Abstract
The synaptonemal complex (SC) is intimately involved in the process of meiotic recombination in most organisms, but its exact role remains enigmatic. One reason for this uncertainty is that the overall structure of the SC is evolutionarily conserved, but many SC proteins are not. Two putative SC proteins have been identified in Drosophila: C(3)G and C(2)M. Mutations in either gene cause defects in SC structure and meiotic recombination. Although neither gene is well conserved at the amino acid level, the predicted secondary structure of C(3)G is similar to that of transversefilament proteins, and C(2)M is a distantly related member of the α-kleisin family that includes Rec8, a meiosis-specific cohesin protein. Here, we use immunogold labeling of SCs in Drosophila ovaries to localize C(3)G and C(2)M at the EM level. We show that both C(3)G and C(2)M are components of the SC, that the orientation of C(3)G within the SC is similar to other transverse-filament proteins, and that the N terminus of C(2)M is located in the central region adjacent to the lateral elements (LEs). Based on our data and the known phenotypes of C(2)M and C(3)G mutants, we propose a model of SC structure in which C(2)M links C(3)G to the LEs.
Keywords: meiosis, recombination, chromosome, immunogold, electron microscopy
In general terms, the structure of the synaptonemal complex (SC) is conserved among diverse organisms with two lateral elements (LEs) that run along the length of each pair of homologous chromosomes, a central element (CE) that is located midway between the two LEs, and transverse filaments (TF) that connect the LEs to the CE (reviewed in ref. 1). However, distinct differences exist among organisms, particularly in the degree of organization of the CE (2). The conditions required for SC assembly also differ, with DNA double-strand breaks being required for SC formation in some species (e.g., budding yeast, mammals, and plants) but not in others (Drosophila and Caenorhabditis elegans) (3–5). These differences may be useful in defining nonconserved features of SC as well as in highlighting conserved functions.
The morphological structures of CEs from a mammal (rat) and two insects (Drosophila and a beetle, Blaps cribrosa) were analyzed at high resolution by using EM tomography (2). In these organisms, the CE structure is essentially the same, but the degree of organization varies considerably. The CE in insects is highly organized, with two (and sometimes more) distinct longitudinal components. These dense longitudinal components appear to be composed of vertical “pillars” that link multiple layers of CE together. In comparison, the CE of mammals is less well organized; multiple layers of CE are not obvious, and the longitudinal components are so discontinuous that they typically appear as a single, rather broad, dark structure midway between LEs (2, 6). Some investigators have suggested that the longitudinal components are formed, at least partially, by the N-terminal domains of TFs (7, 8). Whether this difference among species in the degree of CE organization has any effect on SC formation or on the control of crossing-over is unknown.
TF proteins from budding yeast and mammals (Zip1, SCP1 = SYN1) and putative TF proteins from C. elegans and Drosophila (SYP1, SYP2, and C(3)G, respectively) share little primary amino-acid-sequence homology (9–14). Nevertheless, these proteins are structurally similar, each having a globular N-terminal domain, an extended coiled-coil-rich region, and a globular C-terminal domain (reviewed in ref. 5). EM immunolocalization by using antibodies raised against various TF-protein domains in budding yeast and mammals showed that the N-terminal domains were positioned in the CE, and the C-terminal domains were positioned near or in the LEs (7, 8, 11, 15). Mutants of Zip1 and SCP1 show that the C-terminal domain is required for both SC formation and for its localization to chromosomes and that changes in the length of the coiled-coil-rich region also change the width of the central region (16, 17). These data and protein interactions of isolated Zip1 and SCP1 proteins (7, 8) indicate that TF proteins interact through their coiled-coil domains to form parallel homodimers, that the C-terminal domains associate with the LEs, and that the N termini of homodimers interact in the central region (reviewed in ref. 5). TF proteins are also capable of self-assembly to form polycomplexes, aggregations of parallel LE-like structures alternating with CE-like structures (1, 8, 17, 18). The ability of TF protein to self-assemble into SC-like structures is consistent with the important role of TFs in homologous chromosome synapsis (17).
Two genes, c(3)G and c(2)M, are required for normal SC structure and for wild-type levels of meiotic crossing-over in Drosophila. Mutations in c(3)G eliminate both SC formation and meiotic recombination (refs. 19 and 20; reviewed in ref. 12). By immunofluorescence, the C(3)G protein localizes along meiotic chromosomes during synapsis, and the protein's predicted secondary structure is similar to TF proteins in other species. Therefore, Page and Hawley (12) concluded that C(3)G is a TF protein in Drosophila. The c(2)M gene was recently identified by Manheim and McKim (21). Mutations in c(2)M reduce crossing-over and disrupt SC structure by preventing the assembly of C(3)G protein along meiotic chromosomes. Because C(2)M colocalizes with C(3)G along meiotic chromosomes, Manheim and McKim (21) suggested that C(2)M is a SC protein, possibly a component of LEs. Recent bioinformatic analysis indicates that C(2)M is part of the α-kleisin superfamily of proteins that includes Rec8 (22). Rec8 is a meiosis-specific, Scc1-like protein that interacts with SMC proteins and provides sister-chromatid cohesion during meiosis in yeast (23, 24). However, C(2)M is unlikely to play a role in cohesion during meiosis in Drosophila because c(2)M mutants do not have defects in sister-chromatid cohesion (21) and C(2)M is not cleaved by separase during meiotic divisions (25). Here, using EM immunolocalization, we confirm that both C(3)G and C(2)M are SC proteins and map the location of C(3)G and C(2)M protein domains within SCs.
Materials and Methods
Fly Strains. Flies were reared at 25°C on commercial fruit fly food (Carolina Biological Supply). One strain of flies was wild type for SCs (v/v, vermilion eyes) and the other strain was transgenic [P{GAL4::VP16-nos.UTR}MVD1 P{UASP-c(2)M3XHA}28]. These transgenic flies contain the coding region for c(2)M fused at the N terminus to three copies of the hemagglutinin (HA) tag and cloned into the pUASP vector (21), which is optimized for germ-line expression but requires expression of a GAL4 activator (26). The fusion was turned on in the germ-line with GAL4 under the control of the nanos promoter and 3′ UTR (27).
Antibody Production. A GST-C(3)G6His fusion protein containing amino acids 565–743 of C(3)G (12) was injected into Balb/C mice for the generation of mouse mAb at the University of Florida Hybridoma Core Lab. Western blots of recombinant C(3)G fusion proteins indicated that mAb 1A8 binds to an epitope located in the C-terminal globular domain of C(3)G and mAb 1G5 binds within the coiled-coil-rich domain (data not shown).
A fusion protein containing amino acids 8–135 of the globular N terminus of C(3)G attached to a His6-tag at the C terminus was purified on a nickel column and used as an antigen to produce rabbit polyclonal antibodies. The antibodies were affinitypurified on antigen protein immobilized on poly(vinylidene difluoride) membrane (28).
Immunolabeling and Preparation for EM. Female flies were held with males for 3–6 d posteclosion and fed yeast the day before the experiment. Inseminated female flies were etherized, and their ovaries were dissected in cold Drosophila Ringer's solution. The ovaries were fixed and immunolabeled as described in ref. 12 by using overnight incubation with one of the primary antibodies. The primary antibodies were a mouse mAb (1A8) to the C-terminal globular domain of C(3)G (amino acids 644–743), a mouse mAb (1G5) to the C-terminal end of the central coiled-coil region (amino acids 565–633 of the entire coiled-coil segment that extends from amino acid 158 to amino acid 646), an affinity-purified rabbit polyclonal antibody to the globular N terminus of C(3)G (amino acids 8–185), or a rat mAb directed against the HA epitope (α-HA, Roche clone 3F10). The antibodies were diluted 1:100, 1:100, 1:5, and 1:50, respectively. For EM, ultrasmall (≤1 nm) colloidal-gold-conjugated secondary antibodies to mouse, rabbit, or rat IgGs (Aurion) diluted 1:50 in PBS containing 1% BSA, 0.1% cold-water fish-skin gelatin, and 2% goat serum were used. In some cases, experiments were done in parallel by using FITC-conjugated secondary antibodies (1:200, Jackson ImmunoResearch) to assess the labeling conditions. After 1–2 h of incubation with the secondary antibody, the ovaries were washed with PBS plus 0.2% Tween 20, postfixed for 10 min in 2.5% glutaraldehyde, washed thoroughly with distilled water, and silver-enhanced for 1–5 h at 20°C by using a commercial silver-enhancement kit (Aurion). The ovaries were embedded in LR White or Eponate 12 (Ted Pella). Sections were mounted on Formvar-coated slot grids, poststained with uranyl acetate and lead citrate, and photographed at a setting of ×25,000 by using an AEI 801 electron microscope. The true magnification of EM negatives was determined by using a calibrated diffraction grating replica.
To compare details of SC ultrastructure, ovaries from transgenic flies overexpressing HA-tagged C(2)M protein and from flies wild type for SCs were fixed for 1 h at room temperature in 2.5% glutaraldehyde in 100 mM phosphate buffer (pH 7.4). The ovaries were postfixed with 1% osmium tetraoxide, stained en bloc with 5% uranyl acetate in water, embedded in Eponate 12, sectioned, and poststained as described above.
Measuring the Location of Gold/Silver Particles and Determining Background. Frontal sections of SCs in which the structure of the CE was clear (e.g., the edges of the CE were well defined) and in which the lighter-staining part of the central region was the same width on either side of the CE were chosen for determining the location of gold/silver label. The distance from the middle of the CE to the nearest edge of the LE was set at 100%, and the position of each gold/silver particle was measured as a percentage of this distance. This standardization effectively minimizes differences in SC width that we observed with different preparations.
Results
Immunolabeling Specificity. Primary antibodies to C(3)G and HA-tagged C(2)M were detected by using secondary antibodies conjugated to ultrasmall (≤1 nm) gold particles that easily penetrate tissues (29, 30). This process enabled us to use a preembedding immunolabeling technique in which primary and secondary antibody incubations as well as silver enhancement of the gold particles to a size of ≈5–10 nm were done on intact, fixed ovaries before embedding in resin. The preembedding procedure increases the spatial resolution of labeling and is more sensitive than on-grid labeling of sections (30).
To verify the specificity of labeling and the amount of background labeling due to the secondary antibodies alone, some ovaries were exposed only to goat anti-rabbit or anti-mouse secondary antibodies. The label density on SCs was 10- to 100-fold greater when antibodies to C(3)G or HA-tagged C(2)M were used than when secondary antibodies were used alone (Table 1). This result indicates a high degree of labeling specificity and that few, if any, of the gold/silver particles used to determine the location of each protein domain were due to background labeling.
Table 1. Density of gold/silver particles on immunolabeled Drosophila SCs.
| Antibody | SC length examined, μm | Label density, particles per μm of SC | Label density, adjusted for background* |
|---|---|---|---|
| No primary; gold-conjugated secondary antibodies only | |||
| Goat anti-mouse | 8.5 | 0.2 | 0.0 |
| Goat anti-rabbit | 8.0 | 0.3 | 0.0 |
| Anti-C(3)G | |||
| N terminus | 8.8 | 7.0 | 6.7 |
| C terminus | |||
| 1A8 | 4.1 | 21.7 | 21.5 |
| 1G5 | 6.2 | 18.7 | 18.5 |
| Anti-HA [HA-tagged C(2)M] | 20.2 | 3.1 | 2.8† |
Density of particles was determined by counting the number of gold/silver particles on any part of an identifiable SC in a representative subset (approximately one-fourth) of all the sections analyzed.
Estimated from average level of background observed for gold-conjugated goat anti-mouse and anti-rabbit antibodies.
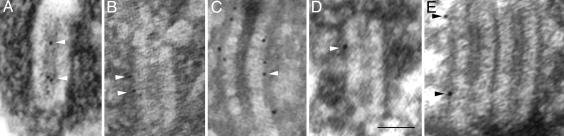
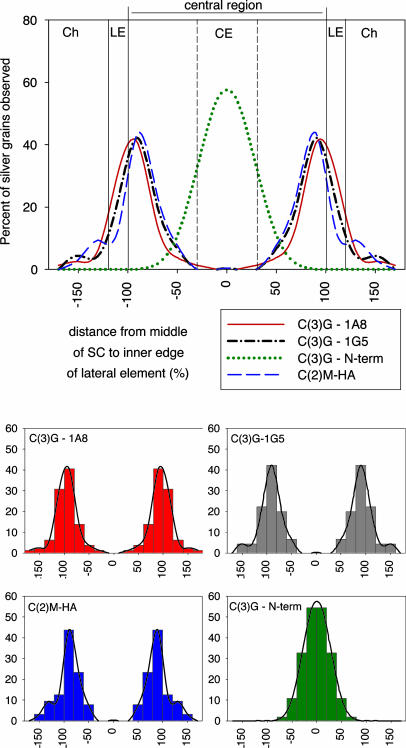
C(3)G Localization. To evaluate the location of C(3)G, we used rabbit antibodies to the N-terminal globular domain, a mouse mAb to the C-terminal end of the coiled-coil-rich domain (1G5), and a mouse mAb to the C-terminal globular domain (1A8) (Fig. 1). The distribution of gold/silver particles for the three antibodies to different parts of the C(3)G protein are shown in Fig. 2. The peak of label for the N-terminal domain of C(3)G is located in the middle of the CE. The peaks of label for both antibodies to the C-terminal portions of C(3)G are located at the very edges of the central region next to the LEs (Table 2). Thus, the orientation of C(3)G within the SC is similar to that of TF proteins from other species with the N terminus in the middle of the CE and the C terminus near or in the LEs (5). Although the peaks of label for the two C-terminal antibodies are statistically indistinguishable (Table 2), the peak for 1A8 is slightly nearer to the LEs than is the peak for 1G5. This labeling pattern is consistent with the orientation of C(3)G molecules and their expected configuration as largely linear homodimers in which the globular C-terminal domain (labeled with 1A8) would be further from the N terminus than is the C-terminal portion of the coiled-coil region (labeled with 1G5).
Fig. 1.
EM micrographs of frontal sections of Drosophila SCs (A–D) and a polycomplex (E) labeled with antibodies to the N-terminal domain of C(3)G (affinity-purified rabbit polyclonal) (A), the C-terminal end of the coiled-coil of C(3)G (mAb 1G5) (B), the C-terminal globular domain of C(3)G (mAb 1A8) (C), or the HA tag on the N terminus of C(2)M (mAb 3F10) (D and E). Some of the dark gold/silver particles are indicated by arrowheads. Although the light formaldehyde fixation necessary for immunolabeling is not as good for cellular preservation as are traditional EM fixation techniques, the basic structure of the SC is retained in these preparations (compare with Fig. 3A). (Scale bar, 100 nm.)
Fig. 2.
Localization of C(3)G and C(2)M protein domains within the SC. (Upper) Smoothed histograms of gold/silver particle distribution after labeling with antibodies to C(3)G and HA-tagged C(2)M. The distance from the middle of the CE to the near edge of the LE was defined as 100%, and the number of gold/silver particles was counted by using 20% intervals. The parts of a SC are indicated by vertical lines (Ch, chromatin). The same gold/silver particle distribution is mirrored from the middle of the CE to show the distribution over a complete SC. (Lower) Four graphs showing the underlying raw data used to generate the smoothed histograms shown above. The titles for the x and y axes on the small graphs are the same as shown for the large graph.
Table 2. Average location of gold/silver particles on Drosophila SCs after immunolabeling with one of four different antibodies.
| Antibody | Total number of gold/silver particles | Mean distance from CE, *% | SD | Median |
|---|---|---|---|---|
| C(3)G N terminus | 64 | 20.7† | 15.4 | 17.8 |
| C(3)G C terminus | ||||
| 1A8 | 79 | 96.5‡ | 23.3 | 100.0 |
| 1G5 | 45 | 94.0‡ | 22.6 | 91.9 |
| C(2)M-HA tag (N terminus) | 64 | 91.4‡ | 22.9 | 89.9 |
Middle of central element to inner edge of LE = 100%.
Mean gold/silver location is different from that of the other three antibodies (ANOVA, P < 0.001, df = 3).
Mean gold/silver locations are statistically indistinguishable (ANOVA, P > 0.42, df = 2).
C(2)M Localization. We attempted to localize the C(2)M protein using an affinity-purified rabbit polyclonal antibody raised to the C-terminal third of the protein. Although we were able to obtain fluorescent labeling using this antibody to C(2)M (21), we were not able to demonstrate gold/silver labeling, even though the conditions for fixation and labeling were exactly the same until the secondary antibody incubation. To overcome this limitation, we used transgenic flies in which three copies of the HA tag were added to the N terminus of C(2)M. The transgenic flies produce high levels of the tagged C(2)M protein when using the UAS/GAL4 system and a germ-line driver (GAL4::VP16-nos.UTR), and the construct almost fully rescues the c(2)MZ0810 mutant phenotype (21). Either the higher expression of HA-tagged C(2)M or the presence of the tag itself (which may be more accessible to antibodies than the rest of the protein) allowed us to localize the protein at the EM level. Because the HA tag is so small (only nine amino acids), even three copies of the tag should not significantly affect the localization results for the N terminus of C(2)M. Using a rat mAb to the HA tag followed by ultrasmall gold-conjugated anti-rat secondary antibodies, we observed that the peak of labeling for the C(2)M protein was at the near edge of the LE (with respect to the CE) (Figs. 1 and 2). The peaks of gold/silver particles for the N-terminal HA tag of C(2)M and the two C-terminal parts of C(3)G are statistically indistinguishable (ANOVA, P > 0.42; Table 2).
In one cell labeled with antibodies to HA-tagged C(2)M, we observed a polycomplex (Fig. 1E). Although we had only one example, the location of C(2)M label on the polycomplex was similar to that for typical SCs, indicating that the interaction between C(2)M and C(3)G in the polycomplex was similar to that of regular SCs.
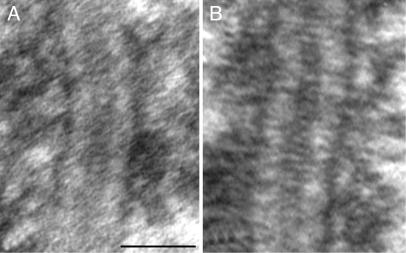
We compared the SC structure of wild-type and transgenic flies using sections from Drosophila ovaries that were prepared by using traditional EM techniques (glutaraldehyde fixation and osmium postfixation) to determine whether overexpression of C(2)M has an effect on SC structure. By EM, the SCs of wild-type and transgenic flies were indistinguishable (Fig. 3), and the average widths of the central region were not significantly different between the two types of flies (wild type, mean = 78 nm; C(2)M-HA, mean = 83 nm; two-sample t test, P = 0.197, df = 14). Thus, overproduction of C(2)M protein does not substantially affect the ultrastructural morphology of the SC.
Fig. 3.
EM micrographs of SCs from conventionally prepared (glutaraldehyde fixation and osmium postfixation) ovaries from flies, wild type for SCs (A) and overexpressing HA-tagged C(2)M (B). The structure of the SC is similar in both types of flies. (Scale bar, 100 nm.)
Discussion
The localization of the N and C termini of C(3)G shows that C(3)G proteins span the central region between the CE and the LE, indicating that C(3)G is indeed a TF protein (12). In addition, the orientation of C(3)G in the central region of Drosophila SC is identical to the orientation reported for TF proteins in budding yeast and mammals (7, 8, 11, 15). The peak of label for the N terminus of C(3)G is located in the middle of the CE (Figs. 1 and 2). This localization pattern, plus the observation by Schmekel et al. (2) that the longitudinal components of the CE of Drosophila form “pillars” that vertically connect different layers of the CE, may indicate that other proteins are associating with C(3)G to form the two longitudinal components. However, without knowledge about the threedimensional structure of the N-terminal domain or the specific epitopes recognized by the polyclonal antibody, the possibility that other protein(s) are present in the CE of Drosophila must remain open to speculation. Whether additional proteins provide the longitudinal portion of the CE in other organisms is difficult to estimate at this point because most CEs are less structured and have only one longitudinal component. Interestingly, SCP1 polycomplexes formed in somatic cells have a longitudinal component that appears comparable to a CE, but, as stated by Öllinger et al. (17), this result does not exclude the possibility that other proteins are involved in the central region in meiotic cells. In any case, it is possible that proteins in addition to C(3)G are involved in forming the intricate CE of Drosophila.
The density of gold/silver label for C(3)G is 2- to 3-fold less for antibodies to the N-terminal domain than for antibodies to the C-terminal domain (Table 1). One would expect these numbers to be equivalent because the antibodies are labeling opposite ends of the same protein. The difference in labeling density is not due to easily controlled variables such as primary and secondary antibody concentrations and silver-intensification procedures. However, the relative affinities of the different primary antibodies to the N and C termini could differ sufficiently to affect the density of gold/silver labeling. Alternatively, the N-terminal region may be less accessible to antibodies than are the C-terminal regions of C(3)G protein.
Our labeling results show that the N terminus of C(2)M is present in the central region, just at the inner edge of the LEs (Figs. 1 and 2). This result, along with the immunofluorescent localization pattern of C(2)M and the effect c(2)M mutants have on C(3)G localization, indicates that C(2)M is likely to be a LE component (21). Overproduction of HA-tagged C(2)M in transgenic flies does not noticeably alter SC structure (Fig. 3). However, Manheim and McKim (21) showed, by immunofluorescence, that the proportion of cells with SCs in the germarium increases in these transgenic flies. If the excess C(2)M is used to create more SC among cells connected by ring canals in the same cyst (31, 32), then C(2)M overproduction in this system may not be sufficient to affect the structure of most SCs.
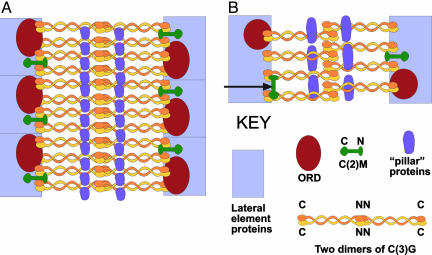
Model of SC Structure in Drosophila. We present a model for Drosophila SC structure that is based on our immunolocalization data (Fig. 4). The model is also consistent with the behavior of mutants that affect SC structure such as ord, c(2)M, and c(3)G (12, 21, 33). ORD is a protein involved in sister-chromatid cohesion (33, 34). Mutants of ord form abnormal SCs with no LEs and rather disorganized CEs (33). In ord mutants, C(2)M and C(3)G proteins initially localize into linear arrays, but these arrays break down precociously (33). Mutants of c(3)G do not form SCs (20, 35), and c(2)M mutants prevent the incorporation of C(3)G and thereby block SC formation (21). In contrast, mutants of c(3)G still incorporate C(2)M into linear arrays (21). These results suggest that both ORD and C(2)M are required for the proper integration of C(3)G and the formation of stable SC.
Fig. 4.
Model of SC structure in Drosophila females based on immunolocalization of C(3)G and HA-tagged C(2)M in frontal (A) and transverse (B) sections. The LEs are probably composed of a number of different proteins (including SMC3 and ORD). The ORD protein is present on the inner side of the LEs and provides sister-chromatid cohesion. The C(2)M protein may be located intermittently along the SC. The N terminus of C(2)M is located near the inner side of the LE close to the C termini of C(3)G proteins. Here, we have shown the C terminus of C(2)M within the LEs, but it is also possible that both N and C termini are positioned along the inner side of the LEs (arrow in B). The N termini of the C(3)G dimers are located in the middle of the CE and are connected by interacting coiled-coil segments to the C termini that are located near or in the LEs. Our data are not sufficient to distinguish whether the N termini of two dimers are overlapping (as shown) or positioned end-to-end. One or more additional proteins may be involved in forming the longitudinal components of the CE.
In the model, the LEs may be at least partially formed by ORD. Drosophila does not have a clear Rec8 homolog, and ORD (and/or the Drosophila Scc1-like Rad21 homolog) may fulfill the sister-chromatid cohesion function of Rec8 in Drosophila (25). ORD, like cohesion proteins in mouse SCP3/mutants (36), is probably one of the earliest contributors to the structure of meiotic chromosomes.
Another likely LE component is C(2)M. C(2)M falls into the α-kleisin (Rec8) family of cohesion proteins, but it is only distantly related and probably does not function in sister-chromatid cohesion during meiosis (see above). Interestingly, C(2)M has been shown to associate with SMC3 in Drosophila (25). SMC3 is another member of the cohesin complex and a component of LEs in mice (23, 37). In ord mutants, the interaction of C(2)M with SMC3 may allow the initial incorporation of C(2)M and C(3)G into linear arrays that become unstable in the absence of ord (33).
The orientation of C(2)M within the SC may be with the C terminus located in the LE and the N terminus extending into the central region. Alternatively, both the N and C termini of C(2)M could be located along the inner edge of the LEs. In either configuration, C(2)M would be available to interact with SMC3 as well as C(3)G. Because C(2)M is required for C(3)G accumulation and organization into TFs, the two proteins may interact through their N and C termini, respectively. However, no such interaction has yet been demonstrated. There seems to be a lower amount of C(2)M compared with C(3)G because C(2)M is limiting for SC formation (21). Therefore, it is possible that C(2)M is located along the SC in an intermittent pattern, perhaps similar to that observed for SMC1 and SMC3 along axial/lateral elements of rat SCs (38). Indeed, in some confocal images, C(2)M staining appears discontinuous (K.S.M., unpublished observations). If this is the case, then only a fraction of C(3)G proteins would be available to interact with C(2)M within the intact SC (Fig. 4).
Like other TF proteins, C(3)G is oriented with the N terminus in the middle of the CE and the C terminus near or in the LEs (5) (Fig. 4). It is unclear from our immunolocalization data whether the N termini of C(3)G proteins interact head-to-head or overlap and interact side-by-side (7, 16). If the N termini do overlap, the amount of overlap must be small compared with the entire length of the C(3)G protein, because the gold/silver label is found predominantly in the middle, not at the edges, of the CE (discussed above). Unfortunately, the question of C(3)G N terminus overlap cannot be addressed by comparing the predicted length of the coiled-coil portion of C(3)G [68 nm (12)] with the widths of the central region measured from EM preparations because the predicted coiled-coil lengths are based on proteins in aqueous solution, whereas the SC-width measurements are based on ovaries that have been fixed, dehydrated, and embedded, treatments that cause a considerable amount of shrinkage. Shrinkage artifacts have little effect on our immunolocalization data, however, because the locations of the gold/silver particles were determined as percentage values. The question of how the N termini of two dimers of C(3)G interact in the middle of the CE awaits the construction and analysis of defined mutations in the C(3)G protein.
Despite little sequence conservation among TF proteins C(3)G, Zip1, and SCP1/SYN1, their predicted secondary structure, orientation within the SC, and role in meiotic synapsis are clearly similar. The features that are retained among TF proteins (and possibly other SC proteins) may contribute to the structural similarity of SCs from different species. The functional role of the SC in mediating synapsis and regulating recombination between homologous chromosomes may be attributable to the unique architecture of the SC that is maintained evolutionarily, even though the primary sequences of the proteins diverge extensively. However, an alternative possibility is that SCs have arisen independently in different phylogenetic groups, and their similar structures are due to convergent evolution in response to comparable functional demands during meiosis.
Acknowledgments
We thank Tim Geary (Stowers Institute for Medical Research) for facilitating the implementation of this collaboration. This work was supported by the Stowers Institute for Medical Research (to R.S.H.), National Institutes of Health Grant GM051444 (to R.S.H.), National Science Foundation Grant MCB-0314644 (to L.K.A.), and American Cancer Society Grant RSG-98-069-04-DDC (to K.S.M.).
Author contributions: L.K.A., S.M.R., and R.S.H. designed research; L.K.A., S.M.R., and A.L. performed research; S.L.P., K.S.M., M.A.L., and R.S.H. contributed new reagents/analytic tools; L.K.A. and R.S.H. analyzed data; and L.K.A. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CE, central element; HA, hemagglutinin; LE, lateral element; SC, synaptonemal complex; TF, transverse filaments.
References
- 1.Zickler, D. & Kleckner, N. (1999) Annu. Rev. Genet. 33, 603–754. [DOI] [PubMed] [Google Scholar]
- 2.Schmekel, K., Skoglund, U. & Daneholt, B. (1993) Chromosoma 102, 682–692. [DOI] [PubMed] [Google Scholar]
- 3.McKim, K. S., Green-Marroquin, B. L., Sekelsky, J. J., Chin, G., Steinberg, C., Khodosh, R. & Hawley, R. S. (1998) Science 279, 876–878. [DOI] [PubMed] [Google Scholar]
- 4.Dernburg, A. F., McDonald, K., Moulder, G., Barstead, R., Dresser, M. & Villeneuve, A. M. (1998) Cell 94, 387–398. [DOI] [PubMed] [Google Scholar]
- 5.Page, S. L. & Hawley, R. S. (2004) Annu. Rev. Cell Dev. Biol. 20, 525–558. [DOI] [PubMed] [Google Scholar]
- 6.Schmekel, K. & Daneholt, B. (1995) Trends Cell Biol. 5, 239–242. [DOI] [PubMed] [Google Scholar]
- 7.Liu, J.-G., Yuan, L., Brundell, E., Björkroth, B., Daneholt, B. & Höög, C. (1996) Exp. Cell Res. 226, 11–19. [DOI] [PubMed] [Google Scholar]
- 8.Dong, H. & Roeder, G. S. (1999) J. Cell Biol. 148, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meuwissen, R. L., Offenberg, H. H., Dietrich, A. J. J., Riesewijk, A., van Iersel, M. & Heyting, C. (1992) EMBO J. 11, 5091–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sym, M., Engebrecht, J.-A. & Roeder, G. S. (1993) Cell 72, 365–378. [DOI] [PubMed] [Google Scholar]
- 11.Dobson, M. J., Pearlman, R. E., Karaiskakis, A., Spyropoulos, B. & Moens, P. B. (1994) J. Cell Sci. 107, 2749–2760. [DOI] [PubMed] [Google Scholar]
- 12.Page, S. L. & Hawley, R. S. (2001) Genes Dev. 15, 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacQueen, A. J., Colaiácovo, M. P., McDonald, K. & Villeneuve, A. M. (2002) Genes Dev. 16, 2428–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colaiácovo, M. P., MacQueen, A. J., Martinez-Perez, E., McDonald, K., Adamo, A., La Volpe, A. & Villeneuve, A. M. (2003) Dev. Cell 5, 463–474. [DOI] [PubMed] [Google Scholar]
- 15.Schmekel, K., Meuwissen, R. L., Dietrich, A. J. J., Vink, A. C. G., van Marle, J., van Veen, H. & Heyting, C. (1996) Exp. Cell Res. 226, 20–30. [DOI] [PubMed] [Google Scholar]
- 16.Tung, K.-S. & Roeder, G. S. (1998) Genetics 149, 817–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Öllinger, R., Alsheimer, M. & Benavente, R. (2005) Mol. Biol. Cell 16, 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth, T. F. (1966) Protoplasma 61, 346–386. [Google Scholar]
- 19.Gowen, M. S. & Gowen, J. W. (1922) Am. Nat. 61, 286–288. [Google Scholar]
- 20.Meyer, G. F. (1964) A Possible Correlation Between the Submicroscopic Structure of Meiotic Chromosomes and Crossing Over, ed. Titlback, M. (Publishing House of the Czechoslovak Academy of Sciences, Prague), pp. 461–462.
- 21.Manheim, E. A. & McKim, K. S. (2003) Curr. Biol. 13, 276–285. [DOI] [PubMed] [Google Scholar]
- 22.Schleiffer, A., Kaitna, S., Maurer-Stroh, S., Glotzer, M., Nasmyth, K. & Eisenhaber, F. (2003) Mol. Cell 11, 571–575. [DOI] [PubMed] [Google Scholar]
- 23.Jessberger, R. (2002) Nat. Rev. Mol. Cell Biol. 3, 767–778. [DOI] [PubMed] [Google Scholar]
- 24.Petronczki, M., Siomos, M. F. & Nasmyth, K. (2003) Cell 112, 423–440. [DOI] [PubMed] [Google Scholar]
- 25.Heidmann, D., Horn, S., Heidmann, S., Schleiffer, A., Nasmyth, K. & Lehner, C. F. (2004) Chromosoma 113, 177–187. [DOI] [PubMed] [Google Scholar]
- 26.Rorth, P. (1998) Mech. Dev. 78, 113–118. [DOI] [PubMed] [Google Scholar]
- 27.Van Doren, M., Williamson, A. L. & Lehmann, R. (1998) Curr. Biol. 8, 243–246. [DOI] [PubMed] [Google Scholar]
- 28.Lammers, J. H., Offenberg, H. H., van Aalderen, M., Vink, A. C., Dietrich, A. J. J. & Heyting, C. (1994) Mol. Cell. Biol. 14, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burry, R. W. (1995) in Immunogold-Silver Staining: Principles, Methods and Applications, ed. Hayat, M. A. (CRC, New York), pp. 217–230.
- 30.Yi, H., Leunissen, J. L. M., Shi, G.-M., Gutekunst, C.-A. & Hersch, S. M. (2001) J. Histochem. Cytochem. 49, 279–283. [DOI] [PubMed] [Google Scholar]
- 31.Carpenter, A. T. C. (1979) Genetics 92, 511–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmekel, K., Wahrman, J. & Daneholt, B. (1993) Chromosoma 102, 396–402. [DOI] [PubMed] [Google Scholar]
- 33.Webber, H. A., Howard, L. & Bickel, S. E. (2004) J. Cell Biol. 164, 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bickel, S. E., Orr-Weaver, T. L. & Balicky, E. M. (2002) Curr. Biol. 12, 925–929. [DOI] [PubMed] [Google Scholar]
- 35.Smith, P. A. & King, R. C. (1968) Genetics 60, 335–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelttari, J., Hoja, M.-R., Yuan, L., Liu, J.-G., Brundell, E., Moens, P., Santucci-Darmanin, S., Jessberger, R., Barbero, J. L., Heyting, C., et al. (2001) Mol. Cell. Biol. 21, 5667–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eijpe, M., Offenberg, H. H., Jessberger, R., Revenkova, E. & Heyting, C. (2003) J. Cell Biol. 160, 657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eijpe, M., Heyting, C., Gross, B. & Jessberger, R. (2000) J. Cell Sci. 113, 673–682. [DOI] [PubMed] [Google Scholar]