Abstract
Previous experimental studies suggest that Mycobacterium tuberculosis inhibits a number of macrophage intracellular processes associated with antigen presentation, including antigen processing, MHC class II expression, trafficking of MHC class II molecules, and peptide-MHC class II binding. In this study, we investigate why multiple mechanisms have been observed. Specifically, we consider what purpose multiple mechanisms may serve, whether experimental protocols favor the detection of some mechanisms over others, and whether alternative mechanisms exist. By using a mathematical model of antigen presentation in macrophages that tracks levels of various molecules, including peptide–MHC class II complexes on the cell surface, we show that mechanisms targeting MHC class II expression are effective at inhibiting antigen presentation, but only after a delay of at least 10 h. By comparison, the effectiveness of mechanisms targeting other cellular processes is immediate, but may be attenuated under certain conditions. Therefore, targeting multiple cellular processes may represent an optimal strategy for M. tuberculosis (and other pathogens with relatively long doubling times) to maintain continuous inhibition of antigen presentation. In addition, based on a sensitivity analysis of the model, we identify other cellular processes that may be targeted by such pathogens to accomplish the same effect, representing potentially novel mechanisms.
Keywords: antigen processing, mathematical model, HLA, cell-mediated immunity
Macrophages play dual roles during tuberculosis (TB) infection (1). On the one hand, they serve as the preferred host for Mycobacterium tuberculosis (Mtb), the intracellular pathogen that causes TB. On the other hand, they also help to alert the immune system to the presence of Mtb, and, if activated, can eliminate it directly. Activation depends on the presentation of antigenic peptide–MHC class II (pMHC) complexes on the macrophage surface that can bind T cell receptors (TCRs) on cognate CD4+ T helper cells. pMHC-TCR binding induces CD4+ T helper cells to secrete IFN-γ, which stimulates macrophages to produce molecules capable of killing Mtb such as nitric oxide (2). This process constitutes an important arm of cell-mediated immunity and may determine infection outcome (3).
The fact that Mtb inhibits antigen presentation in macrophages is now well established (4). Initial studies showed that fewer macrophages infected with mycobacteria express detectable levels of antigen on their surface compared with uninfected macrophages (5, 6). Functional assays later confirmed that infected macrophages are deficient in their ability to signal CD4+ T helper cells by measuring T cell response. The magnitude of T cell response is in turn proportional to pMHC levels, assuming a lower threshold number of pMHC complexes has been exceeded (7, 8). By using such an assay, Gercken et al. (9) found that monocytes cocultured with Mtb for 6 days exhibit a 3- to 10-fold reduction in their ability to stimulate T cell proliferation compared with uninfected controls (9). Furthermore, higher numbers of Mtb bacilli, e.g., with a multiplicity of infection (moi) of 50 versus a moi of 10, correlated with lower T cell response levels. Subsequent studies provided further evidence that an inverse relationship exists between Mtb infectious dose and T cell response (10, 11).
After it was established that Mtb inhibits antigen presentation in macrophages, several intracellular mechanisms were proposed (reviewed in ref. 12). Moreno et al. (13) observed that macrophages cocultured with the Mtb cell wall component lipoarabinomannan fail to present antigen from whole inactivated virus, although presentation of synthesized epitope is unimpaired (13). This observation led to the hypothesis that Mtb inhibits antigen presentation at the stage of antigen processing, a hypothesis also made by Noss et al. (10). Later, based on the observation that Mtb-infected monocytes do not produce stable pMHC complexes and do not localize labeled MHC class II molecules and antigens to the same intracellular compartment, Hmama et al. (14) proposed that Mtb affects MHC class II at a posttranslational stage such as maturation (delivery to the MIIC endosome or invariant chain processing) or peptide loading (14). Finally, based on the observation that infected macrophages express lower levels of MHC class II mRNA than uninfected macrophages, Noss et al. (10) proposed that Mtb inhibits MHC class II mRNA synthesis.
The goal of the present study is to investigate why multiple mechanisms have been proposed to explain how Mtb inhibits antigen presentation. In particular, we address three issues by using a mathematical model: (i) what purpose multiple mechanisms may serve, (ii) whether experimental protocols may have favored the detection of some mechanisms over others, and (iii) whether alternative mechanisms exist that may be used to guide future experiments. Our immediate motivation stems from conflicting data in the literature regarding these mechanisms. Specifically, we refer to the observations by Hmama et al. (14) that MHC class II mRNA levels were unchanged in infected cells, and by Noss et al. (10) that MHC class II mRNA levels decreased in infected cells. Because the two studies differed with respect to experimental conditions (e.g., macrophage cell type, Mtb strain, and degree of IFN-γ-induced activation), it is unclear whether the conclusions hold in general. We seek to help clarify these observations with our model.
Methods
Model Overview. Our mathematical model comprises a set of ordinary differential equations representing the major intracellular processes that contribute to antigen presentation within the context of a single macrophage (Fig. 1). These processes relate to MHC class II expression (at both mRNA and protein levels), antigen processing, and peptide-MHC binding and trafficking, and include the processes hypothesized to be targeted by Mtb. Our model also accounts for the effects of IFN-γ, which is typically added to cultured macrophages during studies on antigen presentation (10, 14).
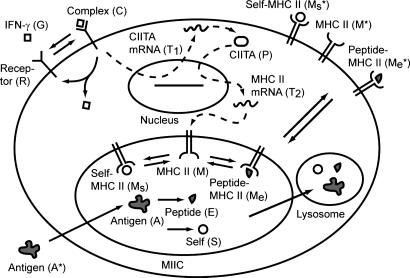
Fig. 1.
Model schematic. Molecular species represented in the model include extracellular IFN-γ (G), IFN-γ receptors (R, free; C, bound), CIITA (T1, mRNA; P, protein), MHC class II mRNA (T2), exogenous antigen (A*, extracellular; A, intracellular; E, peptide), self-peptide (S), free MHC class II molecules (M, intracellular; M*, surface), self peptide-bound MHC class II molecules (Ms, intracellular; Ms*, surface), and exogenous peptide-bound MHC class II molecules (Me, intracellular; Me*, surface). Solid arrows indicate one-step reactions and dashed arrows indicate regulatory interactions. Degradation is represented in the model for the following molecules but not shown: G, T1, P, T2, A*, M, M*, Ms, Ms*, Me, Me*. Up-regulation of M by C directly and contribution of Ms and Ms* to S are also included in the model but are not shown.
To represent these processes, we use ordinary differential equations that allow large numbers of molecules to be tracked. For each molecular species, we derive an equation for the rate of change by using the law of mass action and estimate parameter values by using published experimental data. In total, our model uses 16 equations and 30 parameters to simulate antigen presentation within the context of a single macrophage. Equations and parameter values, as well as details of how equations were derived and parameter values estimated, can be found in Supporting Text, which is published as supporting information on the PNAS web site.
Simulations Using the Mathematical Model. The baseline model comprises Eqs. 1–16, the parameters in Table 3, which is published as supporting information on the PNAS web site, and the initial conditions in Supporting Text and Table 4, which is published as supporting information on the PNAS web site. Protocol-specific parameter values and initial conditions can also be found in Supporting Text. To generate simulations using the mathematical model, we use the NDSolve function of mathematica v4.2 (Wolfram Research) and our own differential equation solver coded in C and run on Sun UNIX machines for confirmation of numerical results. We analyze model output in terms of major features such as relative changes in numbers of molecules, and times at which highest levels are reached. As a marker for antigen presentation, we generally use the number of surface-localized exogenous peptide-bound MHC class II molecules (Eq. 16 in Supporting Text).
Representation of the Inhibitory Effects of Mtb on Intracellular Processes. To represent the inhibitory effect that Mtb is hypothesized to have on an intracellular process, we decrease the corresponding parameter in the model by a factor proportional to experimental infectious dose. We assume that the number of Mtb bacilli does not change significantly on the time scales of the protocols being simulated based on the observation that the doubling time of Mtb is on the order of days (15). We also assume that the inhibitory effect exerted by Mtb on any given intracellular process saturates at high levels of bacilli. Therefore, we represent the inhibitory effect as a multiplicative factor having a value between 0 and 1 (corresponding to complete inhibition and no inhibition, respectively) that approaches 0 as the number of bacilli increases. Further details are provided in Supporting Text.
Sensitivity Analysis. The goal of sensitivity analysis is to correlate variances in parameter values to variances in model output and is useful when parameter values are not known with certainty. Sampling-based sensitivity analysis entails specifying a distribution for each parameter from which values are selected at random and used in model simulations (16). In particular, we use Latin hypercube sampling that allows several parameters to be analyzed simultaneously in a computationally efficient manner. To quantify the correlation of model output with each parameter, we calculate a partial rank correlation coefficient (PRCC) value. PRCC values vary between –1 and 1, corresponding to perfect negative and positive correlations, respectively, and can be further differentiated based on P values derived from Student's t tests. We use the algorithm of Blower and Dowlatabadi (17) implemented in both mathematica and our own differential equation solver. In general, we specify a uniform distribution for each parameter with a range of 10% and 190% of the baseline value, allowing us to examine the effects of both decreases and increases in each parameter.
Results
Baseline Characteristics. In the absence of IFN-γ and antigen, conditions that we used as a negative control, seven molecular species in the model were present in non-zero quantities: free IFN-γ receptors, MHC class II mRNA, free intracellular and surface MHC class II molecules, self-peptides, and intracellular and surface self-peptide–MHC class II complexes. These results are consistent with the finding that cultured macrophages constitutively express several molecules relevant to antigen presentation at basal levels, including IFN-γ receptors and MHC class II molecules (18, 19).
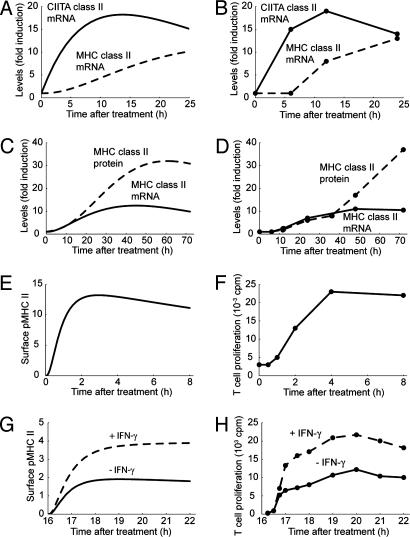
Dynamics of IFN-γ Response. As one positive control, we simulated the addition of IFN-γ to macrophages and compared dynamics of the response to experimental observations. In response to IFN-γ treatment, CIITA mRNA levels in the model increased immediately and reached a maximum ≈14 h later, whereas MHC class II mRNA levels increased more gradually and continued to increase for the first 24 h (Fig. 2A). Pai et al. (20) measured levels of CIITA and MHC class II mRNAs 6, 12, and 24 h after adding IFN-γ and observed highest levels at the 12- and 24-h time points, respectively, in agreement with our model (Fig. 2B). We also compared the coupled dynamics of MHC class II mRNA and protein expression from our model with experimental data. In our simulations, highest MHC class II mRNA and protein levels were attained ≈45 and 60 h after IFN-γ treatment, respectively (Fig. 2C). In comparison, highest MHC class II mRNA and protein levels were observed experimentally 48 and 72 h after IFN-γ treatment, respectively (Fig. 2D and ref. 21). Although MHC class II protein expression reaches its highest levels in the model in less time than observed experimentally, this apparent difference may be attributable to the sparseness of experimental time points.
Fig. 2.
Model testing using various controls. (A and B), Simulation results and experimental data for levels of CIITA mRNA (solid lines) and MHC class II mRNA (dashed lines) in IFN-γ-treated macrophages from Pai et al. (20). (C and D) Simulation results and experimental data for levels of MHC class II mRNA (solid lines) and MHC class II protein (dashed lines) in IFN-γ-treated macrophages from Cullell-Young et al. (21). (E and F) Simulation results for surface pMHC levels (in arbitrary units) and experimental data for T cell response in non-IFN-γ-treated macrophages exposed to antigen from Buus and Werdelin (22). (G and H) Simulation results for surface pMHC levels (in arbitrary units) and experimental data for T cell response in non-IFN-γ-treated macrophages (solid lines) and IFN-γ-treated macrophages (dashed lines) exposed to antigen from Delvig et al. (25). Pretreatment (16 h) with medium or IFN-γ is not shown; hence, the x axis is enumerated from 16 h onward (i.e., when antigen is present).
Dynamics of Antigen Presentation. In the presence of exogenous antigen, the number of surface pMHC complexes in our model rapidly increases, reaches a maximum ≈3 h later, and then decreases over the course of several hours (Fig. 2E). Antigen presentation by macrophages not pretreated with IFN-γ has been found to exhibit similar dynamics experimentally (Fig. 2F and refs. 22 and 23). In such cases, antigen presentation can be detected by T cell hybridoma assay minutes after the addition of antigen (22, 23). These macrophages elicit maximal responses after 1–4 h and remain capable of eliciting responses at the same or slightly decreased levels for several more hours (22, 23). Another feature of our model is dose dependence between exogenous antigen concentration and maximum number of resultant surface pMHC complexes (data not shown), which has also been observed experimentally with T cell responses (8, 24).
Increases in Antigen Presentation Due to IFN-γ Pretreatment. Experimental studies on antigen presentation by macrophages typically use both IFN-γ and exogenous antigen. Timing of IFN-γ treatment may be important, because studies in which IFN-γ is added before antigen show that pretreated macrophages are capable of eliciting T cell responses at levels severalfold higher than untreated macrophages (25). We simulated the addition of IFN-γ 16 h before exogenous antigen and observed a 2-fold increase in surface pMHC levels compared with untreated levels (Fig. 2G). This result is consistent with T cell proliferation data from Delvig et al. (ref. 25 and Fig. 2H). In subsequent simulations, we avoided the issue of pretreatment timing by using the simultaneous addition of IFN-γ and antigen unless stated otherwise.
Simulations of Mtb and Its Hypothesized Mechanisms. After testing the model under the preceding conditions, we used the model to simulate the inhibition of various intracellular processes targeted by Mtb. These processes included: antigen processing (13), MHC class II protein maturation (14), MHC class II peptide loading (14), and MHC class II mRNA synthesis, which we consider MHC class II transcription (10); we designate these hypotheses as H1,H2,H3, and H4, respectively. We then simulated the simultaneous addition of IFN-γ and antigen and recorded surface pMHC levels at time points spanning four orders of magnitude (0.1, 1.0, 10, and 100 h). These results were compared with results from the baseline model in which no processes were inhibited.
In general, inhibiting any particular intracellular process had either immediate or delayed effects on antigen presentation (Table 1). When antigen processing (H1) or MHC class II peptide loading (H3) was inhibited, surface pMHC levels were immediately affected, as indicated at the earliest time point, 0.1 h. The deviation from baseline levels was reduced at intermediate 1- and 10-h time points and then increased by the final 100-h time point. In contrast, inhibition of MHC class II maturation (H2) or MHC class II transcription (H4) resulted in negligible reductions in surface pMHC levels at the 0.1-h time point. However, these levels increasingly deviated from baseline levels at 1-, 10-, and 100-h time points. Both H2 and H4 targeted MHC class II expression and required a delay of at least 10 h to have substantial effects (>25% change in surface pMHC levels). We also simulated the inhibition of pairs of intracellular processes to determine the effect multiple mechanisms may have on antigen presentation when acting together (compare H1 plus H4 and H2 plus H3 in Table 1). Inhibitory mechanisms were synergistic and decreased antigen presentation levels to a greater extent in pairs than singly. In these simulations, each intracellular process was inhibited to the same degree. In a separate set of simulations, we used various degrees of inhibition, further differentiating mechanisms targeting MHC class II expression from other mechanisms (Fig. 5 and Supporting Text, which are published as supporting information on the PNAS web site).
Table 1. Percent changes in surface pMHC levels after inhibition of various intracellular processes hypothesized to be affected by Mtb.
| Hypothesis: affected process | 0.1 h, % | 1.0 h, % | 10 h, % | 100 h, % |
|---|---|---|---|---|
| H1: Antigen processing | ↓ 47 | ↓ 8.4 | ↓ 7.2 | ↓ 43 |
| H2: MHC class II maturation | ↓ 1.4 | ↓ 8.2 | ↓ 49 | ↓ 69 |
| H3: MHC class II peptide loading | ↓ 44 | ↓ 11 | ↓ 12 | ↓ 57 |
| H4: MHC class II transcription | ↓ 0.0026 | ↓ 0.16 | ↓ 26 | ↓ 66 |
| H1 plus H4 | ↓ 47 | ↓ 8.6 | ↓ 31 | ↓ 81 |
| H2 plus H3 | ↓ 45 | ↓ 18 | ↓ 55 | ↓ 86 |
Identical experimental conditions were used in each simulation, and comparisons were made to the baseline model, i.e. when no processes were inhibited.
Simulations of Previous Experimental Protocols. To determine whether previous experimental protocols may have favored the detection of some mechanisms over others and whether any of the four previously hypothesized mechanisms could account for all of the observed changes in macrophages infected with Mtb, we simulated two different experimental protocols under each hypothesized mechanism (10, 14). These protocols differed in several ways that could be accounted for in our model, including duration for which cells were exposed to IFN-γ and Mtb, and concentration of IFN-γ and number of Mtb bacilli used (Fig. 3 A and B). These protocols also differed with respect to the macrophage cell line and Mtb strain used, but these factors fell outside the scope of our model and were not considered.
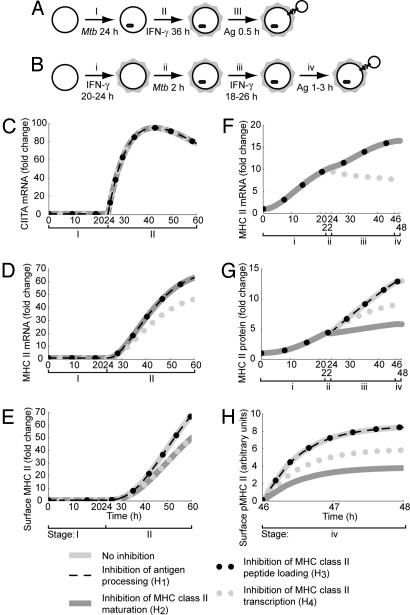
Fig. 3.
Simulation results of two in vitro experimental protocols using four published hypotheses. (A and B) Large circles represent macrophages, highlighted circles represent IFN-γ-treated macrophages, and small circles represent T cell hybridomas. (A) Protocol of Hmama et al. (14). A total of 105 monocytes was infected with Mtb at a moi of 50 for 24 h, treated with 200 units/ml IFN-γ for 36 h, pulsed with 1 mg/ml BSA for 0.5 h, and chased for 0.5, 1, or 4 h. (B) Protocol of Noss et al. (10). A total of 5 × 104 macrophages was treated with 2 ng/ml IFN-γ for 20–24 h, infected with Mtb at a moi of 40 for 2 h, treated with 2 ng/ml IFN-γ for an additional 18–26 h, and pulsed with 0–100 μg/ml hen egg lysozyme or 0–1,000 μg/ml RNase for 1–3 h. (C–E) Simulation results using the protocol of Hmama et al. (14) for levels of CIITA mRNA, MHC class II mRNA, and surface MHC class II protein, respectively. (F–H) Simulation results using the protocol of Noss et al. (10) for levels of MHC class II mRNA, total MHC class II protein, and surface pMHC, respectively.
In our simulations of the experimental protocol of Hmama et al. (14), we found that only an inhibition of H2 was consistent with all of their observations. In the absence of Mtb, the levels of several molecules rose over baseline levels during the course of this protocol, including CIITA mRNA, MHC class II mRNA, and MHC class II protein (Fig. 3 C–E). Only H2 and another hypothesized mechanism, H4, led to reductions in surface MHC class II expression of the same magnitude as those observed by Hmama et al. (ref. 14; 42% and 86%, using heat-killed and live Mtb bacilli, respectively, and Fig. 3E). However, H4 also led to a significant reduction in MHC class II mRNA levels that was not observed by Hmama et al. (14) and could therefore be ruled out as a possible mechanism (Fig. 3D).
When we simulated the experimental protocol of Noss et al. (10), we found that only H4 was capable of producing substantial changes in the levels of all three molecules they monitored. In our simulations, this mechanism reduced levels of MHC class II mRNA, total MHC class II protein, and surface pMHC by 54%, 31%, and 31%, respectively (Fig. 3 F–H). Another mechanism, inhibition of H2, reduced levels of these molecules by 0%, 55%, and 55%, respectively (Fig. 3 F–H). In comparison, Noss et al. (10) measured reductions of 80%, 30%, and between 40% and 80%, respectively, which is consistent with H2 but not H4. Interestingly, in our simulations of this protocol neither H1 nor inhibition of H3 had any significant effect on surface pMHC levels (Fig. 3H).
Sensitivity to Changes in Other Intracellular Processes. Whereas many intermediates of the antigen presentation pathway have been monitored in macrophages after Mtb infection in vitro (14), assays for other processes represented in our model have either not been developed or not been applied to this context. To determine what effect changes in these processes might have on antigen presentation, we varied all of the corresponding rates, rate constants, and scaling factors and experimental conditions in the model over a defined range and tracked surface pMHC levels over time. We then calculated the correlation between these levels and specific parameter values at 1-, 10-, and 100-h time points.
We found that surface pMHC levels correlated significantly with a number of different intracellular processes, including several not previously considered (Table 2). In particular, at times of <10 h after exposure to IFN-γ and antigen, surface pMHC levels correlated positively with rate constants for antigen uptake by pinocytosis and MHC class II trafficking to the cell surface and with the concentration of exogenous antigen. When the concentration of exogenous antigen was sufficiently low, other processes correlated strongly with surface pMHC levels on this time scale, including delivery of antigen to lysosomes and self-peptide production (data not shown). At times of >10 h after exposure to IFN-γ and antigen, surface pMHC levels correlated with factors affecting MHC class II expression, including CIITA transcription and translation and the concentration of IFN-γ in the medium and MHC class II transcription and protein maturation.
Table 2. Additional intracellular processes significantly correlated with surface pMHC levels.
| Time, h | Description (correlation coefficient) |
|---|---|
| 1.0 | MHC class II export (0.79) |
| Antigen concentration in medium (0.41) | |
| Antigen uptake (0.40) | |
| H2 (0.38) | |
| IFN-γ stimulation of translation* (0.33) | |
| 10 | H2 (0.72) |
| IFN-γ stimulation of translation* (0.62) | |
| MHC class II export (0.55) | |
| IFN-γ receptor-ligand binding (0.52) | |
| IFN-γ concentration in medium (0.52) | |
| H4 (0.49) | |
| CIITA translation (0.44) | |
| IFN-γ stimulation of transcription† (0.36) | |
| CIITA transcription (0.36) | |
| 100 | H4 (0.57) |
| H2 (0.56) | |
| CIITA translation (0.56) | |
| CIITA transcription (0.53) | |
| IFN-γ concentration in medium (0.51) | |
| IFN-γ receptor-ligand binding (0.49) | |
| IFN-γ stimulation of transcription† (0.47) | |
| Antigen concentration in medium (0.36) | |
| Antigen uptake (0.33) | |
| MHC class II export (0.32) | |
| IFN-γ degradation in solution (-0.87) | |
| MHC class II degradation (-0.56) | |
| CIITA protein degradation (-0.53) | |
| CIITA mRNA degradation (-0.49) | |
| IFN-γ receptor-ligand dissociation (-0.48) |
One thousand simulation runs were performed using different sampled parameter values. PRCC values determined to be significant (P ≤ 10-30) are shown in parentheses. Intracellular processes considered in previous hypotheses (H1-H4) are italicized.
MHC class II translation.
CIITA transcription.
Discussion
Multiple hypotheses have been offered to explain how Mtb inhibits antigen presentation in macrophages to escape immune surveillance. These hypotheses stem from different experimental protocols that appear in at least one instance to have led to conflicting results. In this study, we address why several mechanisms have been hypothesized by formulating a mathematical model of antigen presentation that accounts for different experimental conditions and can be used to simulate each mechanism.
Mtb Mechanisms Differ in Timing of Effect. We found that hypothesized Mtb mechanisms generally fall into one of two categories: those mechanisms having an immediate effect on the ability of the cell to present antigen, and those mechanisms requiring a delay of ≈10 h to have an effect. The first subset of mechanisms targets intracellular processes involved in the initial formation of pMHC complexes, including antigen processing and MHC class II peptide loading. In our simulations, the effectiveness of these mechanisms in inhibiting antigen presentation decreased after an intermediate length of time (at 1 and 10 h) and later increased (at 100 h). The intermediate decrease resulted from new rounds of pMHC binding resulting from prolonged exposure to IFN-γ and increasing numbers of free MHC class II. The second subset of mechanisms targets intracellular processes necessary for the continued supply of nascent MHC class II molecules, including MHC class II transcription and protein maturation. In our simulations, the effect of these mechanisms on antigen presentation steadily increased over time as a greater proportion of surface pMHC complexes involved nascent MHC class II.
These results are consistent with the intuitive notion that disruptions at different points along the antigen presentation pathway, or any multienzymatic pathway, require different lengths of time to manifest in the end product. These results are also consistent with the interpretation of the experimental data of Noss et al. (10) given by Heldwein and Fenton (26), that substantial inhibition of MHC class II expression requires prolonged (> 18 h) incubation with Mtb. The requirement of a delay of >10 h for inhibition of MHC class II expression to affect antigen presentation was also evident in our sensitivity analysis.
The fact that these four hypothesized mechanisms appear to impair the same cellular function, antigen presentation, raises the following question: do these mechanisms serve the same purpose and act redundantly or do they serve subtly different purposes? Our results suggest that these mechanisms act on different time scales and therefore serve different purposes. As demonstrated in our simulations of pairs of mechanisms, having mechanisms that operate on both shorter and longer time scales may allow Mtb to exert continuous inhibition on antigen presentation despite external sources of IFN-γ. In contrast, having only a single mechanism or multiple mechanisms that act on the same time scale may result in an inhibitory effect that either abates with time (if MHC class II expression increases) or is delayed.
Nascent and recycling MHC class II molecules may have distinct roles in antigen-presenting cells (27), and Mtb may have evolved mechanisms to undermine both sources of MHC class II. T cells require at least 2–4 h of stimulation to become fully activated (28), and mechanisms acting on time scales of both minutes and hours may be physiologically relevant. A recent study by Huppa et al. (29) shows that signaling between an antigen-presenting cell and a T cell has a cumulative effect over a period of 10 h and is sensitive to disruptions that occur even several hours after initial contact (29).
Previous Experimental Protocols Favor the Detection of Mtb Mechanisms Targeting MHC Class II Expression. Our simulations of previous experimental protocols suggest that Mtb mechanisms targeting MHC class II expression may have been responsible for most of the changes observed in levels of various molecules. Specifically, in simulations of the protocols of Hmama et al. (14) and Noss et al. (10), only mechanisms targeting processes associated with MHC class II expression were found to produce changes of the same magnitude as those observed experimentally. Although no single mechanism was found to account for all of the observations, these results do support the individual conclusions of Hmama et al. (14) and Noss et al. (10), who implicated inhibition of MHC class II protein maturation and MHC class II mRNA synthesis, respectively.
Why did one group observe a decrease in MHC class II mRNA levels but not the other (10, 14)? Noss et al. (10) attribute this discrepancy to differences in macrophage cell lines, macrophage activation, and infection lengths and methods. Our model accounts for some of these factors, including one aspect of macrophage activation (IFN-γ-stimulated MHC class II expression) and one consequence of infection length (inhibition of particular intracellular processes), in addition to experimental differences in duration of IFN-γ stimulation and amount of IFN-γ used. In our model, none of these factors accounted for the observed discrepancy in MHC class II mRNA levels.
Hmama et al. (14) and Noss et al. (10) also hypothesized that Mtb inhibits either MHC class II peptide loading or antigen processing. Our simulations show that neither of these mechanisms could have accounted for the observed changes in levels of molecules, given the experimental protocols that were used. On the time scales of both protocols, MHC class II expression is expected to be the limiting factor on antigen presentation as suggested by the half-life of MHC class II and our sensitivity analysis. Indeed, in the protocol used by Noss et al. (10), we predict that the high level of MHC class II expression masks whatever reductions in antigen presentation may result from an inhibition of antigen processing or MHC class II peptide loading.
Because these experimental protocols may have favored the detection of mechanisms targeting MHC class II expression, the actual contribution of mechanisms targeting other processes to the overall ability of Mtb to inhibit antigen presentation may not have been accurately assessed. Without experimental evidence to the contrary, the possibility even exists that mechanisms targeting antigen processing and MHC class II peptide loading are incidental to Mtb infection and do not significantly affect the ability of macrophages to present antigen in the presence of IFN-γ. While an experiment using an Mtb mutant specifically unable to inhibit either intracellular process would quickly answer this question, such a mutant is not yet available, to our knowledge.
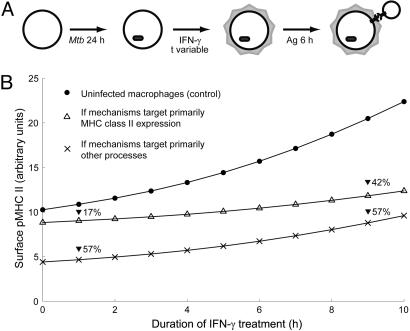
Therefore, we propose an alternative experimental protocol to determine whether mechanisms targeting intracellular processes besides MHC class II expression actually contribute to the ability of Mtb to inhibit antigen presentation (Fig. 4A). In this protocol, macrophages are infected with Mtb in vitro and treated with IFN-γ for various durations before assaying for antigen presentation using model antigen and T cell hybridoma. If mechanisms targeting MHC class II expression are the only means by which Mtb inhibits antigen presentation, the difference in the levels of T cell response (e.g., IL-2 production) elicited by uninfected and infected macrophages should increase as the duration of IFN-γ stimulation increases (Fig. 4B). On the other hand, if mechanisms targeting other intracellular processes play a significant role in the inhibition of antigen presentation, the difference in T cell response should be apparent even with short durations of IFN-γ stimulation and remain relatively constant as the duration of IFN-γ stimulation increases.
Fig. 4.
Proposed experimental protocol to determine the contribution of different mechanisms to inhibition of antigen presentation by Mtb. (A) Protocol schematic using representations of Fig. 3 A and B. (B) Surface pMHC levels expected in uninfected macrophages, Mtb-infected macrophages if mechanisms target primarily MHC class II expression (in this case, MHC class II transcription), and Mtb-infected macrophages if mechanisms target primarily other processes (in this case, antigen processing). Percentage reductions in infected macrophages (relative to uninfected controls) are also shown.
Additional Mechanisms May Target Other Intracellular Processes Strongly Influencing Antigen Presentation. As part of our analysis, we also identified all of the parameters in our model that strongly correlate with the number of pMHC complexes on the macrophage surface. These parameters represent intracellular processes likely to affect antigen presentation if perturbed and may serve as attractive targets to pathogens that evade immune surveillance such as Mtb. Other processes related to MHC class II expression, besides those already considered by previous hypotheses, strongly correlated with surface pMHC levels at long time scales. Recent evidence indicates that one of these processes, CIITA transcription, may be targeted by Mtb (30, 31). It would be interesting to test experimentally whether Mtb also affects any other candidate process such as IFN-γ receptor-ligand binding.
We found that several intracellular processes also negatively correlated with antigen presentation. In contrast to positively correlated processes such as those in Table 1, these processes are expected to inhibit antigen presentation if up-regulated rather than down-regulated. In the presence of low levels of exogenous antigen, one such process is the delivery of antigens (both self and exogenous) and derived peptides to MHC class II-inaccessible lysosomes. Conceivably, an intracellular pathogen such as Mtb could decrease the availability of its own antigens by increasing the rate at which this process occurs, although benefit to the pathogen may be somewhat offset by a concurrent decrease in competing self-antigens (32, 33). Nevertheless, the possibility that some pathogens up-regulate delivery to lysosomes cannot be ruled out because the rate of this process and the concentration of self-peptide have not been carefully measured.
Most of the experimental data on which we base our model originate from studies by using murine cell lines. Therefore, the dynamics of human macrophages infected with Mtb may differ somewhat from those observed in our simulations. However, based on our sensitivity analysis, we believe that our results are robust and can be generalized to the human host.
Supplementary Material
Acknowledgments
We thank Dr. Dan Burns, Dr. Cheong-Hee Chang, Dr. Eugenio de Hostos, Dr. Joanne Flynn, Dr. Malini Raghavan, Mr. Christian Ray, and Dr. David Russell for helpful discussions. This work was supported by National Institutes of Health Grants HL68526 (to D.E.K.) and 062930 (to J.J.L.).
Author contributions: S.T.C., J.J.L., and D.E.K. designed research; S.T.C. performed research; S.T.C., J.J.L., and D.E.K. analyzed data; and S.T.C. wrote the paper.
Abbreviations: p, peptide; moi, multiplicity of infection; PRCC, partial rank correlation coefficient.
References
- 1.Fenton, M. J. (1998) Curr. Opin. Hematol. 5, 72–78. [DOI] [PubMed] [Google Scholar]
- 2.Chan, E. D., Chan, J. & Schluger, N. W. (2001) Am. J. Respir. Cell Mol. Biol. 25, 606–612. [DOI] [PubMed] [Google Scholar]
- 3.Kaufmann, S. H. E. (1999) Curr. Biol. 9, R97–R99. [DOI] [PubMed] [Google Scholar]
- 4.Pancholi, P., Mirza, A., Bhardwaj, N. & Steinman, R. M. (1993) Science 260, 984–986. [DOI] [PubMed] [Google Scholar]
- 5.Kaye, P. M., Sims, M. & Feldmann, M. (1986) Clin. Exp. Immunol. 64, 28–34. [PMC free article] [PubMed] [Google Scholar]
- 6.Mshana, R. N., Hastings, R. B. & Krahenbuhl, J. L. (1988) Immunobiology 177, 40–54. [DOI] [PubMed] [Google Scholar]
- 7.Bekkhoucha, F., Naquet, P., Pierres, A., Marchetto, S. & Pierres, M. (1984) Eur. J. Immunol. 14, 807–814. [DOI] [PubMed] [Google Scholar]
- 8.Demotz, S., Grey, H. M. & Sette, A. (1990) Science 249, 1028–1030. [DOI] [PubMed] [Google Scholar]
- 9.Gercken, J., Pryjma, J., Ernst, M. & Flad, H. D. (1994) Infect. Immun. 62, 3472–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noss, E. H., Harding, C. V. & Boom, W. H. (2000) Cell Immunol. 201, 63–74. [DOI] [PubMed] [Google Scholar]
- 11.Mazzaccaro, R. J., Gedde, M., Jensen, E. R., van Santen, H. M., Ploegh, H. L., Rock, K. L. & Bloom, B. R. (1996) Proc. Natl. Acad. Sci. USA 93, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harding, C. V., Ramachandra, L. & Wick, M. J. (2003) Curr. Opin. Immunol. 15, 112–119. [DOI] [PubMed] [Google Scholar]
- 13.Moreno, C., Mehlert, A. & Lamb, J. (1988) Clin. Exp. Immunol. 74, 206–210. [PMC free article] [PubMed] [Google Scholar]
- 14.Hmama, Z., Gabathuler, R., Jefferies, W. A., de Jong, G. & Reiner, N. E. (1998) J. Immunol. 161, 4882–4893. [PubMed] [Google Scholar]
- 15.Dunn, P. L. & North, R. J. (1995) Infect. Immun. 63, 3428–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helton, J. C. & Davis, F. J. (2001) in Sensitivity Analysis, eds. Saltelli, A., Chan, K. & Scott, E. M. (Wiley, New York), pp. 101–153.
- 17.Blower, S. M. & Dowlatabadi, H. (1994) Int. Stat. Rev. 62, 229–243. [Google Scholar]
- 18.Hume, D. A. (1985) Immunobiology 170, 381–389. [DOI] [PubMed] [Google Scholar]
- 19.Celada, A., Allen, R., Esparza, I., Gray, P. W. & Schreiber, R. D. (1985) J. Clin. Invest. 76, 2196–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pai, R. K., Askew, D., Boom, W. H. & Harding, C. V. (2002) J. Immunol. 169, 1326–1333. [DOI] [PubMed] [Google Scholar]
- 21.Cullell-Young, M., Barrachina, M., Lopez-Lopez, C., Gonalons, E., Lloberas, J., Concepcio, S. & Celada, A. (2001) Immunogenetics 53, 136–144. [DOI] [PubMed] [Google Scholar]
- 22.Buus, S. & Werdelin, O. (1986) Acta Pathol. Microbiol. Immunol. Scand. C 94, 17–24. [DOI] [PubMed] [Google Scholar]
- 23.Ziegler, K. & Unanue, E. R. (1981) J. Immunol. 127, 1869–1875.6795263 [Google Scholar]
- 24.Reske-Kunz, A. B., Spaeth, E., Reske, K., Lohmann-Matthes, M. L. & Rude, E. (1981) Eur. J. Immunol. 11, 745–750. [DOI] [PubMed] [Google Scholar]
- 25.Delvig, A. A., Lee, J. J., Chrzanowska-Lightowlers, Z. M. A. & Robinson, J. H. (2002) J. Leukocyte Biol. 72, 163–166. [PubMed] [Google Scholar]
- 26.Heldwein, K. A. & Fenton, M. J. (2002) Microbes Infect. 4, 937–944. [DOI] [PubMed] [Google Scholar]
- 27.Pinet, V. M. & Long, E. O. (1998) Eur. J. Immunol. 28, 799–804. [DOI] [PubMed] [Google Scholar]
- 28.Weiss, A., Shields, R., Newton, M., Manger, B. & Imboden, J. (1987) J. Immunol. 138, 2169–2176. [PubMed] [Google Scholar]
- 29.Huppa, J. B., Gleimer, M., Sumen, C. & Davis, M. M. (2003) Nat. Immunol. 4, 749–755. [DOI] [PubMed] [Google Scholar]
- 30.Kincaid, E. Z. & Ernst, J. D. (2003) J. Immunol. 171, 2042–2049. [DOI] [PubMed] [Google Scholar]
- 31.Pai, R. K., Convery, M., Hamilton, T. A., Boom, W. H. & Harding, C. V. (2003) J. Immunol. 171, 175–184. [DOI] [PubMed] [Google Scholar]
- 32.Chicz, R. M., Urban, R. G., Gorga, J. C., Vignali, D. A., Lane, W. S. & Strominger, J. L. (1993) J. Exp. Med. 178, 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosloniec, E. F., Vitez, L. J., Buus, S. & Freed, J. H. (1990) J. Exp. Med. 171, 1419–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.