Abstract
Phosphocholine (PC) and total choline (tCho) are increased in malignant breast tumors. In this study, we combined magnetic resonance spectroscopic imaging (MRSI), mass spectrometry (MS) imaging, and pathological assessment of corresponding tumor sections, to investigate the localization of choline metabolites and cations in viable versus necrotic tumor regions in the nonmetastatic MCF-7 and the highly metastatic MDA-MB-231 breast cancer xenograft models. In vivo 3-dimensional MRSI demonstrated that high tCho levels, consisting of free choline (Cho), PC, and glycerophosphocholine (GPC), displayed a heterogeneous spatial distribution in the tumor. MS imaging performed on tumor sections detected the spatial distributions of individual PC, Cho, and GPC, as well as sodium (Na+) and potassium (K+), among many others. PC and Cho intensity were increased in viable compared to necrotic regions of MDA-MB-231 tumors, but relatively homogeneously distributed in MCF-7 tumors. Such behavior may be related to the role of PC and PC-related enzymes, such as choline kinase, choline transporters and others in malignant tumor growth. Na+ and K+ colocalized in the necrotic tumor areas of MDA-MB-231 tumors, whereas in MCF-7 tumors, Na+ was detected in necrotic and K+ in viable tumor regions. This may be attributed to differential Na+/K+ pump functions and K+ channel expressions. Principal component analysis of the MS imaging data clearly identified different tumor microenvironmental regions by their distinct molecular signatures. This molecular information allowed us to differentiate between distinct tumor regions and tumor types, which may, in the future, prove clinically useful in the pathological assessment of breast cancers.
Keywords: breast cancer, choline, phospholipids, mass spectrometry imaging, magnetic resonance spectroscopic imaging
Introduction
Phosphocholine (PC) and total choline-containing metabolite levels (tCho: glycerophosphocholine (GPC) + PC + free choline (Cho)) are elevated in brain (1), prostate (2, 3), colon (4), and lung tumors (5), in which the most aggressive tumors display the highest PC and tCho levels (6). Increases in activity and/or expression of several enzymes in choline metabolism, such as choline transporters (7), choline kinase (8), and phospholipases C (9) and D (10, 11) are responsible for the elevated PC and tCho concentrations in cancers, and provide potential therapeutic targets. Choline kinase has already been developed as a target for treatment (5, 12) and a predictive marker for cancer prognosis (5). Aberrant choline phospholipid metabolism has also been correlated to other diseases, such as neurological disorders including Alzheimer (13, 14), multiple sclerosis (15), inflammation-related pathologies (16), and apoptosis. It is therefore important to develop diagnostic applications to detect choline metabolites that can be translated to the clinic.
Magnetic resonance spectroscopic imaging (MRSI) has been used to detect the extent of invasion of brain, prostatic, and breast tumors (17, 18), and to differentiate between recurrence or necrosis following treatment (1, 19). Magnetic resonance spectroscopy (MRS) studies have demonstrated an elevation of phosphocholine (PC) and tCho-containing metabolites in breast cancer cells and tumors (11, 12, 20–23). However, high spatial resolution in vivo MRSI with the chemical specificity to resolve PC, GPC, Cho is currently unavailable. MS imaging of histological tumor sections is able to detect the spatial localization of these choline metabolites and hundreds of other molecules from the tissue surface. In this study, MS imaging techniques have been applied to investigate the spatial distributions of Cho, PC, and other choline-related phospholipids in microenvironmental regions of breast cancer xenograft tumors to further elucidate the involvement of choline metabolism in cancer growth and spread (21).
MS imaging provides molecular identification of lipids, peptides, and proteins by their molecular weight and determines their spatial distribution on the tissue surface at the same time (24–27). Sectioned samples from biopsies or other biological materials can be directly analyzed without the need for chemical labeling or extended sample preparation, and hundreds of biomolecules can be detected on a single tissue slide. Secondary ion mass spectrometry (SIMS) and matrix assisted laser desorption ionization (MALDI) are able to detect molecular distributions on histological tissue sections (28) at high spatial resolution, reaching spatial resolutions bellow 50 μm with MALDI-MS imaging and in the sub-micrometer range with SIMS (29, 30), while in-vivo MRSI provides 500 μm spatial resolution (31). Here, we have developed an approach using complementary desorption and ionization techniques directly applied on tissue obtained from breast cancer xenograft models.
Our purpose in this study was to further elucidate the spatial and molecular characteristics of the altered choline phospholipid metabolism typically observed in breast cancers. To this end, we have combined MRS studies with newly developed MS imaging approaches, and have analyzed corresponding hematoxylin and eosin stains to differentiate invasive and noninvasive breast cancer models based on their molecular signatures, with a special focus on choline metabolites. We observed increased PC levels in viable tumor regions of a highly metastatic breast tumor model, and a close to homogeneous PC distribution in a nonmetastatic, estrogen-sensitive breast tumor model. MS imaging also detected Cho, sodium and potassium among other molecules, in distinct tumor regions.
Material and Methods
Cell lines, tumor model and inoculations
MDA-MB-231, a highly metastatic human mammary epithelial cancer cell line, and MCF-7, a nonmetastatic, estrogen-sensitive (estrogen-dependent) line, were used for inoculation to generate breast tumor xenograft models. MCF-7 and MDA-MB-231 breast cancer cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and used within six months of obtaining them from ATCC. Both cell lines were tested and authenticated by ATCC by two independent methods: The ATCC cytochrome C oxidase I PCR assay and short tandem repeat (STR) profiling using multiplex PCR. Cell culture of these cell lines was performed as previously described (32). MCF-7 or MDA-MB-231 cells were inoculated in the upper left thoracic mammary fat pad of female severe combined immunodeficient (SCID) mice. For MCF-7 cell inoculations, 0.18 mg 60-day release 17β-Estradiol Pellets (Innovative Research of America, Sarasota, Florida) were subcutaneously implanted near the left shoulder of SCID mice one week prior to inoculation. 2×106 tumor cells were inoculated in a volume of 0.05 ml Hanks balanced solution (HBSS, Sigma, and St. Louis, MO; no matrigel was used). Mice weighed between 19 g – 24 g and tumor sizes ranged between 75 mm3 – 375 mm3 when experiments were performed. All experimental animal protocols were approved by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine.
Sample preparation
Tumors frozen in gelatin were sectioned at 10-μm thickness using a cryo-microtome (Microm Int., Walldorf, Germany) and attached to a cold indium tin oxide (ITO)-coated glass slide (Delta technologies, Stillwater, MN, USA) and stored at −80°C. Prior to analysis, each tumor section was put into a dessicated chamber until it reached room temperature to avoid water film formation on the tissue surface. After the sections reached room temperature, different sample preparation procedures were applied depending on the MS imaging techniques used in this study.
No tissue washing step of tumor sections was required prior to SIMS experiments.
For Metal Assisted (MetA)-SIMS, a thin layer of gold was applied to the tissues surface. The tumor sections were sputter coated with 1 nm gold using a Quorum Technologies (Newhaven, East Sussex, U.K.) SC7640 sputter coater equipped with a FT7607 quartz crystal microbalance stage and a FT7690 film thickness monitor.
For Matrix Enhanced (ME)-SIMS and MALDI-MS, the tissues were covered with 2,5-Dihydroxybenzoic acid (DHB) 30mg/mL in 50% acetonitrile/0.1% aqueous trifluoroacetic acid using air driven thin liquid chromatography sprayer (Sigma). The nitrogen pressure required for efficient nebulization was 0.3–0.4 bar, resulting in matrix crystals of μm size. Alternatively, matrix was deposited on the tissue sections with a vibrational spray coater ImagePrep instrument (Bruker Daltonics, Bremen, Germany). The parameters for each cycle were 1.5 s spray time, 0.3 s incubation time and 0.7 s drying time. The chosen amplitude was of 20% spray power and the membrane offset was optimized with pure ethanol at 10% spray power as recommended by the manufacturer. The membrane was cleaned after every 30 cycles to eliminate clogging effect caused by matrix.
For some MALDI studies and ME-SIMS, a thin layer of gold was applied to the surface after matrix application. The tissues were sputter coated with 1 nm gold as described above for MetA-SIMS.
In vivo/ex vivo MRS measurements
Noninvasive 1H MRSI was performed on a 9.4-T Bruker Biospec Small Animal MR Scanner using a surface coil as previously described (3). Briefly, mice were anesthetized with an i.p. injection of ketamine (25 mg/kg; Phoenix Scientific, Inc.) and acepromazine (2.5 mg/kg; Aveco, Phoenix Scientific) diluted in saline. Body temperature was maintained using a blanket circulated with warm water. Water-suppressed MRSI spectra were acquired of tumors using 3-dimensional (3D) chemical shift imaging (CSI) with VAPOR water suppression. The acquisition parameters were as follows: sweep width 7000 Hz, data size of 512 points, echo time 82 ms, repetition time 1 s, field of view 8 mm × 8 mm × 8 mm, and voxel size of 1 mm × 1 mm × 1 mm, zero filling to 32 × 32 × 32. For spatial referencing of the tumor shape, water-unsuppressed MRSI spectra were acquired with an echo time of 15 ms, and all other parameters were the same as for water-suppressed spectra. Spectra were processed with in-house IDL software, and displayed in AMIRA 5.2.1 (Visage Imaging, Inc., San Diego, CA). High-resolution MRS was acquired of the water-soluble dual-phase extract fractions from the respective breast cancer cell lines as previously described (33). Briefly, approximately 3×107 cells were harvested by trypsinization and dual-phased extraction was performed with methanol/chloroform/water (1:1:1, v/v/v) as previously described (33). Samples were vortexed, incubated overnight at 4 °C, and the phases were separated by centrifugation. Divalent cations were removed from the water-methanol phase using chelex (Sigma-Aldrich, City, Country), methanol was removed by rotary evaporation, and the remaining water phase lyophilized and stored at −80 °C. The samples were dissolved in D2O, containing 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid (TSP; Sigma-Aldrich) as chemical shift reference. Fully relaxed high-resolution 1H MR spectra were acquired on a Bruker Avance 500 spectrometer operating at 11.7 T using a 5-mm HX inverse probe as previously described (33, 34). MR spectra were processed as previously described using Bruker XWIN-NMR 3.5 software (Bruker BioSpin) (33, 34).
Ex vivo MS imaging measurements
SIMS
SIMS imaging experiments (SIMS, MetA-SIMS and ME-SIMS) were performed on a Physical Electronics (Eden Prairie, MN) TRIFT II time-of-flight SIMS (ToF-SIMS) equipped with either 115In+ or Au+ liquid metal ion gun. Experiments with the Indium gun were performed with a primary ion beam current of 450 pA, a primary pulse length of 30 ns, a spot diameter of 500 nm, and a primary ion energy of 15 keV. The raster size (or field of view-FOV) chosen for these SIMS experiments varied from a minimum of 62.5 μm up to 200 μm per tile, and were constant throughout the experiment, covering the entire tumor surface. The FOV is calculated by WinCadence 4.4.0.17 software (ULVAC-PHI Inc.) based on the maximum tile size and the total measurement area defined by the user. The data acquisition and analysis was performed by WinCadence 4.4.0.17 software.
MS microscope mode MALDI
MALDI stigmatic MS imaging was performed on a modified TRIFT-II instrument equipped with a Nd:YAG 355 nm laser and a DMCP-PS CCD camera combination as previously described (35). The sample was completely scanned with a homogeneous laser pulse of 150 × 200 μm and a laser repetition rate of 10 Hz, while the sample stage moved at a constant velocity of 100 μm/s. For MALDI data, single-shot 200 μm Total Ion Count (TIC) MALDI stigmatic images were acquired with DaVis software (LaVision, Göttingen, Germany) and stored as tagged image file format (tiff) files with the corresponding synchronized ADC signals (Acqiris, Geneva, Switzerland). To image an entire section, the sample stage was moved at 100 μm/s at a laser repetition rate of 10 Hz in a line-scan, which was repeated until the entire tumor surface was imaged (36).
MS Microprobe MALDI
MALDI microprobe MS imaging was performed on an Ultraflex-ToF (Bruker, Germany), with an accumulation of 500 shots per position and each position covered by a 150×150 μm area. The acquisition was controlled by FlexControl 2.0 (Bruker Daltonics) and the measurement area selection as well as data visualization was executed with FlexImaging 2.0 software (Bruker Daltonics).
Data processing and principal component analysis (PCA)
The data acquired in microscope mode were processed with our in-house developed imaging software (SIC) which aligned all the acquired data into a Total Ion Image (37).
All MS imaging raw data acquired on different MS instrumentation were converted to the generic MS imaging data format datacube (x, y, m/z) using in-house developed software. Images were generated with publically available software “Datacube Explorer” (www.imzml.org).
Principal component analysis (PCA) was performed using in-house developed ChemomeTricks toolbox for MATLAB version 7.0 (The MathWorks, Natick, MA) (38). PCA is a widely used multivariate data analysis method, described in numerous articles (39, 40). In short, PCA reduces the dimensionality of the dataset by the creation of a new set of variables, the principal components. These principal components are linear combinations of the original variables. Closely correlated variables (i.e. mass channels originating from the same chemical compound) are grouped into the same principal component. The principal components are hierarchically sorted by the amount of the total variance they describe. The first principal component (PC1) represents the largest amount of the total variance; noise-related signals are found in the higher-ranked principal components. Discarding these higher-ranked principal components from further data processing greatly reduces the noise in the data (38).
Results
MS imaging analyses showed a distinct difference in the lipid, and more specifically PC distribution of the highly invasive and metastatic, triple negative (estrogen-, progesterone-, and ErbB2-negative) MDA-MB-231 breast tumor model compared to the nonmetastatic, estrogen- and progesterone-positive and ErbB2-negative MCF-7 breast tumor model. Increased PC and overall choline-containing compound levels in breast tumors have been related to tumor aggressiveness in previous studies (11, 12, 20, 21, 41) and are shown as representative in vivo 1H MR spectrum in Figure 1A and 3D 1H MRSI data in red in Figure 1B. These spatially resolved data provided in vivo information on the tCho distribution, and showed that high tCho levels were typically found only in specific regions within the tumor. The tCho signal consisted of at least three individual compounds: free choline, PC, and GPC, which were resolved by ex vivo MRS at 11.7 T in water-soluble extracts from MDA-MB-231 breast cancer cells as shown in Figure 1C.
Figure 1.
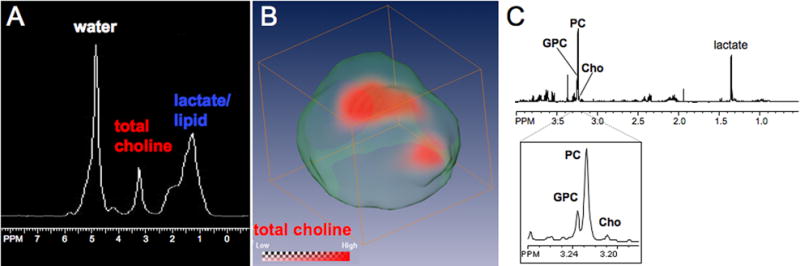
(A) Representative in vivo 1H MR spectrum demonstrating that the total choline signal cannot be spectrally resolved in vivo. (B) Representative 1H MRSI data set showing the spatial distribution of tCho displayed in red within an MDA-MB-231 tumor as described in the experimental section. (C) High-resolution MR spectrum obtained from a representative water-soluble MDA-MB-231 cell extract resolving GPC, PC, and Cho spectrally.
Figure 3.
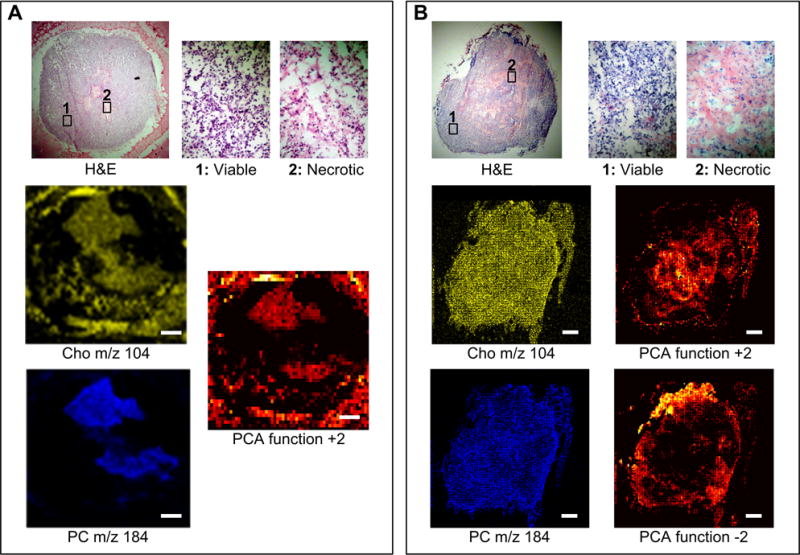
(A) MALDI-MS microscope mode data set of a representative MDA-MB-231 tumor showing the H&E image, the corresponding MS imaging of Cho and PC, and the PCA image of the function +2. (B) SIMS microprobe data set of a representative MCF-7 tumor showing the H&E image, the corresponding MS imaging of Cho and PC, and the PCA images of the functions +2 and −2 providing inverse images, in which tumor regions are defined by masses other than Cho and PC. Scale bar: 1 mm
MS imaging was able to detect Cho, PC, and several other ions and biomolecules in the tumor models, and to localize them to specific tumor regions, as summarized in Table 1. The ratio of elements sodium (Na+) and potassium (K+) changed in two tumor regions (e.g. viable tumor and necrotic region) as shown in Table 2 and Figure 2. Several other molecules were also found in both tumor types and are shown in the overall spectra (m/z 0 to 1000) in Figure 2. The spatial distribution of an array of biomolecules was able to discern several tumor microenvironments in the two tumor models, as shown in Figures 3 and 4. The assignments of molecules were made based on the unique masses of single elements (e.g. Na+ (M.W. 22.989), K+ (M.W. 39.098), the calculated molecular weight of more complex substances, and by measurement of standards. The lipid database Lipid Maps (http://www.lipidmaps.org) was also used in the identification process. In each tumor section, the distinction between these different tumor microenvironments was possible solely based on the mass distribution of these molecules (Figure 3, 4). To correlate substances found by MS imaging within these tumor microenvironments, adjacent tumor sections were hematoxylin and eosin (H&E) stained, and compared to the MS imaging data sets, which enabled the distinction of viable and necrotic tumor regions. Viable tumor regions were characterized by purple hematoxylin-stained nuclei of cancer cells, and necrotic tumor regions by the absence of nuclei. Individual molecules could be visualized at cellular level by high spatial resolution MetA-SIMS (Figure 5). The analyzed sections by MetA-SIMS were H&E stained afterwards.
Table 1.
List of detected compounds.
| Compound | Formula and common name | Designation | Calculated m/z | MALDI | SIMS | MCF-7 predominant region | MDA-MB-231 predominant region |
|---|---|---|---|---|---|---|---|
| Na+ | Na | [M]+ | 22.98 | • | Necrotic region | Necrotic region | |
| K+ | K | [M]+ | 39.09 | • | Viable tumor | Necrotic region | |
| Choline (Cho) | C5H14NO+ | [M]+ | 104.17 | • | • | Homogeneous | Slightly stronger in necrotic region |
| Cho.H2O | C5H14NO.H2O | [M+H2O]+ | 122.18 | • | • | Viable tumor | |
| Phosphocholine (PC) | C5H15PNO4 | [M]+ | 184.07 | • | • | Homogeneous | Viable tumor |
| 1,2-Diacylglycerol | C36H66O5 DG(16:1(9Z)/17:1(9Z)/0:0)[iso2] or DG(16:0/17:2(9Z,12Z)/0:0)[iso2] |
[M]+ | 578.49 | • | Homogeneous | Homogeneous | |
| Phosphatidylcholine (PtdCho) dipalmitoyl | C38H74NO10P PS(16:0/16:0)[U] |
[M]+ | 735.50 | • | Barely detected | Outer part |
Table 2.
Relative intensity scale of Na+ and K+ distributions in MDA-MB-231 and MCF-7 tumors (see Figure 2), measured from 3 by 3 tiles corresponding to about 300 μm by 300 μm. All values were normalized to the values obtained for Na+ in the viable regions of the respective tumor model.
| Compound | Region selection | MDA-MB-231 Relative intensity |
MDA-MB-231 Normalized value |
MCF-7 Relative intensity |
MCF-7 Normalized value |
|---|---|---|---|---|---|
| Na+ | Necrotic region | 26.6×10+5 | 1.73 | 100×10+4 | 5.88 |
| Viable region | 15.4×10+5 | 1.00 | 17×10+4 | 1.00 | |
| K+ | Necrotic region | 57.0×10+5 | 3.70 | 59×10+4 | 3.47 |
| Viable region | 42.0×10+5 | 2.72 | 98×10+4 | 5.76 |
Figure 2.
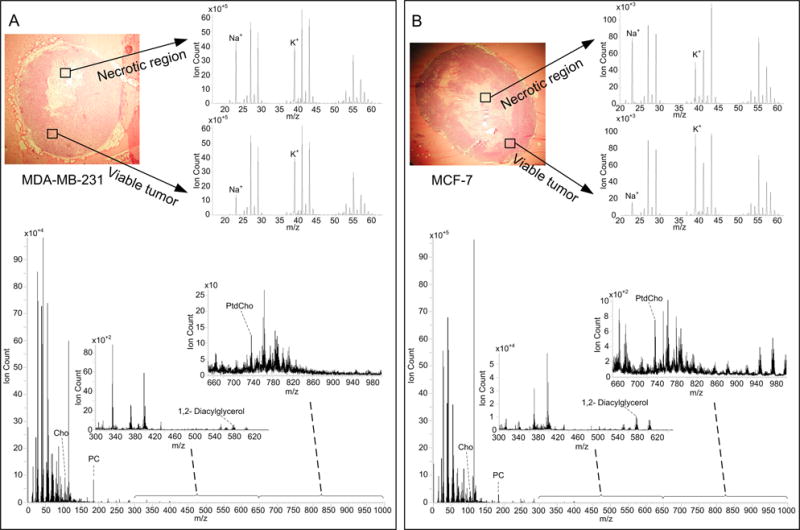
Optical images of H&E stained sections of (A) a representative MDA-MB-231 tumor and (B) a representative MCF-7 tumor. The MS spectra are provided for two selected areas, containing two expanded mass regions (m/z 15 to 65) with peak assignment of Na+ and K+, and the total area spectrum (m/z 1 to 1000) including two expanded spectral regions (m/z 300 to 640, m/z 640 to 1000) with peak assignments for Cho, PC, cholesterol, 1,2-Diacylglycerol and PtdCho.
Figure 4.
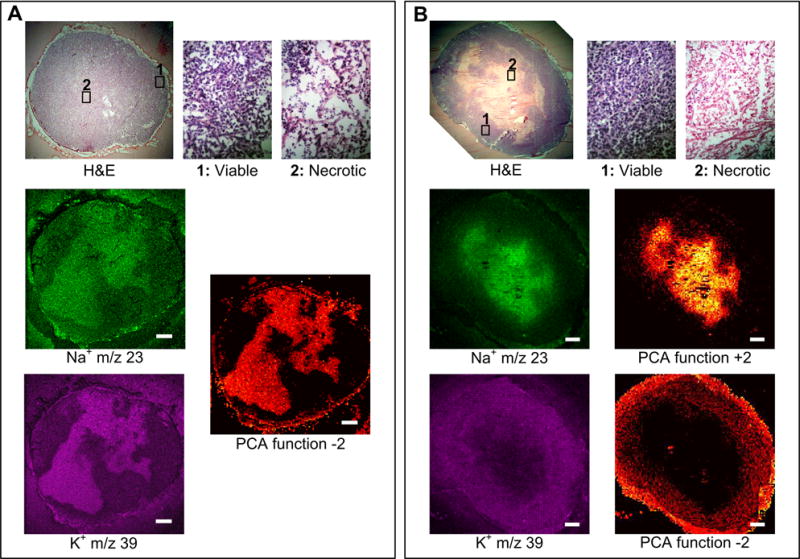
(A) SIMS microprobe data set of a representative MDA-MB-231 tumor showing the H&E image, the corresponding MS imaging of Na+ and K+, and the PCA image of the function −2. (B) SIMS microprobe data set of a representative MCF-7 tumor showing the H&E image, the corresponding MS imaging of Na+ and K+, and the inverse PCA images of the functions +2 and −2. Scale bar: 1 mm.
Figure 5.
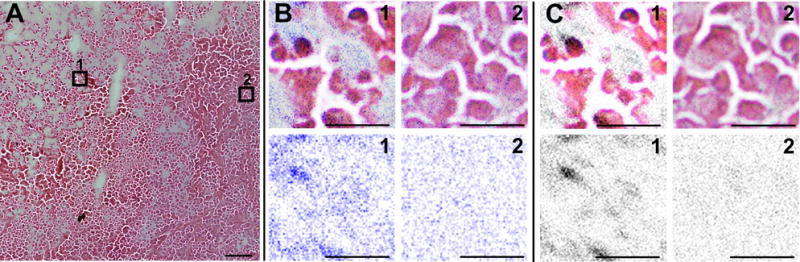
(A) Optical image of a small part of a representative MDA-MB-231 tumor, stained with H&E after concluding a MetA-SIMS experiment with (1) necrotic region and (2) viable tumor areas selected. (B) Na+ shown in blue and (C) K+ distribution shown in gray from MetA-SIMS data and overlay with microscopic image in regions (1) and (2) are shown in the upper panel; the individual ion distributions in areas (1) and (2) are shown in the bottom panel. Due to previous gold coating of the tissue, the hematoxylin staining was less efficient, and therefore the visibility of cell nuclei is less pronounced. Scale bar: (A) 100 μm; (B) and (C) 25 μm.
The distribution of PC in highly metastatic MDA-MB-231 tumors was heterogeneous, as shown by the MALDI-MS data shown in Figure 3A, with high PC concentrations, displayed in blue, localized to distinct tumor regions within viable tumor regions (Figure 3). By applying PCA, PC was identified to be the most important compound in differentiating these tumor microenvironmental regions in MDA-MB-231 tumors, as evident in PCA function +2 (Figure 3A). In contrast, the PC intensity was relatively homogeneously distributed in nonmetastatic MCF-7 tumor models as shown by the SIMS data in Figure 3B, and was not a determining factor in the tumor region assignment using PCA in the MCF-7 tumor model. This was evident from both PCA function +2 and −2, describing 1.6% of the total variation in Figure 3B, which did not match the PC distribution in MCF-7 tumors and used different molecules for the region assignment. PCA function +2 in MCF-7 tumors outlined distinct tumor microenvironmental regions within the hematoxylin-stained viable tumor regions.
Other lipids also demonstrated a heterogeneous spatial distribution in the MDA-MB-231 tumor model in MALDI-MS, SIMS, ME-SIMS (data not shown) and MetA-SIMS modes, such as the distribution of Cho by MALDI-MS, shown in yellow in Figure 3. Cho was present in several regions, and overlapped with PC. In contrast, such heterogeneous spatial distributions were not observed in the MCF-7 tumor model in the case of molecules ranging between m/z 100 to 1000 with MALDI-MS (data not shown), SIMS, ME-SIMS (data not shown) and MetA-SIMS (data not shown), with emphasis on PC, which showed a relatively homogeneous distribution throughout the entire tumor section (Figure 3B, acquired with SIMS).
The spatial distributions of Na+ and K+, detected with SIMS and MetA-SIMS, were also distinctively different between MCF-7 and MDA-MD-231 tumors, as shown in Table 1 and Figure 4. In MDA-MD-231 tumors shown in Figure 4A, Na+ and K+ displayed the same spatial distributions and were positively correlated in PCA. In MCF-7 tumors, Na+ and K+ were present in different regions, negatively correlated, and a determining factor in the PCA (Figure 4B), showing the inverse images of PCA function +2, with Na+ as the determinant region assignment factor and function −2, where K+ provides the determinant region assignment factor. This characteristic finding could be used as a criterion for region assignment in MCF-7 tumors and as a criterion to differentiate between the two tumor types. It is also evident that levels of high Na+ mostly overlap with necrotic regions in the H&E-stained adjacent sections in both tumor models. Levels of high K+ mostly overlap with necrotic regions in MDA-MB-231, and with viable regions in MCF-7 tumors. Experiments performed at high spatial resolution in MDA-MB-231 tumors show that high K+ concentrations localized to hematoxylin-positive bodies in necrotic regions, and that a homogeneous K+-distribution was detected in the viable tumor (Figure 5). Simultaneously, it is possible to visualize the hematoxylin-positive-, K+-colocalized Na+ distribution in the necrotic region, and a homogeneous distribution of Na+ in the viable region (Figure 5). All experiments were repeated in 7 MDA-MB-231 tumors and 6 MCF-7 tumors with consistent reproducibility in both tumor models, and by using different mass spectrometers and tissue preparation methods.
The use of different ionization techniques in both tumor xenograft models provided complementary data regarding the spatial localization of Cho (m/z 104, [M]+, shown in Figure 3) and Cho with a water molecule attached to it (m/z 122, [M+H2O]+), both of which were well visualized with SIMS and MALDI and displayed identical spatial distributions. Cho colocalized with regions were PC was present, but also to regions where PC was absent or below the detection limit (Figure 3). The same behavior was found for GPC (m/z 524, [M]+) (data not shown). The mass values for Na+, K+, Cho, PC and GPC were typically observed in both tumor types, with experiments under the exact same conditions running in parallel.
Discussion
Previous studies have demonstrated the importance of abnormal PC levels and other choline related lipid metabolites in cancer cells (11, 20, 22), but the techniques used in these studies were unable to identify distinct tumor regions containing high PC levels. Spatially resolved MRSI merely provided the tumoral tCho distribution, but was not able to resolve the individual compounds contributing to the tCho signal, such as PC, Cho, and GPC.
In order to validate our experiments, several SIMS, ME-SIMS, MetA-SIMS and MALDI measurements were performed on adjacent slides. The analysis of all samples (with and without any surface treatment) showed similar PC localizations, validating the conclusion that PC localization was not due to artifacts of the sample preparation protocols used.
The measurement time for MS imaging depends on the desired spatial resolution, sample size, and the number of spectra collected. A quick survey scan with low spatial resolution can be combined with a longer analytical scan if more detailed spatial and spectral information are required. The FOV may vary from a few micrometers up to 200 μm per tile, depending on the desired spatial resolution. Both techniques provide extensive molecular information per μm2 of the sample surface.
Our MS Imaging applications detected high PC levels in viable, non-necrotic tumor regions in the invasive/metastatic MDA-MB-231 tumor model, which is in good agreement with previous studies that revealed the importance of PC and related compounds in metastatic tumor growth (11, 20, 22). Free choline (m/z 104) was present in viable tumor with a lower intensity than PC, and it was also detected in necrotic regions, which was confirmed by PCA of this data. PCA demonstrated that PC and Cho positively correlated in active MDA-MB-231 tumor regions and negatively correlated in necrotic areas. With this information, we were able to differentiate tumor microenvironments based on their molecular signatures with precision. The distribution of Na+, K+, Cho and PC in the MDA-MD-231 tumor model revealed tumor microenvironments with distinct molecular compositions, identified by PCA. The molecular distributions of PC and other compounds detected by MS Imaging reveal several sub-regions and features within the viable tumor regions than those determined by H&E staining alone. With the use of PCA, it was also possible to assign complementary regions based on the intensity of certain mass spectral peaks such as PC and Cho. Future MS Imaging fingerprinting applications can potentially identify distinct tumor regions with diagnostic and therapeutic relevance.
The MS Imaging-detected spatial PC distribution was also able to differentiate between the two tumor xenograft models. The borders between necrotic and viable region were sharply defined by PC in highly invasive/metastatic MDA-MB-231 tumors, whereas in MCF-7 tumors, these regions were less defined. In addition, PC was a major principal component in the MDA-MB-231 tumor. However, PC was not relevant for tumor region assignment in MCF-7 tumors, likely due to the lower concentration of PC in MCF-7 compared to MDA-MB-231 tumors (23). Our MS imaging findings confirm the importance of PC in malignant tumor growth (23). Altered expression and activity levels of phosphatidylcholine metabolism-related enzymes, such as choline kinase (5, 8, 34), phospholipases C and D (42) and specific choline transporters (43) may account for the higher MS imaging-detected PC intensity in viable regions of the more aggressive tumor model. A recent studies in prostate tumor models detected increased tCho and PC concentrations in hypoxic tumor regions, and showed that hypoxia inducible factor-1 (HIF-1) signaling increased choline kinase expression, and hence PC production, under hypoxic conditions (3). It is possible that HIF-1 signaling in hypoxic tumor regions increased the PC concentrations in the MDA-MB-231 breast tumor models, thereby potentially leading to a heterogeneous PC distribution in viable tumor regions as observed in our MS imaging data.
Both Na+ and K+ displayed heterogeneous spatial distributions in the breast tumor models investigated in our study. The correlation between Na+ and K+ ions revealed distinct differences between the invasive/metastatic and the nonmetastatic tumor model, as Na+ and K+ were negatively correlated in MCF-7 tumors, and positively correlated in MDA-MB-231 tumors. These different Na+ and K+ correlations may in the future be useful for differentiating breast tumor types. Na+ has previously been related to tumor growth and attributed to alterations in Na+/K+ pump functions (44). Our MS imaging data displayed a heterogeneous spatial distribution of the Na+ intensity in both breast tumor models, which is in good agreement with these Na+ MRSI studies (44). The high Na+ levels in the necrotic regions of both tumor types support previous findings that correlated such high Na+ levels with a decrease in Na+/K+ pump activity and an increase in cell membrane permeability in the necrotic region (45). The same study reported higher K+ levels in viable tumor regions compared to necrotic areas, due to high Na+/K+ pump activity in viable regions. In MCF-7 tumors, the low K+ levels in necrotic regions and high K+ levels in viable regions are in agreement with this study (45). At high spatial resolution, increased Na+ and K+ were observed in hematoxylin-positive bodies in necrotic regions of MDA-MB-231 tumors, which likely originated from dead or dying cells. Both ions were decreased and homogenously distributed in viable regions, opposing the above explanation based on Na+/K+ pump activation, in which high levels of Na+ would be expected in the extracellular space of viable tumor regions and decreased K+ in necrotic regions. Breakdown of voltage-gated K+ channels in necrotic MDA-MB-231 tumor regions may explain the increased K+ in these regions. Voltage-gated K+ channels were reported to be involved in breast cancer cell proliferation (46, 47), and to play an important role in the regulation of cell apoptosis (48). In our study, high K+ levels in necrotic regions of highly metastatic MDA-MB-231 tumors contrasting low K+ levels in necrotic regions of nonmetastatic MCF-7 tumors, suggest that a dysfunction in voltage-gated K+ channels in highly metastatic tumors may contribute to tumor malignancy. Further studies are necessary to elucidate the exact roles of voltage-gated K+ channels and Na+/K+ pump in necrosis, apoptosis, and tumor aggressiveness.
Our MS imaging data demonstrate that several important cancer-related molecules and spatial features can be readily identified in a single MS imaging experiment. In comparison, immunohistochemical stainings of breast biopsies, which are the current clinical standard in diagnostic pathology laboratories, require the use of one tissue section per type of immunostaining, and complex spatial and molecular correlations may be lost. The combination of MS imaging with MRSI data from the same tumors corroborate the validity of results by combining in-vivo and ex-vivo imaging techniques on the same target, with agreeable results. The information that can be obtained by MS imaging of breast tumor sections may in the future provide spatially resolved molecular fingerprinting applications to help guide molecular diagnosis and treatment planning of breast cancer patients.
Acknowledgments
This work is part of research program of the “Stichting voor Fundamenteel Onderzoek der Materie (FOM)”, which is financially supported by the “Nederlandse organisatie voor Wetenschappelijk Onderzoek (NWO)”. We also acknowledge the Dutch BSIK funded programs “Netherlands Proteomics Centre” (NPC) and the Virtual Laboratory for e-Science (VLe). This work was also supported by NIH R01 CA134695. We thank Drs. Zaver M. Bhujwalla, Dmitri Artemov, and Vadappuram P. Chacko for technical assistance with the MRSI and MRS experiments. We thank Mr. Gary Cromwell for maintaining the cell lines.
Abbreviations
- CSI
chemical shift imaging
- DHB
dihydroxybenzoic acid
- Cho
free choline
- GPC
glycerophosphocholine
- HBSS
Hanks balanced solution
- HIF-1
hypoxia inducible factor-1
- ITO
indium tin oxide
- M.W.
molecular weight
- MR
magnetic resonance
- MRSI
magnetic resonance spectroscopic imaging
- MRS
magnetic resonance spectroscopy
- MS
mass spectrometry
- MALDI
matrix assisted laser desorption ionization
- ME-SIMS
matrix enhanced secondary ion mass spectrometry
- MetA
metal assisted
- PC
phosphocholine
- K+
potassium
- PCA
principal component analysis
- SIMS
secondary ion mass spectrometry
- SCID
severe combined immunodeficient
- Na+
sodium
- tCho
total choline-containing metabolite levels
References
- 1.Kurhanewicz J, Vigneron DB, Nelson SJ. Three-dimensional magnetic resonance spectroscopic imaging of brain and prostate cancer. Neoplasia. 2000;2:166–89. doi: 10.1038/sj.neo.7900081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackerstaff E, Artemov D, Gillies RJ, Bhujwalla ZM. Hypoxia and the presence of human vascular endothelial cells affect prostate cancer cell invasion and metabolism. Neoplasia. 2007;9:1138–51. doi: 10.1593/neo.07568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glunde K, Shah T, Winnard PT, et al. Hypoxia regulates choline kinase expression through hypoxia-inducible factor-lot signaling in a human prostate cancer model. Cancer Res. 2008;68:172–80. doi: 10.1158/0008-5472.CAN-07-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bathen TF, Holmgren K, Lundemo AG, et al. Omega-3 fatty acids suppress growth of sw620 human colon cancer xenografts in nude mice. Anticancer Res. 2008;28:3717–23. [PubMed] [Google Scholar]
- 5.de Molina AR, Sormentero-Estrada J, Beldo-Iniesta C, et al. Expression of choline kinase alpha to predict outcome in patients with early-stage non-small-cell lung cancer: A retrospective study. Lancet Oncol. 2007;8:889–97. doi: 10.1016/S1470-2045(07)70279-6. [DOI] [PubMed] [Google Scholar]
- 6.Mori N, Delsite R, Natarajan K, Kulawiec M, Bhujwalla ZM, Singh KK. Loss of p53 function in colon cancer cells results in increased phosphocholine and total choline. Mol Imaging. 2004;3:319–23. doi: 10.1162/15353500200404121. [DOI] [PubMed] [Google Scholar]
- 7.Hara T, Bansal A, DeGrado TR. Choline transporter as a novel target for molecular imaging of cancer. Mol Imaging. 2006;5:498–509. [PubMed] [Google Scholar]
- 8.Gabellieri C, Beloueche-Babari M, Jamin Y, Payne GS, Leach MO, Eykyn TR. Modulation of choline kinase activity in human cancer cells observed by dynamic p-31 nmr. NMR Biomed. 2009;22:456–61. doi: 10.1002/nbm.1361. [DOI] [PubMed] [Google Scholar]
- 9.Spadaro F, Ramoni C, Mezzanzanica D, et al. Phosphatidylcholine-specific phospholipase c activation in epithelial ovarian cancer cells. Cancer Res. 2008;68:6541–9. doi: 10.1158/0008-5472.CAN-07-6763. [DOI] [PubMed] [Google Scholar]
- 10.Al-Saeedi F, Smith T, Welch A. [methyl-h-3]-choline incorporation into mcf-7 cells: Correlation with proliferation, choline kinase and phospholipase d assay. Anticancer Res. 2007;27:901–6. [PubMed] [Google Scholar]
- 11.Glunde K, Jacobs MA, Bhujwalla ZM. Choline metabolism in cancer: Implications for diagnosis and therapy. Expert Rev of Mol Diagn. 2006;6:821–9. doi: 10.1586/14737159.6.6.821. [DOI] [PubMed] [Google Scholar]
- 12.Glunde K, Bhujwalla ZM. Choline kinase alpha in cancer prognosis and treatment. Lancet Oncol. 2007;8:855–7. doi: 10.1016/S1470-2045(07)70289-9. [DOI] [PubMed] [Google Scholar]
- 13.Baykal AT, Jain MR, Li H. Aberrant regulation of choline metabolism by mitochondrial electron transport system inhibition in neuroblastoma cells. Metabolomics. 2008;4:347–56. doi: 10.1007/s11306-008-0125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaeffer EL, Gattaz WF. Cholinergic and glutamatergic alterations beginning at the early stages of alzheimer disease: Participation of the phospholipase a(2) enzyme. Psychopharmacology. 2008;198:1–27. doi: 10.1007/s00213-008-1092-0. [DOI] [PubMed] [Google Scholar]
- 15.Inglese M, Li BSY, Rusinek H, Babb JS, Grossman RI, Gonen O. Diffusely elevated cerebral choline and creatine in relapsing-remitting multiple sclerosis. Magn Reson Med. 2003;50:190–5. doi: 10.1002/mrm.10481. [DOI] [PubMed] [Google Scholar]
- 16.Mori N, Natarajan K, Chacko VP, Artemov D, Bhujwalla ZM. Choline phospholipid metabolites of human vascular endothelial cells altered by cyclooxygenase inhibition, growth factor depletion, and paracrine factors secreted by cancer cells. Mol Imaging. 2003;2:124–30. doi: 10.1162/15353500200303127. [DOI] [PubMed] [Google Scholar]
- 17.Li XJ, Lu Y, Pirzkall A, McKnight T, Nelson SJ. Analysis of the spatial characteristics of metabolic abnormalities in newly diagnosed glioma patients. J Mag Reson Imaging. 2002;16:229–37. doi: 10.1002/jmri.10147. [DOI] [PubMed] [Google Scholar]
- 18.Katz-Brull R, Lavin PT, Lenkinski RE. Clinical utility of proton magnetic resonance spectroscopy in characterizing breast lesions. J Natl Cancer I. 2002;94:1197–203. doi: 10.1093/jnci/94.16.1197. [DOI] [PubMed] [Google Scholar]
- 19.Leach MO, Verrill M, Glaholm J, et al. Measurements of human breast cancer using magnetic resonance spectroscopy: A review of clinical measurements and a report of localized p-31 measurements of response to treatment. NMR Biomed. 1998;11:314–40. doi: 10.1002/(sici)1099-1492(1998110)11:7<314::aid-nbm522>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 20.Glunde K, Jie CF, Bhujwalla ZM. Mechanisms of indomethacin-induced alterations in the choline phospholipid metabolism of breast cancer cells. Neoplasia. 2006;8:758–71. doi: 10.1593/neo.06187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nimmagadda S, Glunde K, Pomper MG, Bhujwalla ZM. Pharmacodynamic markers for choline kinase down-regulation in breast cancer cells. Neoplasia. 2009;11:477–84. doi: 10.1593/neo.81430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glunde K, Jacobs MA, Pathaka AP, Artemov D, Bhujwalla ZM. Molecular and functional imaging of breast cancer. NMR Biomed. 2009;22:92–103. doi: 10.1002/nbm.1269. [DOI] [PubMed] [Google Scholar]
- 23.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80–4. [PubMed] [Google Scholar]
- 24.Chaurand P, Schwartz SA, Caprioli RM. Imaging mass spectrometry: A new tool to investigate the spatial organization of peptides and proteins in mammalian tissue sections. Curr Opin Chem Biol. 2002;6:676–81. doi: 10.1016/s1367-5931(02)00370-8. [DOI] [PubMed] [Google Scholar]
- 25.Herring KD, Oppenheimer SR, Caprioli RM. Direct tissue analysis by matrix-assisted laser desorption ionization mass spectrometry: Application to kidney biology. Semin Nephrol. 2007;27:597–608. doi: 10.1016/j.semnephrol.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz SA, Weil RJ, Johnson MD, Toms SA, Caprioli RM. Protein profiling in brain tumors using mass spectrometry: Feasibility of a new technique for the analysis of protein expression. Clin Cancer Res. 2004;10:981–7. doi: 10.1158/1078-0432.ccr-0927-3. [DOI] [PubMed] [Google Scholar]
- 27.Stoeckli M, Staab D, Schweitzer A, Gardiner J, Seebach D. Imaging of a [beta]-peptide distribution in whole-body mice sections by maldi mass spectrometry. J Am Soc Mass Spectr. 2007;18:1921–4. doi: 10.1016/j.jasms.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Luxembourg SL, Heeren RMA. Fragmentation at and above surfaces in sims: Effects of biomolecular yield enhancing surface modifications. Int J Mass Spectrom. 2006;253:181–92. [Google Scholar]
- 29.Altelaar AFM, Luxembourg SL, McDonnell LA, Piersma SR, Heeren RMA. Imaging mass spectrometry at cellular length scales. Nat Protoc. 2007;2:1185–96. doi: 10.1038/nprot.2007.117. [DOI] [PubMed] [Google Scholar]
- 30.Chughtai K, Heeren RMA. Mass spectrometric imaging for biomedical tissue analysis. Chem Rev. 110:3237–77. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zierhut ML, Ozturk-Isik E, Chen AP, Park I, Vigneron DB, Nelson SJ. 1h spectroscopic imaging of human brain at 3 tesla: Comparison of fast three-dimensional magnetic resonance spectroscopic imaging techniques. J Mag Reson Imaging. 2009;30:473–80. doi: 10.1002/jmri.21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glunde K, Guggino SE, Solaiyappan M, Pathak AP, Ichikawa Y, Bhujwalla ZM. Extracellular acidification alters lysosomal trafficking in human breast cancer cells. Neoplasia. 2003;5:533–45. doi: 10.1016/s1476-5586(03)80037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glunde K, Raman V, Mori N, Bhujwalla ZM. Rna interference-mediated choline kinase suppression in breast cancer cells induces differentiation and reduces proliferation. Cancer Res. 2005;65:11034–43. doi: 10.1158/0008-5472.CAN-05-1807. [DOI] [PubMed] [Google Scholar]
- 34.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004;64:4270–6. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- 35.Luxembourg SL, Mize TH, McDonnell LA, Heeren RMA. High-spatial resolution mass spectrometric imaging of peptide and protein distributions on a surface. Anal Chem. 2004;76:5339–44. doi: 10.1021/ac049692q. [DOI] [PubMed] [Google Scholar]
- 36.Altelaar AFM, Taban IM, McDonnell LA, et al. High-resolution maldi imaging mass spectrometry allows localization of peptide distributions at cellular length scales in pituitary tissue sections. Int J Mass Spectrom. 2007;260:203–11. [Google Scholar]
- 37.Klinkert I, McDonnell LA, Luxembourg SL, et al. Tools and strategies for visualization of large image data sets in high-resolution imaging mass spectrometry. Rev Sci Instrum. 2007;78 doi: 10.1063/1.2737770. [DOI] [PubMed] [Google Scholar]
- 38.Eijkel GB, Kaletas BK, van der Wiel IM, Kros JM, Luider TM, Heeren RMA. Correlating maldi and sims imaging mass spectrometric datasets of biological tissue surfaces. Surf Interface Anal. 2009;41:675–85. [Google Scholar]
- 39.Chou YL. Statistical analysis, with business and economical applications. 2nd. New York Holt: Rinehart and Winston; 1975. [Google Scholar]
- 40.Wall MERA, Rocha LM. A practical approach to microarray data analysis. Kluwer Academic Publishers; 2003. [Google Scholar]
- 41.Bhujwalla ZM, Aboagye EO, Gillies RJ, Chacko VP, Mendola CE, Backer JM. Nm23-transfected mda-mb-435 human breast carcinoma cells form tumors with altered phospholipid metabolism and ph: A p-31 nuclear magnetic resonance study in vivo and in vitro. Magn Reson in Med. 1999;41:897–903. doi: 10.1002/(sici)1522-2594(199905)41:5<897::aid-mrm7>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 42.Iorio E, Mezzanzanica D, Alberti P, et al. Alterations of choline phospholipid metabolism in ovarian tumor progression. Cancer Res. 2005;65:9369–76. doi: 10.1158/0008-5472.CAN-05-1146. [DOI] [PubMed] [Google Scholar]
- 43.Eliyahu G, Kreizman T, Degani H. Phosphocholine as a biomarker of breast cancer: Molecular and biochemical studies. Int J Cancer. 2007;120:1721–30. doi: 10.1002/ijc.22293. [DOI] [PubMed] [Google Scholar]
- 44.Ouwerkerk R, Bleich KB, Gillen JS, Pomper MG, Bottomley PA. Tissue sodium concentration in human brain tumors as measured with 23na mr imaging. Radiology. 2003;227:529–37. doi: 10.1148/radiol.2272020483. [DOI] [PubMed] [Google Scholar]
- 45.Summers RM, Joseph PM, Kundel HL. Sodium nuclear magnetic resonance imaging of neuroblastoma in the nude mouse. Invest Radiol. 1991;26:233–41. doi: 10.1097/00004424-199103000-00007. [DOI] [PubMed] [Google Scholar]
- 46.Brevet M, Ahidouch A, Sevestre H, et al. Expression of k+ channels in normal and cancerous human breast. Histol Histopathol. 2008;23:965–72. doi: 10.14670/HH-23.965. [DOI] [PubMed] [Google Scholar]
- 47.Jang SH, Kang KS, Ryu PD, Lee SY. Kv1.3 voltage-gated k+ channel subunit as a potential diagnostic marker and therapeutic target for breast cancer. BMB Rep. 2009;42:535–9. doi: 10.5483/bmbrep.2009.42.8.535. [DOI] [PubMed] [Google Scholar]
- 48.Szabò I, Zoratti M, Gulbins E. Contribution of voltage-gated potassium channels to the regulation of apoptosis. FEBS Letters. 2010;584:2049–56. doi: 10.1016/j.febslet.2010.01.038. [DOI] [PubMed] [Google Scholar]


