Abstract
Oryza longistaminata originates from African wild rice and contains valuable traits conferring tolerance to biotic and abiotic stress. However, interspecific crosses between O. longistaminata and Oryza sativa cultivars are hindered by reproductive barriers. To dissect the mechanism of interspecific hybrid sterility, we developed a near-isogenic line (NIL) using indica variety RD23 as the recipient parent and O. longistaminata as the donor parent. Both pollen and embryo sac semi-sterility were observed in F1 hybrids between RD23 and NIL. Cytological analysis demonstrated that pollen abortion in F1 hybrids occurred at the early bi-nucleate stage due to a failure of the first mitosis in microspores. Partial embryo sacs in the F1 hybrids were defective during the functional megaspore formation stage. Most notably, nearly half of the male or female gametes were aborted in heterozygotes S40iS40l, regardless of their genotypes. Thus, S40 was indicated as a one-locus sporophytic sterility gene controlling both male and female fertility in hybrids between RD23 and O. longistaminata. A population of 16 802 plants derived from the hybrid RD23/NIL-S40 was developed to fine-map S40. Finally, the S40 locus was delimited to an 80-kb region on the short arm of chromosome 1 in terms with reference sequences of cv. 93-11. Eight open reading frames (ORFs) were localized in this region. On the basis of gene expression and genomic sequence analysis, ORF5 and ORF8 were identified as candidate genes for the S40 locus. These results are helpful in cloning the S40 gene and marker-assisted transferring of the corresponding neutral allele in rice breeding programs.
Introduction
Asian cultivated rice (Oryza sativa L.) was domesticated from wild species (Oryza rufipogon) thousands of years ago (Huang et al., 2012), and during this process, the genetic diversity within this cultivated rice has been gradually reduced. More importantly, owing to the frequent use of a few adapted progenitors in breeding, the decrease of genetic variation has been amplified (Moncada et al., 2001). These challenges limit the increases in yield of new varieties and also make rice more susceptible to disease and insect epidemics (Tanksley and McCouch, 1997; Jarvis et al., 2011). Thus, wild rice with the AA genome containing favorable genes (alleles) could be a valuable resource to overcome these obstacles in rice improvement (Xiao et al., 1996; Xu et al., 2012). Oryza longistaminata Chev. is closely related to O. sativa, as they have a similar AA genome. It is also an excellent gene pool in modern rice breeding because it possesses various outstanding traits, including resistance to bacterial blight disease, weed suppression effects, rhizomes, high biomass production on poor soils and high nitrogen use efficiency (Song and Ronald, 1995; Rodenburg and Johnson, 2009; Yang et al., 2010). Most importantly, transferring genes from O. longistaminata to O. sativa through sexual hybridization can significantly enhance the yield of Asian cultivated rice (Brar, 2004). However, severe reproductive isolation between wild rice (including O. longistaminata) and O. sativa by hybrid sterility and inviability limits extensive utilization of these species and the exploitation of advantageous heterosis effects in related species (Ikeda et al., 2009; Bolaji and Nwokeocha, 2014). Therefore, it is essential to isolate sterility loci and discover wide compatibility varieties containing neutral alleles to overcome reproductive barriers (Wang et al., 2005).
These reproductive barriers also were explained by different models (Oka, 1953, 1974; Kitamura, 1962), among which, one-locus sporophytic sterility model concerned the alleles S1 and S2, and in heterozygotes S1S2, partial gametes deteriorated irrespective of their genotypes, while homozygotes S1S1 and S2S2 were fertile (Oka, 1974). To date, numerous loci controlling rice interspecific or inter-subspecific hybrid sterility have been identified, including female gamete abortion (Wan et al., 1996; Zhao et al., 2007; Chen et al., 2008; Yu et al., 2016), male gamete sterility (Long et al., 2008; Zhang et al., 2011; Zhao et al., 2011) and both in a few cases (Koide et al., 2008, 2012; Chen et al., 2009; Garavito et al., 2010; Shen et al., 2015). These loci, however, were mainly detected between indica (O. sativa) and japonica (O. sativa). Hybrid barriers between O. sativa and O. longistaminata have been little studied. Until now, only three loci (S13, qpsf6 and S44) have been identified between O. longistaminata and O. sativa. A pollen sterility locus S13 was primarily identified on chromosome 1 (Taneichi et al., 2005). qpsf6, causing both male and female sterility (Chen et al., 2009), was supposed to coincide with S1 derived from O. glaberrima (Koide et al., 2008). S44 induced pollen abortion and was fine-mapped to a 1.2 cm interval on the long arm of chromosome 6 (Zhao et al., 2012). None of these three genes have been cloned, nor were those that caused both pollen and embryo sac sterility.
To comprehend the intrinsic mechanism of hybrid sterility between O. sativa and O. longistaminata, a set of near-isogenic lines (NILs) was developed with RD23 as the receptor parent and an accession of O. longistaminata as the donor parent using repeated backcrossing and molecular marker-assisted selection. A new hybrid sterility locus (S40) that affected both pollen and embryo sac development was identified in this study. We intended to delimit male and female factors of S40 and develop closely linked molecular markers to transfer advantageous genes from O. longistaminata in rice breeding programs.
Materials and methods
Plant materials and mapping populations
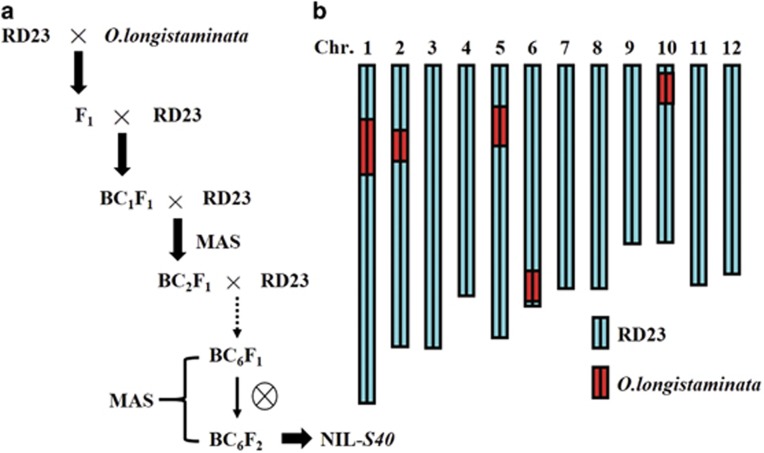
RD23 is an indica variety from Thailand, and an O. longistaminata accession was originally collected from Niger and kindly provided by Hiroshi Hyakutaka (Institute of Physical and Chemical Research, Saitama, Japan, now preserved at Yunnan Academy of Agricultural Sciences, Kunming, Yunnan, China) (Zhao et al., 2012). A backcross between RD23 and O. longistaminata was performed to detect hybrid sterility loci controlling pollen and spikelet fertility. To develop a set of NILs in the RD23 background, both pollen and spikelet semi-sterile individuals were selected as the female parent to conduct continuous backcrosses with RD23 until the BC6F1 generation. Finally, a few plants with pollen and spikelet semi-fertility from the BC6F1 generation were selected and self-pollinated (Figure 1a). In the winter of 2012 in Hainan, China, we planted 202 plants of the BC6F2 generation and identified a hybrid sterility locus controlling both pollen and spikelet fertility and temporarily named it as S40. In the summer of 2013, two parents (RD23 and NIL-S40) and an F2 population containing 16 600 individuals derived from the F1 hybrids accompanied by two BC1F1 populations (RD23//NIL-S40/RD23, NIL-S40/RD23//RD23) were planted in an experimental field at the Food Crops Institute, Jiangsu Academy of Agricultural Sciences. All materials were planted with a spacing of 16.5 × 16.5 cm. The two backcross populations (RD23//RD23/NIL-S40, RD23/NIL-S40//RD23) and another F2 population (368 individuals) were constructed to analyze the genetic patterns of the S40 locus. NIL-S40 carried a locus genotype S40lS40l (l: longistaminata), while RD23 harbored S40iS040i (i: indica).
Figure 1.
A workflow of the NIL construction and graphical genotype of one NIL plant (BC6F2), NIL-S40. Oryza longistaminata (donor) and RD23 (recurrent) were used as the parents. Blue and red bars indicate the RD23 chromosomes and O. longistaminata genomic fragment, respectively. A full color version of this figure is available at the Heredity journal online.
Fertility evaluations of pollen, embryo sac and spikelet
Ten plants from RD23, NIL-S40 and the F1 hybrids were tested to examine the pollen fertility. Six florets were collected randomly from three panicles of each plant and fixed in 70% (v/v) ethanol at room temperature until use. One anther of every floret above was mixed and stained with 1% iodine potassium iodide (I2-KI) solution, and four views were observed by light microscope. Typical type of abortion indicates pollen grains with irregular shape and no starch accumulation so that no pollen grains are stained by I2-KI (Li, 1980). Spherical type of abortion indicates spherical shape pollen grains but smaller than normal pollen grains that are not stained by I2-KI due to the absence of starch accumulation (Tao et al., 2004). Supplementary pollination experiments were conducted as follows: One day before anthesis, panicles of RD23 or F1 hybrid were bagged after removing those flowered spikelets and cutting open a gap at the upper end of non-flowering spikelets to remove anthers. Then bagged panicles were pollinated with pollen of RD23 and F1 hybrid for 4–5 days, respectively. Aniline blue staining of pollen tubes in pistils was performed to observe the behavior of pollen grains on stigma. More than 100 spikelets from RD23 and NIL-S40/RD23 were collected and fixed in FAA solution (100% ethanol: acetic acid: formalin=14:1:2) 30 min after flowering for 16 h to examine the adherence of pollen on stigma. To evaluate pollen germination and pollen tube elongation in vivo, 50 florets were collected and fixed in FAA solution 2 h after pollination. The fixed pistils were treated and stained according to the method of Huang et al. (2013). The stained pistils were observed and photographed with a Leica DM4000B fluorescence microscope. For scanning electron microscopy and transmission electron microscopy observations, mature anthers were prepared by previously reported methods (Dai et al., 2011). 4′,6-diamidino-2-phenylindole (DAPI) staining was performed according to Huang et al. (2013). To observe the embryo sac development of RD23 and F1 hybrids, spikelets at various developmental stages were excised and fixed in FAA solution until use. Thirty fixed pistils at each stage were treated and stained by the method of Dai et al. (2006). Then, these pistils were examined by confocal laser scanning microscopy (Leica TCS SP5, Leica, Wetzlar, German). More than 100 mature pistils were observed by the method above to examine embryo sac fertility. In addition, 10 plants of RD23, NIL-S40 and the F1 hybrids were examined to determine spikelet fertility with the method described by Wan et al. (1996).
Transmission ratio distortion and construction of backcrossing (BC1F1) populations
Two backcrossing (BC1F1) populations mentioned above were developed to measure S40-mediated transmission ratio distortion (TRD). One used RD23 as female parent and F1 hybrid (NIL-S40/RD23) as male parent; the other one used F1 hybrid (NIL-S40/RD23) as female parent and RD23 as male parent. The degree of TRD is frequently measured with regard to a k-value, where k is defined as the ratio of progeny that received the allele exhibiting the preferential transmission from the heterozygote (Koide et al., 2008). k-values vary from 0.5 (Mendelian segregation) to 1.0 (complete elimination of the allelic alternative). On the basis of this system, km and kf were estimated from Ns / (Nf+Ns) backcrossing data using heterozygotes as the male and female parents. Ns represents the number of semi-sterile plants (heterozygotes S40i/S40l) and Nf denotes the numbers of fertile plants (homozygotes S40i/S40i).
Development of molecular markers and sequencing analysis
To develop new simple sequence repeat (SSR) markers, a specific genomic sequence was attained from the International Rice Genome Sequencing Project (IRGSP; http://rgp.dna.affrc.go.jp/IRGSP/), and online SSR searching software SSRIT (http://www.gramene.org/microsat/) was used to screen these sequences. Then, appropriate microsatellite sequences were subjected to SSR primer design using Primer Premier 5.0 software (PREMIER Biosoft, Palo Alto, CA, USA). Additionally, insertion–deletion) markers were developed (insertion or deletion ⩾5-bp) by analyzing the sequence difference between O. longistaminata and indica variety 93-11 in the delimited region using BLASTN (http://www.ncbi.nlm.nih.gov/BLAST/). PCR primers of products ~100–250 bp were designed at ~10-kb intervals based on the sequence difference. To determine the distance between ES-2 and ES-60 on RD23 and NIL-S40 (O. longistaminata) chromosome, we developed a series of primers overlapping this region using 93-11 as reference sequence. Then these primers were used for resequencing of RD23 and NIL-S40.
Real-time PCR analysis
Fresh roots, stems and leaves at seedling and maturity, coupled with pistils and anthers at various developmental stages from RD23, were prepared for RNA extraction using an RNA Prep Pure Plant kit (Tiangen Co., Beijing, China). Real-time PCR was carried out using a SYBR Premix Ex Taq kit (Takara, Dalian, China) on an ABI Prism 7900 Real-Time PCR System after reverse transcription using a SuperScript II kit (Takara). Relative changes of gene expression were measured with the 2−△△CT method (Livak and Schmittgen, 2001). The rice ubiquitin gene (Os03g0234200) was used as a reference in the experiment (primer pair Ubq). The primers used in real-time PCR were designed online (http://quantprime.mpimp-golm.mpg.de/).
Results
Construction of near-isogenic line and fertility evaluation of parents and F1 hybrids
A total of 301 SSR markers evenly distributed on 12 chromosomes of rice were chosen to determine the genetic background of NIL-S40 and survey substituted segments. The result indicated that substituted segments was identified on chromosomes 1, 2, 5, 6 and 10 (Figure 1b).
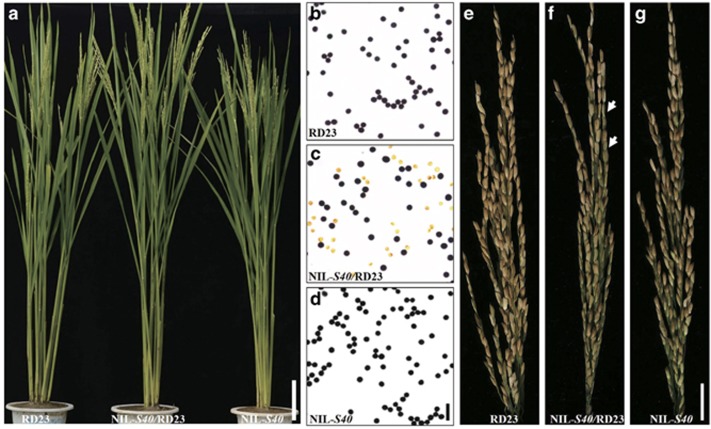
During the tillering and heading stages, F1 hybrids exhibited no obvious differences from the parents, RD23 and NIL-S40 (Supplementary Figure 1; Figure 2a). Stain I2-KI revealed that pollen grains of F1 hybrids exhibited typical semi-sterility, whereas both RD23 and NIL-S40 were fertile with pollen fertility >90% (Figure 2b–d; Table 1). Among three types of pollen abortion, that is, typical, spherical and stained abortion, the former two were the main forms observed in F1 hybrids (Figure 2c). At anthesis, anthers of the F1 hybrids demonstrated partial dehiscence (Supplementary Figures 2a–f). In vivo pollen germination test showed that pollen grains adhering to stigma of RD23, F1 hybrid and NIL-S40 could germinate efficiently and pollen tube could grow normally (Supplementary Figures 2g–l). But due to partial anther dehiscence, fewer numbers of pollen grains were poured on stigma of F1 hybrids than those of RD23 and NIL-S40 (Supplementary Figures 2g–i and m). In addition, spikelet fertility was much lower in the F1 hybrids than in RD23 and NIL-S40 (Figures 2e–g; Table 1). We conducted three supplementary pollination cross experiments to determine the main causes that lowered spikelet fertility in F1 hybrids. When RD23, as a female parent, was pollinated with the pollen of F1 hybrids (NIL-S40/RD23), the setting rate of RD23 was ~50.34±2.11%, but when pollinated with pollen of RD23 itself, the setting rate was higher, at 84.37±2.32% (Table 1). This suggested that partial anther dehiscence in F1 hybrids did affect the setting rate. When female F1 hybrids were pollinated with pollen from RD23, the setting rate of F1 hybrids was only ~35.6±1.7% (Table 1), which implied that female development in F1 hybrids was also impaired.
Figure 2.
The phenotype and fertility comparison of RD23, F1 hybrid and NIL-S40. (a) The plant architecture showed no clear differences among RD23 (left), F1 hybrid (middle) and NIL-S40 (right) after heading. Bar=15 cm. RD23 (b) showed normal pollen fertility, but the pollen of F1 hybrid (c) was sterile. The pollen fertility of NIL-S40 (d) was also normal. Bar=200 μm. (e) Spikelet fertility of RD23 was normal. (f) Spikelet fertility of the F1 hybrid was semi-sterile. (g) Spikelet fertility of NIL-S40 was normal. The arrows indicate shriveled grains. Bar=2 cm.
Table 1. The fertility-related traits of two parents and their F1 hybrids.
| Parents and hybrid | Pollen fertility (%) | Embryo sac fertility (%) | Spikelet fertility (%) | Supplementary pollinationa (%) | Supplementary pollinationb (%) |
|---|---|---|---|---|---|
| RD23 | 95.78±1.25 | 94.44±3.32 | 92.19±3.18 | 84.37±2.32 | 50.34±2.11 |
| NIL-S40 | 96.23±2.12 | 93.41±5.02 | 94.33±2.17 | — | — |
| NIL-S40/RD23 | 52.16±2.08c | 47.77±8.15c | 29.18±2.56c | 35.6±1.7 | — |
Supplementary pollination with RD23.
Supplementary pollination with NIL-S40/RD23.
Statistically significant difference with respect to their parents (P<0.001).
Cytological observation of pollen and embryo sacs in RD23 and F1 hybrids
Male gamete development has been divided into eight stages in rice (Zhang and Wilson, 2009). To reveal when microspores in F1 hybrids started to become abnormal, acetic acid carmine staining was used to observe the pollen development of RD23 and the F1 hybrids. The results demonstrated that pollen development in F1 hybrids was normal from microspore mother cell formation stage to microspore stage in RD23 (Supplementary Figures 3a–i and m). Thereafter, in the microspores of F1 hybrids and RD23 appeared clear differences at the early bicellular pollen stage: some microspores of F1 hybrids contained only one nucleus, while nearly all microspores of RD23 contained one vegetative nucleus and one generative nucleus (Supplementary Figures 3j and n). At the late bicellular pollen stage, defective microspores in F1 hybrids were easily observed due to failures in starch accumulation (Supplementary Figures 3k and o). By the mature pollen stage, typical abortive and spherical abortive pollen grains in the F1 hybrids were clearly recognizable (Supplementary Figures 3l and p). Furthermore, DAPI staining of pollen development confirmed that the defective microspores emerged at the early bicellular stage (Supplementary Figures 4a, b, d and e). At the mature pollen stage, pollen grains of RD23 contained three nuclei: two bright, intensely stained sperm nuclei and one diffuse, weakly stained vegetative nucleus (Supplementary Figures 4c and g), while half of pollen grains in F1 hybrids contained only one or no nucleus, and the other half were the same as RD23 (Supplementary Figures 4f and g). Thus, pollen abortion in the F1 hybrids occurred at the early bicellular pollen stage as a result of the failure in the first mitosis that prevented the formation of a functional generative nucleus. Transmission electron microscopy and scanning electron microscopy scanning of pollen grains at maturity revealed that defective pollen grains of F1 hybrids had irregular shape, incomplete pollen walls and little starch accumulation compared to those of RD23 (Supplementary Figure 5). No apparent defects were observed in tapetum degradation or other anther wall structures in F1 hybrids (Supplementary Figure 6). Above all, half of the pollen grains in F1 hybrids were aborted because of failure during the first mitosis, little starch accumulation and defects in pollen wall structure.
To investigate the developmental defects of embryo sacs in the F1 hybrids, confocal laser scanning microscopy was used to observe the embryo sac development in RD23 and F1 hybrids. In general, the parental (RD23) sac demonstrated normal megasporogenesis and megagametogenesis. Meiosis of a diploid megasporogenesis consisted of two successive divisions that developed to form a megagametophyte (Supplementary Figures 7a and b). After the first mitosis in the functional megaspore, the daughter nuclei were clearly observed (Supplementary Figure 7c), and a second mitosis followed to form the tetra-nucleate embryo sac (Supplementary Figure 7d). Megasporogenesis in F1 hybrids appeared to be normal (Supplementary Figures 7e and i), but in some hybrid ovaries, megagametogenesis was blocked at the uninucleate stage by the failure to form functional megaspores (Supplementary Figures 7f and j). As a result, only some hybrid ovaries could complete subsequent mitosis to form the bi-nucleate and tetra-nucleate embryo sacs (Supplementary Figures 7g, h, k and l). Finally, after the third mitosis, the embryo sacs in RD23 formed a normal style of seven cells with eight nuclei (Supplementary Figure 7m). Hybrid tetra-nucleate embryo sacs could also conduct the third mitosis to develop into mature embryo sacs as RD23 (Supplementary Figure 7n). The defective embryo sacs that failed to form functional megaspores were characterized by a lack of vacuole differentiation and less or withered cytoplasm (Supplementary Figures 7o and p). Therefore, the developmental defect at the functional megaspore formation stage resulted in the abortion of embryo sacs in F1 hybrids.
Genetic analysis of S40
TRD associated with preferential gametic dysfunction has been frequently detected in interspecific and inter-subspecific hybrids of rice (Chen et al., 2008; Koide et al., 2008, 2012; Garavito et al., 2010; Zhang et al., 2011; Zhao et al., 2011; Shen et al., 2015; Yu et al., 2016). To analyze genetic patterns of the S40 locus, we constructed two backcross populations (RD23//RD23/NIL-S40, RD23/NIL-S40//RD23). NIL-S40 carried a locus genotype S40lS40l (l: longistaminata), while RD23 harbored S40iS40i (i: indica). The segregation ratio of both genotypes and phenotypes in progeny fitted Mendel’s law when pollinating RD23 and RD23/NIL-S40 with RD23/NIL-S40 and RD23 pollen grains, respectively. In these two populations, the number of homozygotes S40iS40i was almost equal to the number of heterozygotes S40iS40l (Table 2). On the basis of the sex-independent TRD (siTRD) system, the parameters km and kf of the S40i and S40l alleles transmitted efficiently through male and female gametes were 0.51 and 0.49, respectively, which indicated that both male and female gametes carrying the S40i or S40l allele could be transmitted equally (Table 2). In addition, in the F2 population (368 individuals) derived from RD23/ NIL-S40, all homozygotes, S40iS40i and S40lS40l, were fertile, while the heterozygotes S40iS40l demonstrated typical pollen and embryo sac semi-sterility, which resulted in a 1:1 segregation ratio of fertile and semi-sterile plants (Tables 1 and 2). These results suggested that in heterozygous S40iS40l plants, male and female gametes deteriorated in a sporophytic manner.
Table 2. Segregation patterns of S40 locus for male and female gametes in the BC1F1 populations or F2 population.
|
Genetic crosses |
Progeny phenotype |
Expected segregation ratio | P-value | TRDk-value | ||||
|---|---|---|---|---|---|---|---|---|
| Female phenotype | Male phenotype | S40i/S40i | S40i/S40l | S40l/S40l | Total | |||
| S40i/S40i | S40i/S40l | 118 | 122 | 240 | 1:1 | 0.8 | km=0.51 | |
| S40i/S40l | S40i/S40i | 96 | 91 | 187 | 1:1 | 0.71 | kf=0.49 | |
| S40i/S40l | S40,i/S40l | 89 | 184 | 95 | 368 | 1:2:1 | 0.91 | |
Abbreviation: TRD, transmission ratio distortion. i and l indicate indica and longistaminata, respectively.
All homozygotes, S40iS40i and S40lS40l, were fertile (both pollen and spikelet), while the heterozygotes S40iS40l demonstrated typical pollen and embryo sac semi-sterility; P-value was obtained using the χ2-test under the hypothesis of Mendelian segregation.
Fine mapping of S40
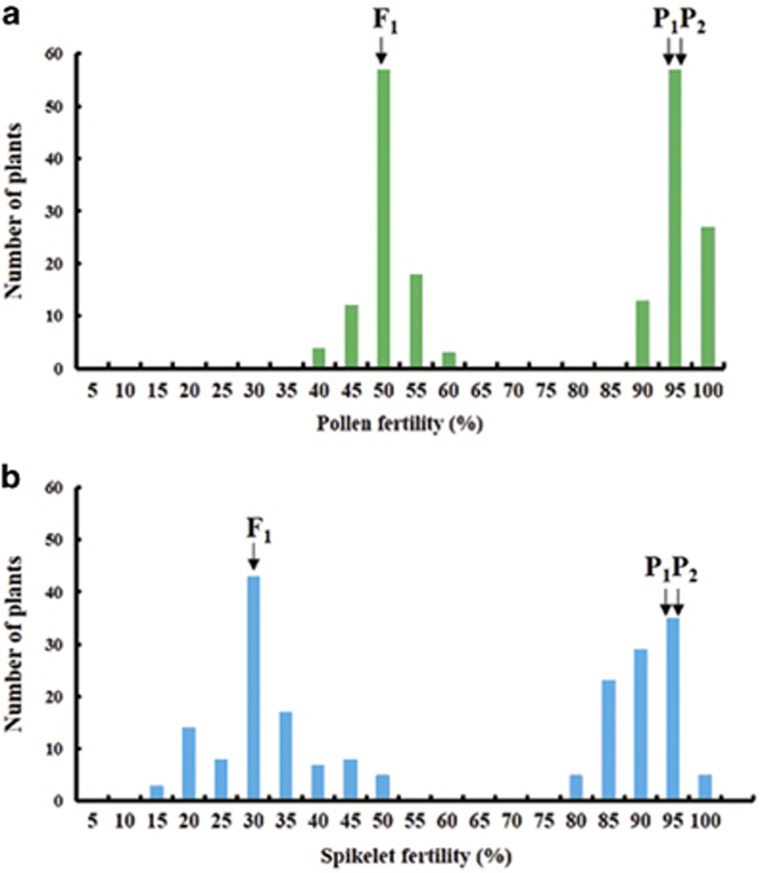
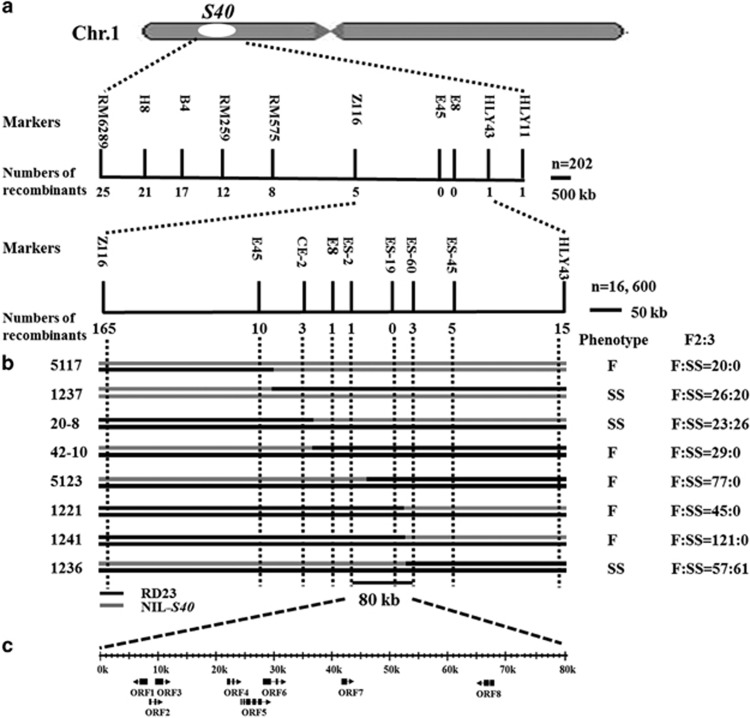
A total of 202 plants from the BC6F2 generation were used to detect the distribution of pollen and spikelet fertility and perform the primary mapping of S40 locus. The ratio of fertile plants and sterile plants (both pollen and spikelet sterility) followed the segregation ratio of 1:1 (χ2=0.15<χ20.05,1=3.84, P=0.05) (Figure 3). Twenty fertile (both pollen and spikelet full fertility) and 20 sterile (both pollen and spikelet sterility) plants were used for linkage analysis. Only markers distributed on chromosome 1 showed linkage with pollen and spikelet fertility (Supplementary Table 1). Then, the S40 locus was located to an interval of ~1.3 Mb between molecular markers Z116 and HLY43, spanning the short arm of chromosome 1 (Figure 4a). To further map the S40 locus, the flanking markers Z116 and HLY43 were used to screen a total of 16 600 individuals from the F2 population, and a total of 203 recombinants were obtained (Figure 4a). Subsequently, seven newly developed primer pairs from the interval between Z116 and HLY43 (Supplementary Table 2) were used to assay these recombination events. Finally, the S40 locus was delimited to the interval flanked by ES-2 and ES-60, with one recombinant event between ES-2 and S40 and three events between ES-60 and S40. In addition, one marker, ES-19, was determined to co-segregate with the S40 gene (Figure 4a). On the basis of reference sequences of an indica variety 93-11, the genomic region containing the S40 locus was ~80 kb in length. Through resequencing, the distance between ES-2 and ES-60 was also determined ~80 kb on the O. longistaminata chromosome. F2:3 populations of eight key recombinants (between markers E45 and ES-60) validated the fine mapping results (Figure 4b).
Figure 3.
Distribution of pollen and spikelet fertility of 202 plants in RD23/NIL-S40 F2 population. (a) Frequency distribution of pollen fertility. (b) Frequency distribution of spikelet fertility. Pollen fertility was tightly linked to spikelet fertility in the F2 population. The segregation pattern of highly fertile and partly sterile plants conformed to the expected 1:1 ratio (χ2=0.15<χ20.05,1=3.84, P=0.05). P1, P2 and F1 represent RD23, NIL-S40 and RD23/NIL-S40, respectively.
Figure 4.
Fine mapping of S40. (a) The physical map of S40. S40 was delimited to the interval flanked by ES-2 and ES-60 based on recombination events among F2 progeny (202 +16 600 plants) derived from RD23/NIL-S40. (b) The key recombinant individuals for fine mapping from the F2 population (left) and results of the corresponding F2:3 reconfirmation (right). F, male and female full fertility and SS, male and female semi-sterility. (c) Eight ORFs were predicated by Gramene system in the 80 kb DNA fragment of cv. RD23.
Analysis of candidate genes
Gene prediction of the 80-kb DNA sequence containing S40 using the online Gramene database (http://www.gramene.org/) identified eight putative open reading frames (ORFs). ORF1 encodes a putative LINE subclass retrotransposon protein; ORF2 encodes a putative uncharacterized protein; ORF3, ORF4 and ORF6 encode putative flavin monooxygenase; ORF5 encodes an ubiquitin-like SUMO protease 1 (ulp1) containing a C-terminal catalytic domain; ORF7 encodes putative expansion precursor and ORF8 encodes a putative R2R3 MYB family transcription factor (Figure 4c; Table 3).
Table 3. Putative genes at the S40 locus for RD23 based on BAC clones of cv. 93-11 and Nipponbare.
| ORF | Gramene locus ID | MSU locus ID | Predicted protein function |
|---|---|---|---|
| 1 | BGIOSGA001908 | LOC_Os01g16700 | Retrotransposon protein, putative, LINE subclass, expressed |
| 2 | BGIOSGA003243 | Expressed protein | |
| 3 | BGIOSGA003244 | LOC_Os01g16714 | Flavin monooxygenase, putative, expressed |
| 4 | BGIOSGA003245 | LOC_Os01g16714 | Flavin monooxygenase, putative, expressed |
| 5 | BGIOSGA003246 | LOC_Os01g16730 | Ulp1 protease family, C-terminal catalytic domain-containing protein, expressed |
| 6 | BGIOSGA003247 | LOC_Os01g16750 | Flavin monooxygenase, putative, expressed |
| 7 | BGIOSGA003248 | LOC_Os01g16770 | Expansin precursor, putative, expressed |
| 8 | BGIOSGA001907 | LOC_Os01g16810 | MYB family transcription factor, putative, expressed |
Abbreviations: MSU, Michigan State University; ORF, open reading frame; Ulp, ubiquitin-like SUMO protease 1.
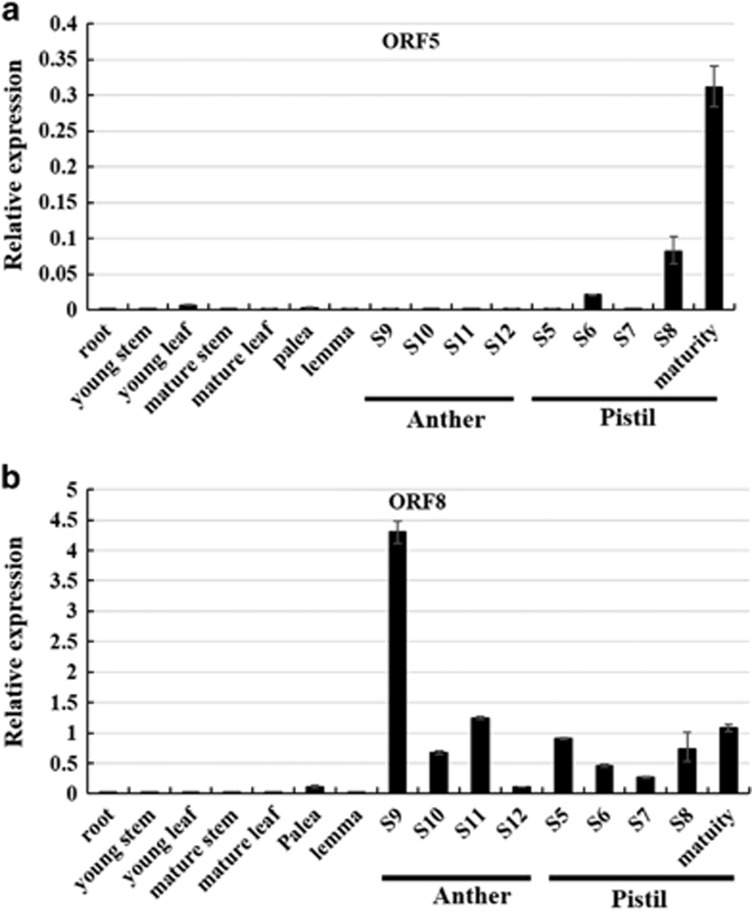
To identify candidate gene(s) for S40, we first sequenced the genome of seven ORFs (ORF2, 3, 4, 5, 6, 7 and 8) that had functional annotation from RD23 and NIL-S40. Because the amino-acid sequences of ORF2, ORF6 and ORF7 were the same as those of RD23 and NIL-S40, we excluded them as candidate genes. The amino-acid sequences of ORF3, ORF4, ORF5 and ORF8 had differences from those of RD23 and NIL-S40, but ORF3 and ORF4 might not be the candidate genes either because they were barely expressed in mature anthers or pistils (Supplementary Figure 8). Interestingly, ORF5 was highly expressed in pistils from stage 6 to the mature stage (Figure 5a), while ORF8 was predominantly expressed in anthers from stages 9 to 11 (Figure 5b). Therefore, we focused on studying ORF5 and ORF8. On the basis of genomic and protein sequence analysis, RD23-ORF5 (LOC_Os01g16730), encoding an ulp1 protein with 406 amino acids, contains four introns and five exons (Supplementary Figures 9a–c). Twenty-three single-nucleotide polymorphisms or insertion–deletions were detected in the genomic sequence of RD23-ORF5 and NIL-S40-ORF5, three of which were located in the third and fourth exon (Supplementary Figures 9a and b). An A742G mutation located in the third exon caused one variant amino acid at amino-acid position 189 in NIL-S40, which has asparagine (N) instead of aspartic acid (D) in RD23 (Supplementary Figures 9b and c). RD23-ORF8 (LOC_Os01g16810), containing only one intron and two exons, encodes a R2R3 MYB family transcription factor with 340 amino acids (Supplementary Figures 10a–c). RD23 has a 3 bp deletion mutation and a silent mutation, A940G, in the genomic sequence of ORF8 compared to NIL-S40, and these two variations are both located in the coding region of ORF8 (Supplementary Figures 10a and b). Because of the 3 bp deletion, RD23 has an amino-acid deletion (alanine) between amino acids 235 and 236 compared to NIL-S40 (Supplementary Figure 10c). Taken together, these results suggested that both ORF5 and ORF8 might be the candidate genes for S40.
Figure 5.
Expression patterns of ORF5 and ORF8 in RD23. (a) ORF5 was primarily expressed in pistil. (b) ORF8 was mainly expressed in anther.
Discussion
In this study, we identified a hybrid sterility locus S40 that was involved in both male and female development in hybrids between RD23 and O. longistaminata. Cytological observation indicated that microspores in the F1 hybrids were hindered from the first mitosis at the early bicellular pollen stage. Consequently, the proportion of aborted pollen grains with one or no nucleus in the F1 hybrids was significantly higher than those in RD23 at mature stage. Despite the normal function of tapetum, the pollen wall structure in F1 hybrids was incomplete. In addition, partial embryo sacs were arrested at the functional megaspore formation stage, and subsequent mitosis was thus unable to continue. The F1 hybrids also showed partial anther dehiscence. Above all, partial dehiscence of anthers and semi-sterile embryo sacs together led to the lower spikelet fertility of F1 hybrids. Until now, only three loci—S1 (qpsf1), S6 and S37—have been suggested to affect both pollen and embryo sac fertility, among which, S1, causing hybrid sterility between O. sativa and O. glaberrima, was the most studied (Koide et al., 2008; Garavito et al., 2010). In the S1S1a heterozygote, the second mitotic division in half of the microspores carrying the S1a genotype was defective, and abnormal polarization of the egg during megagametogenesis was the main cause of the abortion of embryo sacs (Koide et al., 2008). S1 is a complex locus, with at least two components controlling pollen and embryo sac fertility separately. The male factor causing pollen sterility was delimited to a 45-kb interval on chromosome 6 of Nipponbare (Koide et al., 2008). Thereafter, the female component was fine-mapped to a physical distance of 27.8 kb in O. sativa and 50.3 kb in O. glaberrima by Garavito et al. (2010). Shen et al. (2015) demonstrated that in F1 hybrids (DJY1/NIL-S37), nearly half of the microspores were hindered at the bicellular stage, and partial embryo sacs remained undifferentiated until the mature stage. Interestingly, neither S1 nor S37 displayed defects in anther dehiscence. Some hybrid loci did affect anther dehiscence in F1 hybrids of rice (Yao et al., 2004; Zhang et al., 2004; Mi et al., 2016). Though anther indehiscence was frequently observed in abnormal pollen of interspecific or inter-subspecific F1 hybrid, its mechanism was still little studied. Further study of S40 would be helpful to understand the mechanism of hybrid anther indehiscence in rice.
On the basis of genomic sequence analysis and real-time PCR, ORF5 and ORF8 are the most likely candidate genes for S40 (Figure 5; Supplementary Figures 9 and 10). ORF5 encodes a protein belonging to the ulp1 protease family, which has been demonstrated to be essential for processing the Small Ubiquitin-like Modifier (SUMO) precursor protein and substrate deconjugation in Arabidopsis (Colby et al., 2006). SUMOylation is characterized as an ubiquitin-like protein (UBL) conjugation process due to the fact that SUMO and the catalyzing mechanism are quite similar to those of ubiquitin (Park et al., 2011). The complex SUMOylation system regulates several biological processes in plants (Miura et al., 2007; Park et al., 2011). Most notably, SUMO modification could coordinate gene expression essential for growth and hormonal responses in plants by regulating transcription factor activity (Miura et al., 2007; Park et al., 2011). Interestingly, Sa, controlling pollen fertility in hybrids between indica and japonica, is comprised of two adjacent genes, SaM and SaF, which encode a SUMO E3 ligase-like protein and an F-box protein separately. It was implied that a SUMOylation signaling pathway may participate in the sterility process of microspores in the SaiSaj heterozygote (Long et al., 2008). In our study, ORF5 was highly and specifically expressed in pistils, and there was an amino-acid sequence difference between RD23 and NIL-S40. Thus, ORF5 may be at least one of the female components of S40 controlling female fertility in hybrids between RD23 and NIL-S40 through the SUMOylation signaling pathway.
ORF8 encodes a R2R3 MYB family transcription factor. It had been proved that several R2R3-type MYB transcription factors, such as DUO POLLEN1 (DUO1), DUO POLLEN3 (DUO3), MYB21 and MYB24 in Arabidopsis and carbon-starved anther (CSA) in rice, were indispensable for pollen development. In Arabidopsis, DUO1 and DUO3 were specifically expressed in generative cells and played a crucial role in pollen formation by activating a germline-specific differentiation procedure (Twell, 2011). MYB21 and MYB24 were supposed to affect pollen maturation, anther dehiscence and filament elongation in Arabidopsis (Song et al., 2011). CSA is necessary for sugar partitioning from leaf to anther in rice, and the second mitosis seemed to be impaired in CSA pollen grains (Zhang et al., 2010). Surprisingly, ORF8 is the CSA gene, and among eight ORFs, ORF8 was the only gene highly expressed in anthers that demonstrated differences in the amino-acid sequence between RD23 and NIL-S40. ORF8 was most highly expressed at Stage 9 during male reproductive development when microspores underwent the first mitosis. Half of the microspores in F1 hybrids became defective at this stage. Taken together, it seemed reasonable that ORF8 was the most likely candidate gene for the male component of S40.
Various genetic mechanisms have been proposed to illustrate hybrid sterility in rice (Oka, 1974). Among these models, one-locus sporophytic sterility model proposed by Oka (1974) is still an untested theory, as no such gene has been identified in any plant species to date. In this study, no specific type of male or female gametes was preferentially aborted in heterozygotes S40iS40l. This pattern was clearly distinct from the most studied hybrid sterility loci Sa (Long et al., 2008) and S5 (Chen et al., 2008) that caused gametes carrying japonica genotype preferential dysfunction rather those carrying indica genotype. Furthermore, the selfing of a S40iS40l plant produced S40iS40i (fertile), S40iS40l (sterile) and S40lS40l (fertile) plants in a ratio of 1:2:1 (Table 2), and backcrossing of S40iS40l with S40iS40i in both female and male directions produced fertile and sterile plants in a 1:1 ratio (Table 2). Therefore, the genetic mechanism of S40 was consistent with one-locus sporophytic model, and these results provide the first evidence to confirm this hypothetical model. But the underlying molecular mechanism is still an open question. Thus, by transformation of putative genes, more knowledge regarding S40 will be understood, as well as the molecular mechanism underlying hybrid sterility between Asian and African rice species. In addition, the close linkage of the S40 locus with many flanking molecular markers would be very useful for transferring the S40 fragment to different varieties in rice breeding programs.
Acknowledgments
This research was supported by the grants from the National Key Research and Development Project (2016 TFD 0101107), China, National Transform Science and Technology Program (2016ZX08001004-002), the Chinese National High Technology Research and Development Program (‘863’ Program, Nos. 2014AA10A604).
Author contributions
HC carried out major parts of all experiments. ZZ performed the primary mapping of S40. WK, LL, SY, WB, YX, HZ helped in designing the work on gene fine mapping and preparation of the manuscript. DT and JW designed the study and prepared the manuscript.
Data archiving
There were no data to deposit.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
The authors declare no conflict of interest.
Supplementary Material
References
- Bolaji AO, Nwokeocha CC. (2014). Issues concerning reproductive isolation in a rice hybrid swarm involving Oryza sativa Linn., O. longistaminata A. Chev. et Roehr. and Oryza glaberrima Steud. located in Jebba Nigeria. Int J Biol Chem Sci 7: 2040–2049. [Google Scholar]
- Brar DS (2004). Broadening the gene pool of rice through introgression from wild species. In: Toriyama K, Heong KL, Hardy B (eds). Rice is Life: Scientific Perspectives for the 21st Century, Proceedings of the world rice research conference International Rice Research Institute (IRRI): Tsukuba, Japan, pp 157–159.
- Chen J, Ding J, Ouyang Y, Du H, Yang J, Cheng K et al. (2008). A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc Natl Acad Sci USA 105: 11436–11441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hu F, Xu P, Li J, Deng X, Zhou J et al. (2009). QTL analysis for hybrid sterility and plant height in interspecific populations derived from a wild rice relative Oryza longistaminata. Breed Sci 59: 441–445. [Google Scholar]
- Colby T, Matthäi A, Boeckelmann A, Stuible HP. (2006). SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol 142: 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XM, Huang QC, Qin GY, Li GP. (2006). Observation on particular embryo sac of rice using confocal microscopy. Acta Agric Boreali Sinica 21: 26–29. [Google Scholar]
- Dai X, You C, Chen G, Li X, Zhang Q, Wu C. (2011). OsBC1L4 encodes a COBRA-like protein that affects cellulose synthesis in rice. Plant Mol Biol 75: 333–345. [DOI] [PubMed] [Google Scholar]
- Garavito A, Guyot R, Lozano J, Gavory F, Samain S, Panaud O et al. (2010). A genetic model for the female sterility barrier between Asian and African cultivated rice species. Genetics 185: 1425–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhao X, Cheng K, Jiang Y, Ouyang Y, Xu C et al. (2013). OsAP65, a rice aspartic protease, is essential for male fertility and plays a role in pollen germination and pollen tube growth. J Exp Bot 64: 3351–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, Wang Z, Wang A, Zhao Q et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R, Sokei Y, Akintayo I. (2009). Seed fertility of F1 hybrids between upland rice NERICA cultivars and Oryza sativa L. or O. glaberrima Steud. Breed Sci 59: 27–35. [Google Scholar]
- Jarvis DI et al. (2011). Damage, Diversity and Genetic Vulnerability: the Role of Crop Genetic Diversity in the Agricultural Production System to Reduce Pest and Disease Damage. Proceedings of an International Symposium; Rabat, Morocco, pp 15-17.
- Kitamura E. (1962). Genetic studies on sterility observed in hybrids between related varieties of cultivated rice. Bull Chgoku Agr Exp Stat 8: 141–205. [Google Scholar]
- Koide Y, Onishi K, Nishimoto D, Baruah AR, Kanazawa A, Sano Y. (2008). Sex-independent transmission ratio distortion system responsible for reproductive barriers between Asian and African rice species. New Phytol 179: 888–900. [DOI] [PubMed] [Google Scholar]
- Koide Y, Shinya Y, Ikenaga M, Sawamura N, Matsubara K, Onishi K et al. (2012). Complex genetic nature of sex-independent transmission ratio distortion in Asian rice species: the involvement of unlinked modifiers and sex-specific mechanisms. Heredity 108: 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Li ZB. (1980). A preliminary discussion about the classification of male sterile lines of rice in China. Acta Agron Sin 6: 17–26. [Google Scholar]
- Long Y, Zhao L, Niu B, Su J, Wu H, Chen Y et al. (2008). Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc Natl Acad Sci USA 105: 18871–18876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi J, Li G, Huang J, Yu H, Zhou F, Zhang Q et al. (2016). Stacking S5-n and f5-n to overcome sterility in indica-japonica hybrid rice. Theor Appl Genet 129: 563–575. [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Hasegawa PM. (2007). Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol 10: 495–502. [DOI] [PubMed] [Google Scholar]
- Moncada P, Martinez CP, Borrero J, Chatel M, Hjr G, Guimaraes E et al. (2001). Quantitative trait loci for yield and yield components in an Oryza sativa × Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor Appl Genet 102: 41–52. [Google Scholar]
- Oka HI. (1953). Phylogenetic differentiation of cultivated rice. The mechanism of sterility in the intervarietial hybrid. Jpn J Breed 2: 217–224. [Google Scholar]
- Oka HI. (1974). Analysis of genes controlling F1 sterility in rice by the use of isogenic lines. Genetics 77: 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Kim WY, Park HC, Sang YL, Bohnert HJ, Yun DJ. (2011). SUMO and SUMOylation in Plants. Mol Cells 32: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenburg J, Johnson DE. (2009). Weed management in rice-based cropping systems in Africa. Adv Agron 103: 149–218. [Google Scholar]
- Shen Y, Zhao Z, Ma H, Bian X, Yu Y, Yu X et al. (2015). Fine mapping of S37, a locus responsible for pollen and embryo sac sterility in hybrids between Oryza sativa L. and O. glaberrima Steud. Plant Cell Rep 34: 1885–1897. [DOI] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C et al. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Ronald P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene Xa21. Science 270: 1804–1806. [DOI] [PubMed] [Google Scholar]
- Taneichi T, Koide Y, Nishimoto D, Sano Y. (2005). Hybrid Sterility Gene S13 found in a Distantly Related Rice Species, O. longistaminata. In: 77th Annual Meeting of the Genetics Society of Japan; Tokyo, Japan.
- Tanksley SD, Mccouch SR. (1997). Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Tao DY, Xu P, Li J, Hu FY, Yang YQ, Zhou JW et al. (2004). Inheritance and mapping of male sterility restoration gene in upland japonica restorer lines. Euphytica 138: 247–254. [Google Scholar]
- Twell D. (2011). Male gametogenesis and germline specification in flowering plants. Sex Plant Reprod 24: 149–160. [DOI] [PubMed] [Google Scholar]
- Wan J, Yamaguchi Y, Kato H, Ikehashi H. (1996). Two new loci for hybrid sterility in cultivated rice (Oryza sativa L.). Theor Appl Genet 92: 183–190. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xue Y, Li J. (2005). Towards molecular breeding and improvement of rice in China. Trends Plant Sci 10: 610–614. [DOI] [PubMed] [Google Scholar]
- Xiao JH, Grandillo S, Ahn SN, Mccouch SR, Tanksley SD, Li JM et al. (1996). Genes from wild rice improve yield. Nature 384: 223–224. [Google Scholar]
- Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X et al. (2012). Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol 30: 105–111. [DOI] [PubMed] [Google Scholar]
- Yang H, Hu L, Hurek T, Reinholdhurek B. (2010). Global characterization of the root transcriptome of a wild species of rice, Oryza longistaminata, by deep sequencing. BMC Genomics 11: 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Lu YG, Feng JH, Liu XD, Zhang GQ. (2004). Anther culture characteristics of Japonica rice Taichung65 and its F1 sterile near isogenic lines. J South China Agric Univ 1: 1–4. [Google Scholar]
- Yu Y, Zhao Z, Shi Y, Tian H, Liu L, Bian X et al. (2016). Hybrid sterility in rice (Oryza Sativa L.) involves the tetratricopeptide repeat domain containing protein. Genetics 203: 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DB, Wilson ZA. (2009). Stamen specification and anther development in rice. Sci Bull 54: 2342–2353. [Google Scholar]
- Zhang H, Liang W, Yang X, Luo X, Jiang N, Ma H et al. (2010). Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22: 672–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao Z, Zhou J, Jiang L, Bian X, Wang Y et al. (2011). Fine mapping of a gene responsible for pollen semi-sterility in hybrids between Oryza sativa L. and O. glaberrima Steud. Mol Breed 28: 323–334. [Google Scholar]
- Zhang ZS, Lu YG, Feng JH, Liu XD, Zhang GQ. (2004). Studies on the anther dehiscence in F1 of hybrids between Taichung65 and its F1 sterile near isogenic lines. J Trop Subtrop Bot 12: 521–527. [Google Scholar]
- Zhao J, Li J, Xu P, Zhou J, Hu F, Deng X et al. (2012). A new gene controlling hybrid sterility between Oryza sativa and Oryza longistaminata. Euphytica 187: 339–344. [Google Scholar]
- Zhao ZG, Jiang L, Zhang W, Yu C, Zhu S, Xie K et al. (2007). Fine mapping of S31, a gene responsible for hybrid embryo-sac abortion in rice (Oryza sativa L.). Planta 226: 1087–1096. [DOI] [PubMed] [Google Scholar]
- Zhao ZG, Zhu SS, Zhang YH, Bian XF, Wang Y, Jiang L et al. (2011). Molecular analysis of an additional case of hybrid sterility in rice (Oryza sativa L.). Planta 233: 485–494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.