of the delta domain normally precedes expansion, so for these mutants, the conversion to the expanded conformation occurs prematurely. We also compared the tubes made by the E153 mutants to the tubes made by one of the few previously reported tube-producing HK97 mutants, E363H, which changes a capsomer interface residue [12]). The E153 mutant tubes (Figure 4A–4D) were generally wider and more abundant than those produced by the E363H mutant (Figure 4F), which makes mostly proheads [12].
Figure 3. Mutants that disrupt the E153-R210 salt bridge assemble abnormally.
A. Agarose gel analysis. Crude supernatant (supern.), and pellet fractions from autoinduced lysates were run on a 0.9% agarose gel. The E153 mutants showed no distinct bands. Controls K92A and E363H each produced strong proheads and head bands, which are mixtures of Prohead I, Prohead II and the Head II produced by the freeze/thaw step during preparation. Asterisks (“*”) mark the stainable material that appeared in the wells. B. SDS gel analysis. Pellets and supernatants were denatured and analyzed on SDS gels. Control mutants K92A and E363H make Prohead I (PI) and Prohead II (PII), so they produce control bands of 42 kDa uncleaved major capsid protein (in PI) and of 31 kDa cleaved major capsid protein (in PII), and a ladder crosslinked bands induced by the freeze/thaw step during preparation. E153 mutants produced uncleaved major capsid protein and a ladder of high-molecular-weight bands (crosslinked oligomers of uncleaved major capsid protein that indicate that abnormal assembly has occurred).
Figure 4. Electron micrographs of assemblies produced by E153 mutants.
Undiluted pellet fractions from experiments shown in Figure 3 were mixed thoroughly, applied directly onto grids, rinsed, and stained with 1% uranyl acetate. A. E153Q B. E153A C. E153R D. E153V E. Example of the rare proheads found in the PEG fraction of mutant E153R. F. Narrow tubes and proheads produced by mutant E153H.
Sheets of major capsid protein form when the E153 - R210 interaction is inverted or replaced with a hydrogen-bonding pair
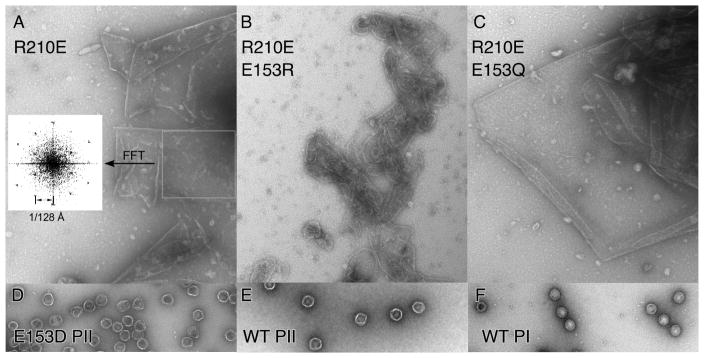
To observe what occurs when the positions of the two residues involved were exchanged, we made the double mutant E153R-R210E. At the same time, we tested whether a related residue pair capable of forming a hydrogen bond (Q-E) might possibly substitute for the E153-R210 pair, by creating an additional double mutant E153Q-R210E. We also made a single substitution, R210E, to include as the necessary control. The other single-mutant controls were included in the first set of E153 mutants (Figure 5C and D). None of the new substitutions were able to form functional phage in complementation tests (Table 1) or able make proheads in our biochemical assays (Figure 5E and F), with the exception of the very few proheads made by E153R (see below). The R210E mutation by itself had the interesting property of making large sheets (Figure 6A), often found folded over on themselves to make multiple layers. For R210E we were able to confirm the assumed hexagonal character of the sheets by simple image processing. When we selected an area of these sheets and calculated the Fourier transform, we observed a hexagonal pattern typical of a regular hexagonal array (inset in Figure 6.A) [23]. The lattice constant for R210E sheets exactly matched the dimensions of our model of a hexagonal sheet (See figure 8.A). The E153Q-R210E mutant also yielded large sheets of major capsid protein (Figure 6C), which often seemed to be fairly flat and much larger than those produced by the R210E mutant. The Fourier transform of the sheet in (Figure 6C) produced a hexagonal pattern similar to those observed in Figure 6.A, except that the spots were doubled (not shown), indicating that this sheet has two layers that arose from folding of the sheet. The charge-swap double mutant, E153R-R210E, yielded many gently curved assemblies which were distinctly different than the sheet-like appearance of R210E and E153Q-R210E (Figure 6B), as if the charge swap was partly successful in restoring some aspect of normal assembly. These charge-swap experiments may have failed to restore normal assembly because we did not take into account the additional ionic interactions involving E153, or because the precise orientations of the interactions are also important [24].
Figure 5. Gel analysis of double and single mutants modifying the E153-R210E interaction.
A. Agarose gel. Double mutants E153R-R210E and E153Q-R210E and single mutant controls failed to assembly correctly. For mutants containing E153R, E153Q or R210E, the majority of the protein was found in the wells of the pellet fraction. E153R, which produced faint prohead and head-like bands. E153D and control K92A produced proheads and heads. B. SDS gel. Analysis of pellet fractions of double mutants showed results similar to the E153 mutants shown in Figure 3, but nearly all of the major capsid protein was cleaved for E153D, indicating normal assembly.
Figure 6. Sheets and proheads made by E153 and R210 mutants.
Electron micrographs of pellet fractions from experiments shown in Figure 5 were prepared as described in the legend to Figure 4. A. R210E. Inset: Simulated diffraction pattern of area boxed on right (using FFT (fast Fourier transform) function in ImageJ [51]) showing a pattern typical for a hexagonal array with the lattice constant indicated. B. R210E/E153R and C. R210E/E153Q. The rest of the samples were diluted from purified stocks and stained as above. D. E153 Prohead II. E. WT Prohead II. F. WT Prohead I.
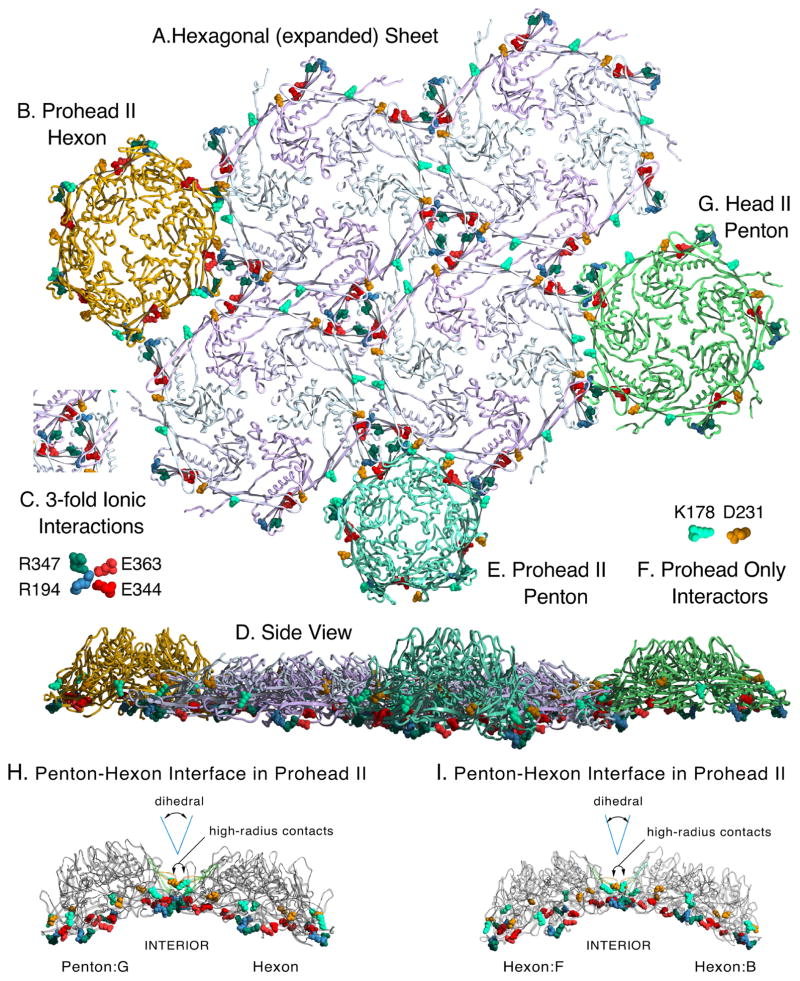
Figure 8. Modeled sheet of expanded and crosslinked mutant HK97 hexons and docking of related structures.
A. A model of a hexagonal sheet of HK97 major capsid protein made by rigid-body manipulations of six copies of chain A from the HK97 Head II (PDB ID: 1OHG). Several important charged contact residues at the interfaces of adjacent capsomers are shown in space-filling mode, color coded as indicated in insets C. and F. Those in C. are known to be preserved in all HK97 capsid structures, from prohead to head. We also believe that all of the highlighted residues provide electrostatic steering to align capsomer interfaces during assembly. We highlight the same residues in the following panels, which show docking of capsomers (from Prohead II and Head II atomic models) to the sheet model. The crosslink residues N356, K169 are in a position to form covalent bonds between hexon sheet subunits, but are hidden. B. Docking of a Prohead II hexon (chains A–F from PDBID: 3E8K), showing a poor fit for binding. C. Key to four residues involved in inter-capsomer ionic interactions are shown with color-coding as labeled. D. A side view of the entire array of capsomers shown in panels A. B. E. and G. E. Docking of a Prohead II penton (5 chains G from PDBID: 3E8K) F. Key to coloring of residues K178 and D231 which form a salt bridge in proheads, but not in heads. G. Docking of a Head II penton (5 chains G from PDBID: 1OHG). (protruding N-arms of Head II subunits were truncated when they would interfere or obscure the dockings). H. Side views of the contacts between a penton and a hexon in Prohead II. D231 and K178, which determine the dihedral angle between subunits on adjacent capsomers, are shown in space-filling mode to show that they are on the “hillsides” above the main contacts. P-domain ionic contact residues are also shown in space-filling mode. I. Same as (H) except contacts between two hexons are shown.
Mutant E153R produces a few proheads via a gain of function
Mutant E153R was included in our experiments as an extreme mutation that converts a negatively charged salt-bridge partner residue to a positively charged one, and also to serve as a control for the charge-swapped double mutants (above). Surprisingly, this was the only one of the mutants described so far that produced normal appearing proheads at detectable levels (Figure 4E). The level of proheads produced was low and thus not readily visible in the gel presented in Figure 3; however, E153R Prohead II and Head II bands were detectable in most experiments (Figure 5A, lanes 2, 3). Both the head and prohead bands from the E153R mutant exhibited upward shifts of electrophoretic mobility in the native gel relative to their wild-type counterparts. We believe these mobility shifts most likely result from a change in charge state due to the mutation. Cleavage of the HK97 major capsid protein from 42 kDa to 31 kDa is known to be dependent on assembly [4], so the appearance of the 31 kDa cleaved form of the major capsid protein in the SDS gel of the E153R extract (Figure 5B, lanes 7, 8, 9) confirms that what was made is Prohead II. We suspect that the proheads that do assemble represent a gain of function in which the mutant R153 side chains bind the E-loops to new partners in neighboring subunits, allowing a few proheads to form, but not fast enough to prevent the bulk of the E153R protein from assembling into tubes. Modeling the mutation into HK97 Prohead II hexamers and pentamers (PDB ID:3E8K) revealed that there are plausible potential partners for such a new interaction with R153. The best-positioned candidate is His 188, located nearby on the β-strand below the helix containing R210; a less likely alternative, Tyr 206, is adjacent to R210 on the same helix (see Figure 2A for relative locations).
Mutant E153D is viable and produces proheads
A highly conservative mutation in the E153-R210 residue pair is E153D, which moves the carboxylate ion only 1.5 Å closer to the protein backbone. The phenotypes of all other mutations affecting this salt bridge were so severe that we expected that the E153D protein would also be defective. However, the E153D mutant protein was able to make viable phage in complementation tests (Table 1). Biochemical tests (Figure 5) showed that E153D produced a clear prohead band in an agarose gel (Figure 5A, lanes 14, 15). We also found the cleaved form of the major capsid protein in an SDS gel (Figure 5B, lanes 5, 6), indicating successful assembly and processing to fully cleaved Prohead II. The appearance of an additional slower migrating band in the agarose gel shows that the particles are also capable of expanding to form heads. An unexpected characteristic of the proheads and expanded particles made by the E153D mutant is that they both migrate slower than normal HK97 heads (Figure 5A), even though there is no expected change of charge due to the mutation. The unusual mobility of the E153D particles suggests that they are in an unusual conformation, which correlates with a different unusual property of the particles - they do not crosslink fully in vitro, as we show below.
E153R and E153D mutant proheads show altered expansion and incomplete crosslinking
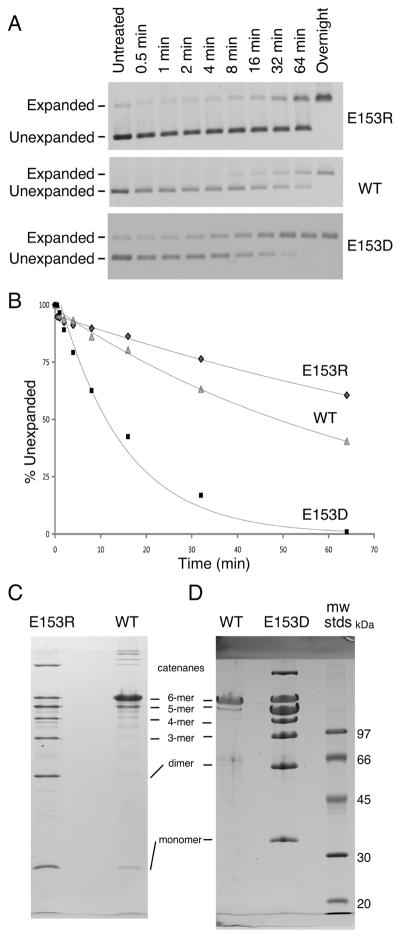
We tested the abilities of E153D and E153R proheads to expand and crosslink in vitro using two different conditions defined previously [12, 13, 15]. Expansion induced by low pH was monitored by agarose gel electrophoresis (Figure 7A) and plotted (Figure 7B). Under these conditions (pH 4.3 in 0.4 M KCl), the E153R proheads expanded at one-half (0.007 min−1) the wild-type rate (0.015 min−1), while the E153D proheads expanded 5 times faster (0.07 min−1) than wild type. After overnight incubations, both wild-type and mutant proheads had expanded fully, as judged by the band shifts in the agarose gels. However, only the wild-type sample had fully crosslinked (not shown). To confirm these unusual crosslinking results, we induced expansion using an alternative and gentler treatment with dimethyl formamide (DMF) [12]. It can clearly be seen that neither DMF-treated sample crosslinked to the same extent as the wild-type samples (Figure 7C). The pattern of crosslinking for both mutants resembles that seen for HK97 Expansion Intermediate III [7], which is a transient particle that occurs during expansion at low pH [15] and has about 60% of possible crosslinks. These results suggest that the altered E153 interactions of the two mutants somehow physically constrain the proheads from expanding completely in vitro, and incomplete crosslinking is a consequence.
Figure 7. E153R and E153D proheads have different expansion properties than wild type.
Expansion of E153R, E153D and wild-type proheads was induced by low pH treatment [12, 21]. A. Samples were neutralized at the indicated times and run on an agarose gel at the experiment’s end. Note the larger gap between expanded and unexpanded particles for E153R than for wild type. B. Expansion graphed relative to the amount of prohead present at the beginning. C. and D. Expansion was induced using dimethyl formamide (DMF) [12]. DMF treatment induced wild-type proheads to expand fully and the resulting crosslinks prevented most of the major capsid protein from entering the gel, so only small amounts of hexamers and pentamers appeared in the wild-type samples in panels C and D. Neither E153R proheads (C) nor E153D proheads (D) crosslinking fully, but instead make a ladder of crosslinked bands that resemble those seen for partially expanded HK97 particles [13, 15].
We have previously shown that crosslinking is required to make a viable HK97 phage particle [25], but complementation tests showed that the E153D protein is able to support phage growth in vivo (Table 1), even though it cannot crosslink to completion in vitro. This is a phenotype we had not encountered before. However, we have also shown that a mutant that expands ~30 times slower than wild-type in vitro (E363D [12]) is not viable in vivo, even though it eventually crosslinks completely in vitro. These previous results show that rapid crosslinking is needed to produce a viable phage in vivo. In our current experiments, E153D expanded more rapidly than wild type, so the rapid-crosslinking criterion may be met, even though in vitro crosslinking was incomplete. We suggest that DNA packaging in vivo could provide the extra driving force needed for E153D particles to expand both rapidly and fully, so that complete crosslinking can produce viable phage.
Discussion
E153-R210 mutants both lose and gain assembly abilities
We have shown that most mutants that perturb the E153-R210 salt bridge in the HK97 capsid protein lose their ability to assemble normal capsids while at the same time gaining an abnormal ability to assemble into sheets or tubes. Below, we show that the location of the E153-R210 links within assembled capsomers suggests how the changes in assembly properties in these mutants arise and some implications for the mechanism of capsid assembly. We have previously proposed an induced-fit assembly model [4] which requires that capsomers adjust their shape while binding to an assembling capsid. As originally presented, the model lacked any specific suggestion as to how a capsomer could act as a unit to accomplish such changes. The E153-R210 interactions that we have disrupted in this current work appear to be perfectly positioned to play that role in normal prohead assembly, because they connect every subunit to two neighboring subunits, and they occur exclusively within capsomers. The E153-R210 interactions also seem to play a role in keeping capsomers from adopting a flattened, mature conformation that leads to abnormal assembly and provides an additional rationale for why a prohead stage is needed at all.
E153-R210 interactions provide a supporting role in an induced-fit model of capsid assembly
The previously proposed induced-fit model for capsid assembly [4] is based on the essential character of the transient interactions that occur between residue K178 on the outer strand of each E-loop with residue D231 on the G-loop of the adjacent capsomer (the G-loop is shown in yellow in Fig. 2A). These salt bridges occur between the “hillsides” of dome-shaped neighboring capsomers (Figure 8H and I) and are far above the other extensive contacts between neighboring capsomers. Their locations suggests that they control the dihedral angle between the adjacent capsomers (Figure 8H and I) which could influence the size of assembling capsids because the dihedral angle between capsomers is greater in smaller capsids. HK97 major capsid proteins with substitution at D231 or K178 misassembled or failed to assemble, suggesting that altering the dihedral angles between subunits on adjacent capsomers has a drastic influence on assembly. We argue that this intra-subunit dihedral angle is set during the normal docking of each added capsomer during assembly, as outlined below.
We believe that the E153-R210 salt bridges play at least two key roles in the induced-fit model of assembly. The first is to anchor each E-loop firmly to the capsomer so it can participate in setting the dihedral angles between subunits. The second is to create the network of contacts that tie the subunits of each capsomer into a five-or-six-membered unit that confers to capsomers an ability to change shape in a programmed manner during assembly. We have illustrated how this could occur in Supplemental Movie S2, where we show how the subunits of a hypothetical symmetric hexamer change conformation to make the critical contacts needed for assembly, and how that produces the peculiar asymmetric hexon shape found in proheads. The movie also shows how the preserved E153-R210 salt bridges act as pivots that hold the connected parts of individual subunits at a fixed distance from each other and enables them to maintain the dome-shape needed to build proheads. This anchoring allows subunits to bend or move only along the simple paths needed to change capsomer shape in response to binding. The fact that pivoting of an identical nature occurs during the even larger conformational changes that occur during maturation (see Supplemental Movie S1) suggests that the mechanism we propose is plausible.
The E153-R210 connections allow conformational changes to be mechanically transmitted from one side of each assembling hexamer to the adjacent and distal sides, setting the orientation of its long axis and the angles between the distal subunits and the adjacent capsomers’ subunits. Those angles in turn specify whether a penton or a hexon can bind at each newly created site and also determine the orientation of an asymmetric hexon’s long axis as it binds. For T=7 procapsids, where all hexons are identical, regulating the placement of pentons and oriented hexons in a simple symmetric pattern will create a shell with only one size and shape. This is the core of the induced-fit assembly model, and we believe that the interconnecting network of E153-R210 interactions mediates the adjustment of hexamer conformations required for that model to work.
Assembly without E153-R210 connections
We can explain how mutant subunits lacking E153-R210 links are able to assemble into capsomers and larger assemblies, if we extend our pathway for HK97 capsid assembly (Figure 1) to include earlier steps. Our working model for HK97 assembly suggests that major capsid proteins are first brought together by their delta domains, which position the C-terminal parts of the subunits so they can then associate via the oppositely-charged borders of their wedge-shaped A-domains (see Figure 2A and Figure 4 in ref [14]) to form a nascent capsomer. In a non-mutant situation, the E153 of each E-loop in this nascent capsomer can then find its partner R210 on the neighboring subunit and lock into place to facilitate normal assembly. In mutants without E153-R210 links, nascent capsomers must also be made; but these mutant capsomers are unable to adopt the dome-shaped conformations needed to assemble correctly without the E153-R210 salt bridges to constrain them. Instead, mutant capsomers flatten out and prematurely adopt conformations similar to those of the expanded hexons of Head II (Figure 8A) and form sheets and tubes instead of proheads. Once assembly is initiated, a tube or sheet of flattened mutant hexons will likely only incorporate more flattened hexons, and we have tried to illustrate why this occurs in Figure 8. Pentons in either Prohead II (Figure 8E) or Head II (Figure 8G) conformation can only mate with the hexon sheet lattice weakly along one edge at a time. So, in order for mutant pentons lacking the E153-R210 link to bind by two edges and be stably incorporated, they would have to switch to an open and flattened conformation. A flattened penton is essentially an expanded hexon with a missing subunit and would be unstable unless it could acquire an additional subunit. We conclude that most of the pentons that are made in mutants with a missing E153-R210 link eventually convert to hexamers, because we found that most of the mutant proteins assembled into sheets and tubes, which are normally built exclusively from hexons. The curved sections or tube ends containing caps observed in some mutants may still have pentons, but their presence is only inferred. The number of pentons required to form these curved sections is drastically fewer than would occur in normal proheads.
Mutants that break E-loop-backbone helix interactions assemble using an alternate pathway
The abnormal structures made by the E153-R210 mutants that we described above appear to have assembled directly into a mature and flattened conformation without a prohead-like intermediate and with the delta domain still attached. If so, this is a drastic departure from the normal assembly mechanism in which proheads are made first and only transform into the mature conformation after assembly is complete and the delta domain is removed. We suggest that a major driving factor for the existence of a prohead stage with a distinctly different capsid protein conformation in bacteriophages and herpesviruses is to avoid that mature and flattened conformation until a complete particle is built. If the mature conformation could be attained easily, it would most likely lead to runaway assembly into sheets or tubes, just as we see with the E153/R210 mutants. We further suggest that it would be impossible to build a capsid of defined size and shape from proteins that are already in the mature conformation. We also believe that a major function of scaffolding proteins and delta domains is to help maintain capsid proteins in the precursor “prohead” conformation during assembly. In HK97, conversion of Prohead I particles to the mature state appears to be suppressed by the delta domain. The evidence for this is that Prohead II (with no delta domain) readily expands [21], while Prohead I only expands in vitro after treatments that cause the delta domains to unfold [26, 27] without destroying this fragile particle. We conclude that in HK97 the E153/R210 interactions provide constraints from the outside of the prohead, which act together with delta domain constraints from the inside to suppress conversion to the mature conformation (“expansion”), since the mutant major capsid proteins lacking the E153-R210 salt bridges appear to spontaneously convert to the expanded conformation.
The E-loop may be conserved because of its role in regulating assembly
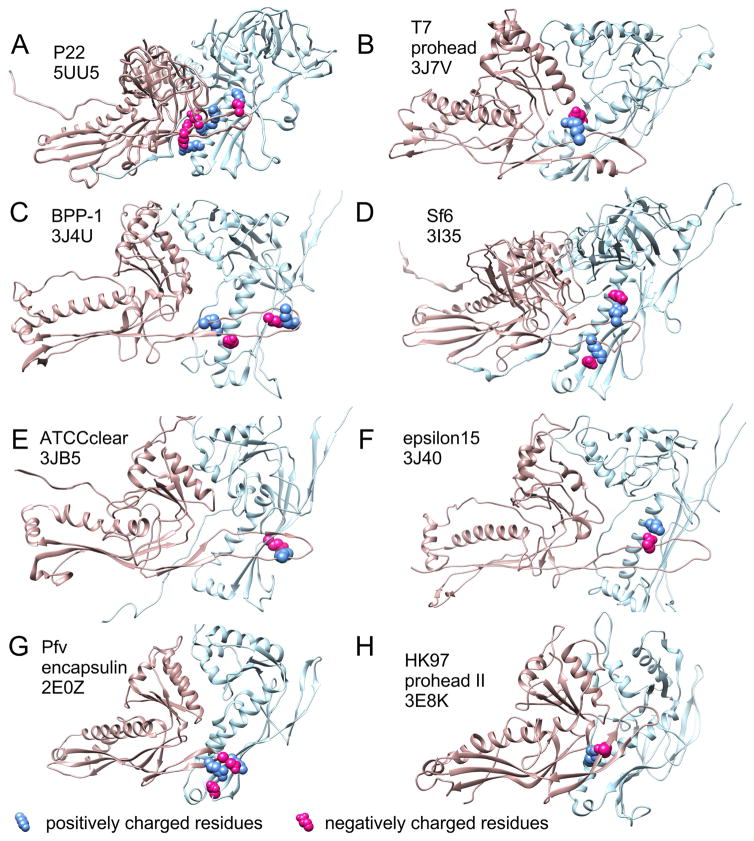
E-loop-like structural elements occur in many capsids which range in size from T=1 to T=16 and include the well-studied bacteriophages P22 [28–30], T4 [31], T5 [32], T7 [33], as well as epsilon15 [34], BPP-1 [35], Sf6 [36], three bacterial encapsulins [37–39], Herpesvirus nucleocapsids [40] and have been inferred in many others, including ϕ29[41] and P2[42]. While the E-loop was first revealed as a structural element involved in forming the covalent bonds that stabilize the HK97 capsid [16], none of the examples above use such covalent crosslinks, so crosslinking is probably only a minor, late-acquired role for the E-loop. The E-loop’s persistence in so many other distantly related capsid proteins suggests that it plays other important roles. Those roles may often include controlling size and shape during assembly, as we outlined above for HK97. We compiled a table (Supplemental Table 1) of published structural models of examples of the HK97 fold from phages and encapsulin proteins in the Protein Data Bank that show a clear E-loop in order to see if we could identify interactions analogous to the E153-R210 salt bridge in HK97. Of the 13 different organisms represented in the table, all but the T=1 encapsulin 3DKT show clear overlaps of the E-loop with the adjacent subunit of the same capsomer. In six of the ten HK97 fold examples from phages, (including P22, T7, Sf6, epsilon 15, BPP-1, ATCCclear) and in one encapsulin there is evidence for one or more ionic interactions between the E-loop and the P-domain of the overlapped adjacent subunit (Figure 9). One of these, the P22 major capsid protein appears to have up to four such connections between the E-loop and the backbone helix of the adjacent capsomer in the mature capsid shell. The rest have fewer. Mutational studies will be needed to test if any of these indicated E-loop interactions are important for regulating capsid assembly.
Figure 9. Capsid protein PDB models with E-loops overlapping the adjacent subunits and potential analogs of the E153-R210 interaction in HK97.
The PDB files listed in Supplemental Table 1 were opened in Chimera [50] and (in those with side chains) searched for potential ionic interactions between the E-loop and the adjacent subunit. For those with such interactions, figure panels showing two subunits from the model’s hexon were prepared with residues involved in potential side chain interactions shown in space-filling mode with positively charged residues in blue and negatively charged residues in red. HK97 is shown in panel H for comparison. The source name and the PDB ID’s are indicated in the figure panels. The chains used for each panel are as follows: A, (F and A), B, (D and C), C, (B and A), D, (C and B), E, (F and A), F, (C and D), G, (A and B), H, (F and A).
Materials and Methods
Plasmids and mutagenesis
Mutations of the HK97 major capsid protein were made in a wild type HK97 expression plasmid, pV0-SacI, containing HK97 gp5 and gp4 under the control of a T7 promoter and an ampicillin resistance gene [21]. Plasmid pV0-SacI, contains silent change in the third codon of gene 5 that introduces a SacI restriction site, but is otherwise identical to the plasmid pT7-Hd2.9 [21]. In most cases, site-directed mutagenesis was carried out as described previously [43], but using Phusion polymerase to carry out the required PCR reactions. Each mutagenesis used four primers: two phosphorylated primers, one of which was mutagenic, and two flanking primers. Each final mutation-containing PCR product overlapped two unique restriction sites in the wild-type plasmid and was digested appropriately and used to replace the wild-type DNA between the two sites. The PCR-generated segments between the flanking restriction sites in resulting plasmids were sequenced (GeneWiz, South Plainfield, NJ) to confirm that each mutation was present in an otherwise wild-type context. In a few cases, a Q5 site-directed mutagenesis kit (New England Biolabs, Ipswich, MA) was used to make mutations. In those cases, a fully sequenced fragment containing the mutations was subcloned into a wild-type plasmid backbone to ensure that only desired mutations were present.
Expression
Plasmid constructs were co-transformed into expression strain BL21(DE3) cells along with pLysS [44]. Small-scale expression experiments were done in 1.5 ml cultures of TYM5052 autoinduction media using the recipes of Studier [45] and containing 50 μg/ml ampicillin and 25 μg/ml chloramphenicol. High-level expression of the cloned HK97 genes was achieved by incubation of such cultures for 24 hrs under vigorous aeration conditions. The cultures were collected, lysed and fractionated as described previously [22]. This procedure yields thee fractions: a crude insoluble pellet fraction, a soluble supernatant fraction and a PEG (polyethylene glycol) precipitation fraction. These were analyzed using native agarose, SDS polyacrylamide gel electrophoresis and electron microscopy.
Large-scale purification of proheads were done essentially as described previously [12, 46]. Large cultures of BL21(DE3) cells containing pLysS and the appropriate expression plasmid were grown in LB plus antibiotics at 28°C to late log phase, induced with IPTG at 0.4 mM, grown for an additional 16 hrs and harvested by centrifugation. Cell pellets were processed and proheads purified by a combination of PEG precipitation, high-speed pelleting in an ultracentrifuge, velocity sedimentation in glycerol gradients and chromatography on a Poros HQ20 ion exchange column. The final prohead-containing column fractions were pooled and concentrated by ultracentrifugation.
Gel Electrophoresis
Agarose-gel electrophoresis was performed using standard DNA horizontal mini-gel equipment and TAMg buffer (40 mM Tris-HCl, 20 mM Acetic Acid pH 8.1, and 1 mM MgSO4). Samples were mixed with a solution of dyes and glycerol (50% (v/v) glycerol, 0.025% (w/v) bromphenol blue, 0.025% (w/v) xylene cyanol XFF) and run at 120V on 0.9% agarose gels. Gels were stained with ~0.002 % (w/v) Coomassie Brilliant Blue R250 in ~4.5 % (v/v) methanol, ~9 % (v/v) acetic acid for 4–16 hrs, destained in 10% acetic acid, and photographed wet.
Samples for sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis [47] were TCA (trichloroacetic acid) precipitated prior to sodium dodecyl sulfate (SDS) polyacrylamide gel analysis to prevent spontaneous cross-linking of the HK97 major capsid protein during sample preparation [21]. Deoxycholate was added to 0.8 mg/ml (final) with the TCA for the concentrated samples produced in small scale expression experiments to improve recovery. The separating gels were made using an atypical low-crosslinker acrylamide recipe [48]. Gels were stained with Coomassie brilliant Blue R250 and photographed wet.
Electron Microscopy
Samples were adsorbed to glow-discharged carbon/Parlodion or carbon/Formvar support films on 400 mesh copper grids, rinsed with water, negatively stained with 1% uranyl acetate and visualized using a Morgagni 268(D) EM FEI (Eindhoven, Netherlands) at 56,000X nominal magnification with images recorded digitally using a 10 megapixel ORCA camera (Hamamatsu, Bridgewater, NJ).
Complementation Assays
Complementation tests were used to assay the ability of mutant proteins expressed from plasmids to replace the missing functions of phages in which specific genes are inactivated by amber (am) mutations. Operationally, serial dilutions of phage were spotted on lawns of bacteria expressing wild-type or mutant HK97 genes and the results used to compute a crude relative efficiency of plating (EOP) of the mutant relative to the wild type control. A value of 10 or 3 was given for each fully or partly cleared spot, multiplied together to give an estimate of the phage titer and divided by the titer obtained using the wild-type-expressing strain.
In vitro expansion reactions
HK97 proheads undergo expansion during maturation, but can be induced to expand in vitro [21]. Low pH/high salt expansion was induced as outlined previously [12]. Samples of purified proheads were diluted to ~2 mg/ml in expansion buffer (50 mM sodium acetate pH 4.1 with 400 mM KCl) to initiate expansion. The reaction was stopped at the indicated times by removing aliquots and neutralized by 4-fold dilution into 100 mM Tris-HCl (pH 8.0). Samples were mixed with dyes and glycerol and run on 1% agarose gel in TAMg buffer. Relative intensity of gel bands was measured using EDAS 290 camera system and 1D Image Analysis Software from Kodak (Rochester, NY).
Molecular visualization
All modeling was done using SPDBV [49] aided by the POV ray tracer (http://wiki.povray.org/), except for that described in Supplemental Table 1 and Figure 9, which was done using Chimera [50].
Supplementary Material
Highlights.
HK97 capsid assembly requires E-loop-to-backbone-helix links within capsomers
These salt bridge links persist despite large conformation changes during assembly
Capsomers flatten and assemble abnormally when these salt bridges are missing
These links enable capsomer morphing needed for an induced-fit assembly model
The E-loop is conserved suggesting it similarly regulates assembly in many systems
Acknowledgments
This work was supported by NIH grant GM47795 from the U.S. Public Health Service to RWH and RLD. We thank our many colleagues and especially A. Huet and C. Peebles for valuable discussions and critiques of the ongoing work and manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symposia on Quantitative Biology. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 2.Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol Mol Biol Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steven AC, Heymann JB, Cheng N, Trus BL, Conway JF. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Current opinion in structural biology. 2005;15:227–36. doi: 10.1016/j.sbi.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tso DJ, Hendrix RW, Duda RL. Transient contacts on the exterior of the HK97 procapsid that are essential for capsid assembly. J Mol Biol. 2014;426:2112–29. doi: 10.1016/j.jmb.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steven AC, Bauer AC, Bisher ME, Robey FA, Black LW. The maturation-dependent conformational change of phage T4 capsid involves the translocation of specific epitopes between the inner and the outer capsid surfaces. J Struct Biol. 1991;106:221–36. doi: 10.1016/1047-8477(91)90072-5. [DOI] [PubMed] [Google Scholar]
- 6.Steven AC, Couture E, Aebi U, Showe MK. Structure of T4 polyheads. II. A pathway of polyhead transformation as a model for T4 capsid maturation. J Mol Biol. 1976;106:187–221. doi: 10.1016/0022-2836(76)90307-7. [DOI] [PubMed] [Google Scholar]
- 7.Gan L, Speir JA, Conway JF, Lander G, Cheng N, Firek BA, et al. Capsid conformational sampling in HK97 maturation visualized by X-ray crystallography and cryo-EM. Structure. 2006;14:1655–65. doi: 10.1016/j.str.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Wikoff WR, Conway JF, Tang J, Lee KK, Gan L, Cheng N, et al. Time-resolved molecular dynamics of bacteriophage HK97 capsid maturation interpreted by electron cryo-microscopy and X-ray crystallography. J Struct Biol. 2006;153:300–6. doi: 10.1016/j.jsb.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Huang RK, Khayat R, Lee KK, Gertsman I, Duda RL, Hendrix RW, et al. The Prohead-I structure of bacteriophage HK97: implications for scaffold-mediated control of particle assembly and maturation. J Mol Biol. 2011;408:541–54. doi: 10.1016/j.jmb.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gertsman I, Gan L, Guttman M, Lee K, Speir JA, Duda RL, et al. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–50. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, et al. Virus maturation involving large subunit rotations and local refolding. Science. 2001;292:744–8. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- 12.Dierkes LE, Peebles CL, Firek BA, Hendrix RW, Duda RL. Mutational analysis of a conserved glutamic acid required for self-catalyzed cross-linking of bacteriophage HK97 capsids. J Virol. 2009;83:2088–98. doi: 10.1128/JVI.02000-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gan L, Conway JF, Firek BA, Cheng N, Hendrix RW, Steven AC, et al. Control of crosslinking by quaternary structure changes during bacteriophage HK97 maturation. Molecular cell. 2004;14:559–69. doi: 10.1016/j.molcel.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 14.Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 A resolution. J Mol Biol. 2003;334:885–99. doi: 10.1016/j.jmb.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 15.Lata R, Conway JF, Cheng N, Duda RL, Hendrix RW, Wikoff WR, et al. Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell. 2000;100:253–63. doi: 10.1016/s0092-8674(00)81563-9. [DOI] [PubMed] [Google Scholar]
- 16.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–33. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 17.Lee KK, Gan L, Tsuruta H, Moyer C, Conway JF, Duda RL, et al. Virus capsid expansion driven by the capture of mobile surface loops. Structure. 2008;16:1491–502. doi: 10.1016/j.str.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duda RL, Martincic K, Hendrix RW. Genetic basis of bacteriophage HK97 prohead assembly. J Mol Biol. 1995;247:636–47. doi: 10.1006/jmbi.1994.0169. [DOI] [PubMed] [Google Scholar]
- 19.Xie Z, Hendrix RW. Assembly in vitro of bacteriophage HK97 proheads. J Mol Biol. 1995;253:74–85. doi: 10.1006/jmbi.1995.0537. [DOI] [PubMed] [Google Scholar]
- 20.Gertsman I, Fu CY, Huang R, Komives EA, Johnson JE. Critical salt bridges guide capsid assembly, stability, and maturation behavior in bacteriophage HK97. Molecular & cellular proteomics: MCP. 2010;9:1752–63. doi: 10.1074/mcp.M000039-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duda RL, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix RW. Structural transitions during bacteriophage HK97 head assembly. J Mol Biol. 1995;247:618–35. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- 22.Oh B, Moyer CL, Hendrix RW, Duda RL. The delta domain of the HK97 major capsid protein is essential for assembly. Virology. 2014;456–457:171–8. doi: 10.1016/j.virol.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harburn G, Taylor CA, Welsberry TR. Atlas of Optical Transforms. London, UK: G. Bell & Sons, LTD; 1975. [Google Scholar]
- 24.Donald JE, Kulp DW, DeGrado WF. Salt bridges: geometrically specific, designable interactions. Proteins. 2011;79:898–915. doi: 10.1002/prot.22927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross PD, Cheng N, Conway JF, Firek BA, Hendrix RW, Duda RL, et al. Crosslinking renders bacteriophage HK97 capsid maturation irreversible and effects an essential stabilization. Embo J. 2005;24:1352–63. doi: 10.1038/sj.emboj.7600613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardone G, Duda RL, Cheng N, You L, Conway JF, Hendrix RW, et al. Metastable intermediates as stepping stones on the maturation pathways of viral capsids. MBio. 2014;5:e02067. doi: 10.1128/mBio.02067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross PD, Conway JF, Cheng N, Dierkes L, Firek BA, Hendrix RW, et al. A free energy cascade with locks drives assembly and maturation of bacteriophage HK97 capsid. J Mol Biol. 2006;364:512–25. doi: 10.1016/j.jmb.2006.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1355–60. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizzo AA, Suhanovsky MM, Baker ML, Fraser LC, Jones LM, Rempel DL, et al. Multiple functional roles of the accessory I-domain of bacteriophage P22 coat protein revealed by NMR structure and CryoEM modeling. Structure. 2014;22:830–41. doi: 10.1016/j.str.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hryc CF, Chen DH, Afonine PV, Jakana J, Wang Z, Haase-Pettingell C, et al. Accurate model annotation of a near-atomic resolution cryo-EM map. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:3103–8. doi: 10.1073/pnas.1621152114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, et al. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7163–8. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Effantin G, Boulanger P, Neumann E, Letellier L, Conway JF. Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. J Mol Biol. 2006;361:993–1002. doi: 10.1016/j.jmb.2006.06.081. [DOI] [PubMed] [Google Scholar]
- 33.Guo F, Liu Z, Fang PA, Zhang Q, Wright ET, Wu W, et al. Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4606–14. doi: 10.1073/pnas.1407020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker ML, Hryc CF, Zhang Q, Wu W, Jakana J, Haase-Pettingell C, et al. Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12301–6. doi: 10.1073/pnas.1309947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai W, Hodes A, Hui WH, Gingery M, Miller JF, Zhou ZH. Three-dimensional structure of tropism-switching Bordetella bacteriophage. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4347–52. doi: 10.1073/pnas.0915008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao H, Li K, Lynn AY, Aron KE, Yu G, Jiang W, et al. Structure of a headful DNA-packaging bacterial virus at 2.9 A resolution by electron cryo-microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:3601–6. doi: 10.1073/pnas.1615025114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akita F, Chong KT, Tanaka H, Yamashita E, Miyazaki N, Nakaishi Y, et al. The crystal structure of a virus-like particle from the hyperthermophilic archaeon Pyrococcus furiosus provides insight into the evolution of viruses. J Mol Biol. 2007;368:1469–83. doi: 10.1016/j.jmb.2007.02.075. [DOI] [PubMed] [Google Scholar]
- 38.McHugh CA, Fontana J, Nemecek D, Cheng N, Aksyuk AA, Heymann JB, et al. A virus capsid-like nanocompartment that stores iron and protects bacteria from oxidative stress. EMBO J. 2014;33:1896–911. doi: 10.15252/embj.201488566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutter M, Boehringer D, Gutmann S, Gunther S, Prangishvili D, Loessner MJ, et al. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nature structural & molecular biology. 2008;15:939–47. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- 40.Huet A, Makhov AM, Huffman JB, Vos M, Homa FL, Conway JF. Extensive subunit contacts underpin herpesvirus capsid stability and interior-to-exterior allostery. Nature structural & molecular biology. 2016;23:531–9. doi: 10.1038/nsmb.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Molecular cell. 2005;18:149–59. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 42.Dearborn AD, Laurinmaki P, Chandramouli P, Rodenburg CM, Wang S, Butcher SJ, et al. Structure and size determination of bacteriophage P2 and P4 procapsids: function of size responsiveness mutations. J Struct Biol. 2012;178:215–24. doi: 10.1016/j.jsb.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adereth Y, Champion KJ, Hsu T, Dammai V. Site-directed mutagenesis using Pfu DNA polymerase and T4 DNA ligase. BioTechniques. 2005;38:864, 6, 8. doi: 10.2144/05386BM03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods in enzymology. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 45.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein expression and purification. 2005;41:207–34. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Conway JF, Cheng N, Steven AC, Hendrix RW, Duda RL. Control of Virus Assembly: HK97 “Whiffleball” Mutant Capsids Without Pentons. J Mol Biol. 2005;348:167–82. doi: 10.1016/j.jmb.2005.02.045. [DOI] [PubMed] [Google Scholar]
- 47.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 48.Dreyfuss G, Adam SA, Choi YD. Physical change in cytoplasmic messenger ribonucleoproteins in cells treated with inhibitors of mRNA transcription. Mol Cell Biol. 1984;4:415–23. doi: 10.1128/mcb.4.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 50.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.